Abstract
RNA amplification methods have been used to facilitate making probes from small tissue samples for microarray studies. Our original amplification technique relied on driving the first reverse transcription with oligo(dT) with a T7 RNA polymerase promoter (T7dT) on the 5′ end, and subsequent transcriptions with random 9mers with a T3 RNA polymerase promoter (T3N9). Thus, initially, poly(A)+ RNA is amplified. This creates a potential problem: amplifications based on oligo(dT) priming could be sensitive to RNA degradation; broken mRNA strands should give rise to shorter cDNAs than those seen when intact templates are used. This would be especially troublesome when targets other than those corresponding to the 3′ ends of transcripts are printed on an array. To solve this problem, we elected to prime cDNA synthesis with T3N9 at the beginning of each amplification cycle. Following two rounds of amplification, the resulting probes were comparable to those obtained with our original protocol or the Arcturus RiboAmp kit. We show below that as many as four rounds of amplification can be performed reliably. In addition, as predicted, the method works well with degraded templates.
INTRODUCTION
DNA microarrays have been used to study the expression of thousands of genes at the same time in a variety of cells and tissues (1–4). In the past few years, techniques have been developed to label probes for microarray studies that are much less RNA intensive than the original direct labeling methods (5,6). Recently we described a technique for preparing probes from 0.5–1.0 µg of RNA without signal or template amplification (7). We were unable to use this method to study cells harvested by needle biopsy, cell sorting or laser-capture microdissection, however. To do this, we would have had to amplify the RNA template first. The most commonly used methods failed to produce as much product as we needed after one or two rounds of amplification (8–13). To solve this problem, we developed and validated a new protocol (14). This method, like all of the others in the literature, relied initially on reverse transcription driven by oligo(dT) with a T7 RNA polymerase recognition sequence on the 5′ end and, as noted above, it has certain limitations. To amplify partially degraded RNA [and poly(A)– RNA species], we felt that it might be useful to prime cDNA synthesis with T3N9 from the very beginning of the procedure. Below, we show that doing so allows us to make probes that are comparable in performance to those produced with our original method or the Arcturus RiboAmp kit, that four rounds of amplification can safely be performed, and that there are indeed advantages in using the modified method that we describe.
MATERIALS AND METHODS
Microarray fabrication
cDNA microarrays with a total of 10 816 elements were printed on poly-l-lysine-coated slides. The cDNAs used were provided by Bento Soares, University of Iowa. Plasmids were extracted from the bacteria using QiaPrep Turbo kits (Qiagen, Valencia, CA) and a BioRobot 8000 (Qiagen, Valencia, CA). The cDNA inserts were amplified with modified M13 primers (M13F 5′-GTTGTAAAACGACGGCCAGTG-3′ and M13R 5′-CACACAGGAAACAGCTATG-3′) and purified with MultiScreen PCR plates (Millipore, MA). The PCR products were diluted in 50% DMSO to an average concentration of 200 ng/µl. These products (5 µl each) were transferred to 384-well plates (Genetix, St James, NY) and then printed using an OmniGrid arrayer (GeneMachines, San Carlos, CA). The printed slides were aged for a week, and then post-processed before hybridization. For the detailed descriptions of coating glass with poly-l-lysine and post-processing the printed slides, please visit the following web site: http://cmgm.stanford.edu/pbrown/mguide/index.html.
RNA sample preparation and amplification
Total RNA preparation. Total RNA was extracted from mouse C2 and NIH 3T3 cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
RNA base hydrolysis. Mouse C2 total RNA (2.5 µg) in a volume of 2.5 µl was mixed with 2.5 µl of 5× first strand cDNA synthesis buffer (250 mM Tris–HCI pH 8.3, 375 mM KCI, 15 mM MgCl2) from Invitrogen (Carlsbad, CA). The RNA was then heated to 80°C for 5, 15 or 30 min. 500 ng of each product was analyzed using a Bioanalyzer 2100 and the RNA 6000 LabChip kit (Agilent, Palo Alto, CA).
First strand cDNA synthesis. One microgram (2 µl) of intact or base-hydrolyzed total RNA was used in the first round of amplification. The RNA templates were added to a 0.2 ml PCR tube containing 1 µl of T7dT (100 pmol/µl, 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGTTTT TTTTTTTTTTTTTTTT-3′, Operon, Alameda, CA) or T3N9 primer (100 pmol/µl, 5′-GCGCGAAATTAACCCTCACTAAAGGGAGANNNNNNNNN-3′, Operon, Alameda, CA),1 µl of RNasin (Promega, Madison, WI), and 6 µl of RNase-free water. The RNA was denatured at 70°C for 10 min, and chilled on ice for 10 min. Then 4 µl of 5× first strand buffer (Invitrogen, Carlsbad, CA), 1 µl of 10 mM dNTPs, 2 µl of 100 mM DTT (Invitrogen, Carlsbad, CA), 1 µl of RNase-free water, and 2 µl of SuperScript II RT (Invitrogen, Carlsbad, CA) were added to the RNA solution and the RT reaction was carried out at 42°C for 2 h in a Peltier thermal cycler (MJ. Research, Waltham, MA).
Second strand cDNA synthesis. For second strand cDNA synthesis, 43 µl of RNase-free water, 20 µl of 5× second-strand buffer (Invitrogen, Carlsbad, CA), 2 µl of 10 mM dNTPs, 1 µl of Escherichia coli DNA ligase (Invitrogen, Carlsbad, CA), 3 µl of E.coli DNA polymerse I (Invitrogen, Carlsbad, CA), and 1 µl of RNase H (Invitrogen, Carlsbad, CA) were added to the RT reaction mix. The resulting solution was incubated at 16°C for 2 h. At the end of this time, 2 µl of T4 DNA polymerase (Invitrogen, Carlsbad, CA) were added and the samples were incubated at 16°C for 5 min. The reaction was stopped with 10 µl of 0.5 M EDTA. MinElute columns (Qiagen, Valencia, CA) were used to clean up the RT product. The cDNAs were dried down to 8 µl in a SpeedVac (Thermo Savant, Holbrook, NY).
In vitro RNA transcription. RNA was transcribed from the DNA template with MEGA Script T7 or MEGA Script T3 reagents (Ambion, Austin, TX) according to the manufacturer’s instructions, and purified with an RNeasy Mini kit (Qiagen, Valencia, CA).
Second and subsequent rounds of amplification. T3N9 was used to prime first strand cDNA synthesis from 1 µg of amplified RNA in RNase-free water in the second and subsequent rounds. The procedures are identical to those described in steps 3–5.
When the Arcturus RiboAmp kit was employed to amplify purified samples of total RNA, the reagents were used according to the manufacturer’s directions.
Probe labeling with amine-modified random primers
Probes were synthesized from 5 µg of unamplified total RNA, 2.5 µg of template from T7dT primed amplifications, or 10 µg of template from T3N9 primed amplifications. Amine-modifed random primers were used to drive the labeling reaction as described in detail elsewhere (7,14).
Array scanning and data analysis
The arrays were scanned with a GenePix 4000A scanner (Axon, Foster City, CA) at 10 µm resolution. The PMT voltage settings were varied to obtain the maximum signal intensities with <1% probe saturation. The resulting images were analyzed using IPLab (Fairfax, VA) and ArraySuite (NHGRI, Bethesda, MD) software. In all the experiments described, ratios were assigned quality scores (7,15), and those with scores of <1 were removed from the data sets. The log2 mean intensity was calculated to generate scatter plots for the self/self comparisons of probes prepared from amplified C2 RNA. To compare our amplification methods to the Arcturus RiboAmp method, and to study the amplification of degraded RNA samples, a list of genes that are differentially expressed in mouse C2 versus 3T3 cells was compiled. This was done by labeling unamplified, intact total RNA from these two cell lines with Cy3 and Cy5, respectively, combining the labeled products, and using them to develop two arrays. The C2/3T3 ratios obtained were then ordered from lowest to highest, and the top and bottom 10% of ranked genes were tabulated. As a matter of convenience, those that were concordant in duplicate experiments were called ‘true outliers’ or differentially expressed genes. We treated the data from each amplification experiment the same way, and the top and bottom 10% of ranked genes were compared with the master list of differentially expressed ones. In this way, we could determine whether the amplifications permitted us to detect differences in expression that should have been present.
RESULTS AND DISCUSSION
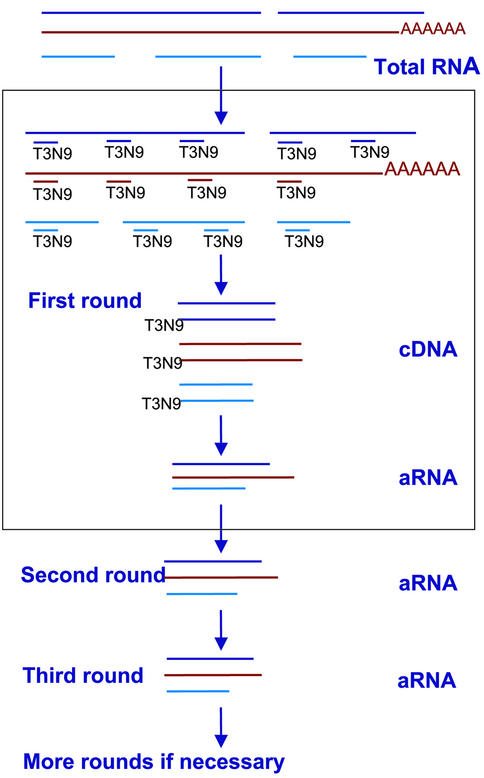
Our new amplification method is diagrammed in Figure 1. Complementary DNA synthesis from total RNA is primed by random 9mers with a T3 polymerase promoter sequence on their 5′ ends (T3N9). The RNA strands are replaced with DNA by RNase H, DNA polymerase I, and E.coli DNA ligase. Antisense RNA is then transcribed from the double stranded DNA template with T3 RNA polymerase. The T3N9 primer is used to drive first strand cDNA synthesis in the second and subsequent rounds of amplification. Double stranded DNA is then prepared as above, and RNA is generated with T3 RNA polymerase. Note that the resulting RNA products should not be 3′-end biased.
Figure 1.
RNA amplification strategy. First-strand synthesis of the cDNA used for the first round of amplification is primed with T3N9. This is followed by second-strand replacement reactions (see Materials and Methods). For the second and subsequent rounds of amplification, reverse transcription of the first strand is primed with same T3N9 primer. This step can be repeated several times.
In a previous paper (14), we introduced an amplification method in which T7dT was used to prime the first reverse transcription, and T3N9 was used to prime the second and subsequent ones. We compared our new method to the one we described earlier and the RiboAmp kit (Arcturus, Mountain View, CA), which is recommended by the manufacturer for amplifying RNA from laser-captured cells. First we prepared probes from unamplified C2 (Cy3) and 3T3 (Cy5) RNA samples. These were hybridized to 10 816 element cDNA arrays. After the data were filtered and normalized, 544 genes were identified as differentially expressed, ‘true outliers’ based on the results of two experiments (see the ‘Array scanning and data analysis’ section for details). Since the instructions provided with the Arcturus kit indicate that it can be employed to perform a maximum of two rounds of amplification, we next used Arcturus’ method, and our original and new methods to amplify C2 and 3T3 RNAs one or two times, and then we used the products to make probes. We could detect 445, 456 and 446 of the differentially expressed genes, respectively, when probes were prepared with RNA amplified by means of our original method, our new method and the Arcturus kit. Using probes prepared from RNAs that had undergone two rounds of amplification, we were able to detect 394 of the differentially expressed genes with our original method, 429 with new T3N9-based method, and 397 with Arcturus’ RiboAmp kit. Thus, the method based exclusively on T3N9 priming seemed as reliable as those based on oligo dT priming in the first round of amplification.
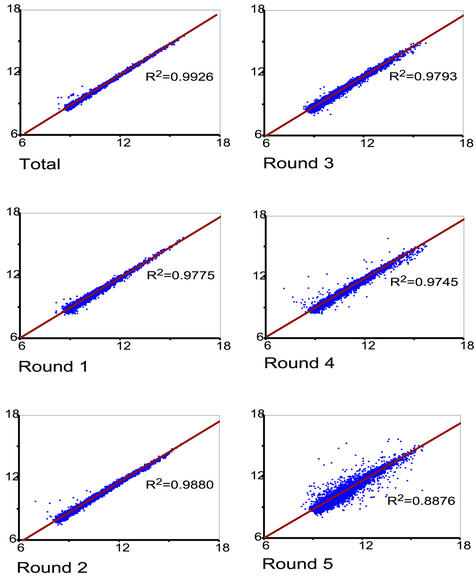
We showed earlier (14) that our original amplification method could be used for multiple rounds of template amplification, and wanted to know whether this was still the case when T3N9 replaced T7dT in the first stage of the procedure. To answer this question, we amplified two samples of C2 RNA in parallel, labeled probes with Cy3 or Cy5 from the products at each stage, and hybridized arrays with these probes. We have found that the results of such self-on-self experiments correlate well with our ability to detect differentially expressed genes in two RNA samples. As Figure 2 shows, correlation coefficients of 0.9926, 0.9775, 0.9880, 0.9793 and 0.9745 were found when unamplified RNA and RNA amplified one to four times, respectively, were used for probe preparation. Following a fifth round of amplification, however, the correlation coefficient dropped to 0.8876. M versus A plots, which are available at our web site (http://intramural.nimh.nih.gov/research/log/pubs/pubs.html), also illustrate the high quality of probes prepared from RNA amplified as many as four times. We have learned empirically that three stages of amplification allow us to produce enough template from 10–100 cells to profile them. In fact, we have recently been able to profile single hypothalamic magnocellular neurons following three rounds of template amplification (data not shown). To compare two or more samples to one another, we recommend using the same number of rounds of amplification for each one.
Figure 2.
Reproducibility of RNA amplification with T3N9: self versus self experiments. A single RNA sample was divided in half and amplified up to five rounds. Following each round, the samples were used to prepare Cy3- or Cy5-labeled probes, which were combined and applied to a 10 816-element array. DNA used as a template for the first round of amplification was synthesized from 1 µg of mouse C2 total RNA. The reverse transcription was primed by T3N9 in each of the five rounds of amplification (see Materials and Methods and Fig. 1). Probes made from unamplified total RNA (R2 = 0.9926), and RNA that had been amplified one, two, three and four times gave excellent correlation coefficients: R2 = 0.9775, 0.9880, 0.9793 and 0.9745, respectively. Following the fifth round, the scatter increased and the correlation coefficient dropped to 0.8876.
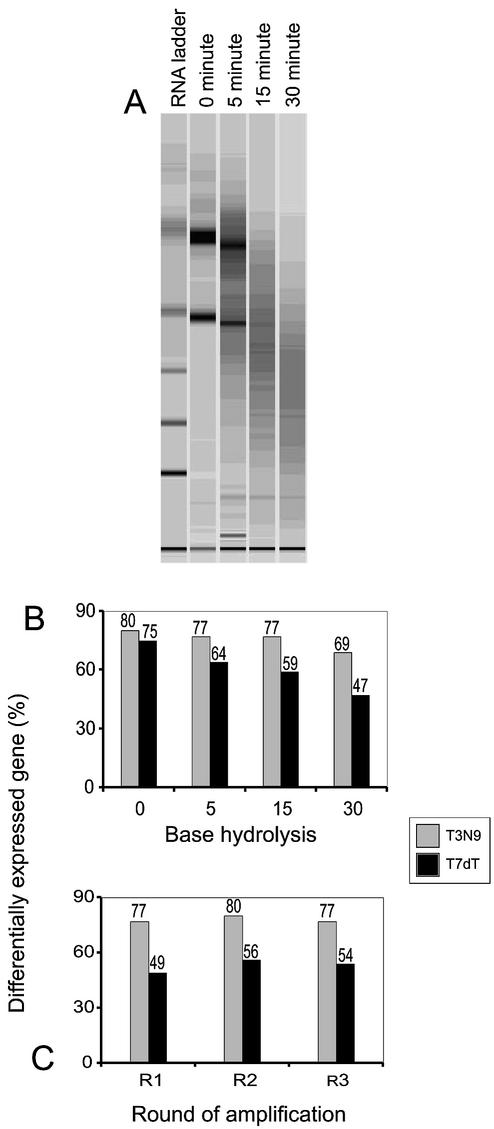
To determine whether our new method could be employed to amplify degraded RNA templates, we heated C2 and 3T3 total RNA samples in a pH 8.3 solution containing calcium and magnesium. As Figure 3A shows, this results in a time-dependent breakdown of the RNA. Using the RNAs collected after 0, 5, 15 and 30 min of base hydrolysis at 80°C as templates, we performed single-stage amplifications, driving first-strand synthesis with either T7dT or T3N9. We then labeled the C2 products with Cy3 and the 3T3 products with Cy5, combined them, and hybridized them to 10 816 element arrays. The expression profiles were compared with those obtained when intact C2 and 3T3 RNA samples were labeled without amplification, and used to assemble a list of differentially expressed genes (see Array scanning and data analysis). The fraction of differentially expressed genes detected with degraded RNA was remarkably constant when T3N9 primed amplification was used (see Fig. 3B). It only seemed to fall off in the case of the most damaged sample—one that had been incubated at 80°C for 30 min. When T7dT priming was done, on the other hand, there was a marked reduction in our ability to detect differentially expressed genes, especially in the badly degraded samples.
Figure 3.
RNA degradation and the reliability of microarray data. (A) Incubating total RNA from C2 cells in a basic solution at 80°C resulted in a time-dependent breakdown of the sample. (B) Probes made from T7dT- or T3N9-amplified samples of degraded RNA from C2 and 3T3 cells were compared to those made from intact, unamplified templates. Even with badly degraded RNA, the T3N9-amplification method could be used to detect most of the differentially expressed genes found with intact, unamplified templates. (C) Using total RNAs hydrolyzed in a basic solution for 15 min as templates, we could still detect 80 and 77% of differentially expressed genes after two and three rounds of amplification starting with T3N9. In contrast, only 56 and 54% of the differentially expressed genes could be detected with T7dT.
We selected the RNA samples that had been base-hydrolyzed for 15 min for further study, amplifying them three times with our original method (T7dT in round 1) or our new one (T3N9 in round 1). As Figure 3C shows, we could detect 77% of differentially expressed genes in the two samples with the new method after three rounds of amplification even though the templates were quite degraded. Using our original method, we could only detect 54% of the differentially expressed transcripts.
Unfortunately, our arrays have few elements representing histone transcripts on them, and those that are there correspond to the ‘replacement variant genes’ which are not cell-cycle regulated and which are polyadenylated (16). For this reason, we could not test the hypothesis that T3N9 priming would permit us to study histone gene transcription, but there is no reason to doubt this.
In conclusion, random primers with a phage RNA promoter sequence on their 5′ ends (T3N9) generate high fidelity amplified templates that can be used to study gene expression with high-density DNA microarrays. This method for amplifying RNA should be especially useful for studying degraded samples. Multiple rounds of amplification can be performed using the same primer and reagents.
Acknowledgments
ACKNOWLEDGEMENTS
This project was partly supported by federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400, and by a grant from Merck and Co., Inc. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed]
- 2.DeRisi J., Penland,L., Brown,P.O., Bittner,M.L., Meltzer,P.S., Ray,M., Chen,Y., Su,Y.A. and Trent,J.M. (1996) Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet., 14, 457–460. [DOI] [PubMed]
- 3.Lashkari D.A., Derisi,J.L., McCusker,J.H., Namath,A.F., Gentile,C., Hwang,S.Y., Brown,P.O. and Davis,R.W. (1997) Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl Acad. Sci. USA, 94, 13057–13062. [DOI] [PMC free article] [PubMed]
- 4.Baelde H.J., Cleton-Jansen,A.-M., van Beerendonk,H., Namba,M., Bovee,J.V.M.G. and Hogendoorn,P.C.W. (2001) High quality RNA isolation form tumors with low cellularity and high extracellular matrix component for cDNA microarrays: application to chondrosarcoma. J. Clin. Pathol., 54, 778–782. [DOI] [PMC free article] [PubMed]
- 5.Stears R.L., Getts,R.C. and Gullans,S.R. (2000) A novel, sensitive detection system for high-density microarrays using dendrimer technology. Physiol. Genomics, 3, 93–99. [DOI] [PubMed]
- 6.Wong K.-K., Cheng,R.S. and Mok,S.C. (2001) Identification of differentially expressed genes from ovarian cancer cells by MICROMAX cDNA microarray system. Biotechniques, 30, 670–675. [DOI] [PubMed]
- 7.Xiang C.C., Kozhich,O.A., Chen,M., Inman,J.M., Phan,Q.N. and Brownstein,M.J. (2002) An improved method to label probes for DNA microarray work: amine-modified random primers. Nat. Biotechnol., 20, 738–742. [DOI] [PubMed]
- 8.Phillips J. and Eberwine,J.H. (1996) Antisense RNA amplification: a linear amplification method for analyzing the mRNA population from single living cells. Methods, 10, 283–288. [DOI] [PubMed]
- 9.Wang E., Miller,L.D., Ohnmacht,G.A., Liu,E.T. and Marincola,F.M. (2000) High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol., 18, 457–459. [DOI] [PubMed]
- 10.Baugh L.R., Hill,A.A., Brown,E.L. and Hunter,C.P. (2001) Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res., 29, e29. [DOI] [PMC free article] [PubMed]
- 11.Pabon C., Modrusan,Z., Ruvolo,M.V., Coleman,I.M., Daniel,S., Yue,H. and Arnold,L.J.,Jr (2001) Optimized T7 amplification system for microarray analysis. Biotechniques, 31, 874–879. [DOI] [PubMed]
- 12.Puskas L., Zvara,G.A., Hackler,L.,Jr and Van Hummelen,P. (2002) RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques, 32, 1330–1340. [DOI] [PubMed]
- 13.Zhao H., Hastie,T., Whitfield,M.L., Borresen-Dale,A.-L. and Jeffrey,S.S. (2002) Optimization and evaluation of T7 based RNA linear amplification protocols for cDNA microarray analysis. BMC Genomics, 3, 31–45. [DOI] [PMC free article] [PubMed]
- 14.Xiang C.C., Chen,M., Kozhich,O.A., Phan,Q.N., Inman,J.M., Chen,Y. and Brownstein,M.J. (2002) A new method to make probes directly from small numbers of cells for DNA microarray studies. Biotechniques, 34, 386–393. [DOI] [PubMed]
- 15.Chen Y., Kamat,V., Dougherty,E.R., Bittner,M.L., Meltzer,P.S. and Trent,J.M. (2002) Ratio statistics of gene expression levels and applications to microarray data analysis. Bioinformatics, 18, 1207–1215. [DOI] [PubMed]
- 16.Marzluff W.F., Gongidi,P., Woods,K.R., Jin,J. and Maltais,L.J. (2002) The human and mouse replication-dependent histone genes. Genomics, 80, 487–498. [PubMed]