Abstract
Contiguous gene syndromes (CGS) are a group of disorders associated with chromosomal rearrangements of which the phenotype is thought to result from altered copy numbers of physically linked dosage-sensitive genes. Smith-Magenis syndrome (SMS) is a CGS associated with a deletion within band p11.2 of chromosome 17. Recently, patients harboring the predicted reciprocal duplication product [dup(17)(p11.2p11.2)] have been described as having a relatively mild phenotype. By chromosomal engineering, we created rearranged chromosomes carrying the deletion [Df(11)17] or duplication [Dp(11)17] of the syntenic region on mouse chromosome 11 that spans the genomic interval commonly deleted in SMS patients. Df(11)17/+ mice exhibit craniofacial abnormalities, seizures, marked obesity, and male-specific reduced fertility. Dp(11)17/+ animals are underweight and do not have seizures, craniofacial abnormalities, or reduced fertility. Examination of Df(11)17/Dp(11)17 animals suggests that most of the observed phenotypes result from gene dosage effects. Our murine models represent a powerful tool to analyze the consequences of gene dosage imbalance in this genomic interval and to investigate the molecular genetic bases of both SMS and dup(17)(p11.2p11.2).
Contiguous gene syndromes (CGS) are a group of disorders associated with chromosomal rearrangements (deletions or duplications) of which the phenotype is thought to result from altered copy numbers of physically linked dosage-sensitive genes. Several CGS have been described, each of them presenting a complex and specific phenotype (28). However, to date only a few contiguous gene deletion syndromes [e.g., DiGeorge and velocardiofacial syndromes, associated with del(22)(q11.2q11.2)] have been partially modeled in mouse (10, 13, 14, 19). The molecular mechanism for several contiguous gene deletion syndromes is nonallelic homologous recombination (NAHR) or unequal crossing over, between flanking low-copy-number repeats (LCRs) (9, 32, 33), and thus the vast majority of patients with any given CGS harbor a common deletion encompassing a specific genomic interval. This molecular model predicts a reciprocal duplication product for each NAHR event (16). Although the specific rearranged intervals for a number of CGS are known, which gene(s) is susceptible to dosage remains largely unknown. Since the rearrangements are due to the recombination of specific LCRs, most patients have the same genomic region rearranged. Thus, identifying patients with unusually sized rearrangements to enable the discovery of the responsible gene(s) has met with limited success. The ability to model such conditions in mice is essential for understanding the dosage-sensitive genes and the phenotypic consequences of gene copy number imbalances.
Smith-Magenis syndrome (SMS) is a microdeletion syndrome associated with a deletion within chromosome 17 band p11.2. First described in 1986 (31), SMS has a birth prevalence estimated at 1 in 25,000 (7). The clinical phenotype has been well described and includes craniofacial abnormalities, brachydactyly, self-injurious behavior, sleep abnormalities, and mental retardation. Less commonly reported are cleft palate, congenital heart defects, seizures, hearing impairment, and urinary tract anomalies (8). Hypercholesterolemia has also recently been reported in children with SMS (30). Molecular studies (5) revealed a common deletion region of ∼4 Mb in more than 90% of SMS patients. Three copies of an LCR, termed SMS-REP, were identified in the common deletion region (5, 22); the flanking SMS-REP copies provide substrates for homologous recombination and mediate NAHR, resulting in the rearrangement of this chromosomal segment. Recently, the predicted reciprocal duplication product has been described for humans (23). The phenotype for this independent syndrome associated with the duplication rearrangement seems to be less severe than SMS, with mild to borderline mental retardation and behavioral difficulties in the few reported patients.
Human chromosome 17p11.2 is syntenic to the 32-to-34-centimorgan (cM) region of murine chromosome 11. The numbers and orders of the genes are highly conserved (3). The 28 genes that map within or around the common deletion region of SMS have homologous counterparts on the syntenic region of mouse chromosome 11. The ∼1.0-Mb SMS critical interval, defined as the smallest region of overlapping deletions in SMS patients with rarely identified unusually sized deletions, reveals 19 genes with the same gene order and orientation as the murine counterpart (3).
Chromosome engineering is a strategy recently developed to introduce defined chromosomal rearrangements into the mouse genome to delineate haploinsufficiency effects (haploid genetics) (20, 37). The rearrangement is engineered in embryonic stem (ES) cells by serial targeting of the specific endpoints of the human deletion in mouse with the Cre-loxP site-specific recombination system (26, 39). Using this methodology, and taking advantage of the significant conserved synteny between human and mouse in this genomic interval, we were able to create a deletion and reciprocal duplication in the mouse chromosome region syntenic to the SMS critical interval. The phenotypes of the mice with these rearrangements were analyzed in order to assess phenotypic consequences of gene dosage imbalances in this region of the mouse genome and make inferences regarding the human syndromes.
MATERIALS AND METHODS
Construction of vectors and targeting of endpoints.
A previously described system (26, 39) for generating defined chromosomal rearrangements was utilized. Briefly, the method utilizes two mouse genomic libraries, the 5′ hprt library and the 3′ hprt library, containing in each vector backbone one loxP site, one half of the Hprt minigene, exons 1 to 2 and exons 3 to 9, respectively, a coat color marker (tyrosinase minigene and K14Agouti transgene, respectively), and a resistance marker for selection (neomycin [Neo] and puromycin [Puro], respectively). For targeting of the mouse genome, the two serially inserted loxP sites can be recombined by Cre recombinase and the Hprt minigene is reconstituted so that cells with rearranged chromosomes can be selected by using hypoxanthine-aminopterin-thymidine (HAT) media. To prepare the vector for insertion into the wild-type chromosome, a gap is introduced in the genomic insert of the vector before electroporation of the clone into ES cells. The insertion of the vector and the duplication of the insert area are achieved by gap repair.
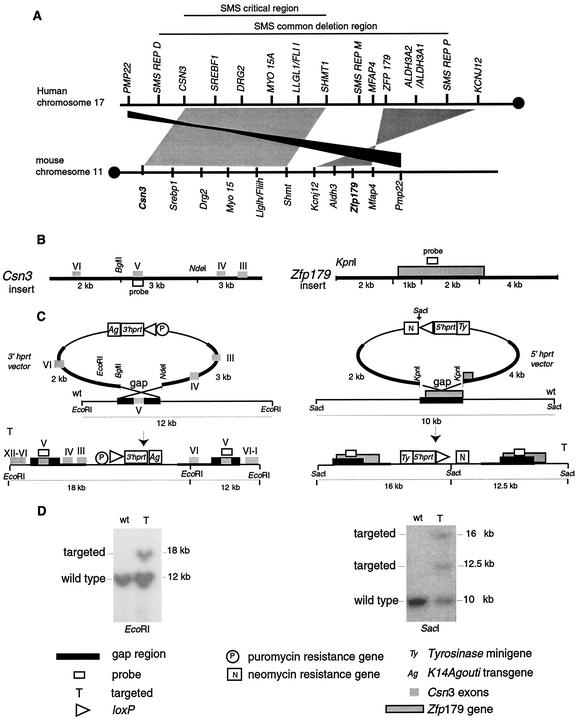
The AB2.2 ES cell line derived from Hprt-deficient 129S5/SvEvBrd mice (17) was used. We screened the 5′ hprt library for Zfp179, and one clone was selected that contained an ∼9-kb genomic insert encompassing most of the Zfp179 gene except for the last exon (exon 16), which is not included in the translated protein (38). A 3-kb gap was created between two KpnI sites. This enzyme was then used for linearization of the clone prior to electroporation. The 3′ hprt library was screened for Csn3. One clone was selected that contained an ∼8-kb genomic insert. This clone contained exons II± to VI of the Csn3 gene. A 3-kb gap between BglII and NdeI was introduced, the BglII site was restored after religation, and this site was used for linearization of the clone prior to electroporation into ES cells. ES cell growth, electroporation, drug selection, targeting detection, and germ line transmission were performed as described previously (17, 25). The targeting of both vectors was performed sequentially. For insertional vectors, targeting events create a miniduplication of the insert region, and as a result a disruption of Csn3 is obtained while a duplication of the whole gene is achieved for Zfp179 (see Fig. 1 and 2). Cells electroporated with the Zfp179 vector were selected with G418 for 7 to 9 days, 96 ES cell clones were isolated and screened by Southern blotting by using a Zfp179 probe, and of the clones screened, 15% showed the expected pattern for targeted insertion when the DNA was digested with SacI. Three ES cell clones were tested for germ line transmission, and clone 3 was selected and electroporated with the Csn3 vector. ES clones were selected for puromycin resistance, 96 of them were analyzed by Southern blotting, and 19% of these cells presented the anticipated pattern for targeted insertion when the DNA was digested with EcoRI.
FIG. 1.
Synteny and targeted endpoints. (A) The 17p11.2 region of human chromosome 17 and the 32-to-34-cM region of syntenic mouse chromosome 11 are shown. Circles depict centromeres. The horizontal lines (above) denote the genomic regions encompassing the SMS common deletion region and the SMS critical region. Note that the numbers, orientations, and relative orders of the genes in these syntenic genomic intervals are extremely conserved. Schematic representations of the insertion of loxP sites for chromosomal engineering of the rearrangements are shown below. SMS REP, copy of an LCR (D, distal; M, middle; P, proximal). (B) Csn3 and Zfp179 were selected as proximal and distal anchor points, respectively. The 3′ hprt Csn3 and the 5′ hprt Zfp179 plasmid vectors are shown, and the restriction maps of the inserts (bold lines) are indicated. The Csn3 insert contains exons III to VI of the gene and can be cut into three fragments (3, 3, and 2 kb) when codigested with NdeI and BglII, while the Zfp179 insert contains the whole Zfp179 gene except for the last exon that is not transcribed into protein and can be cut into two flanking fragments of 3 and 2 kb and two internal fragments of 1 and 2 kb (shown as 3 kb for simplicity) when digested with KpnI. The internal fragments are removed by one step of subcloning in order to create a gap in the region of homology to obtain the gapped targeting vector. (C) The gapped targeting vectors are linearized at the BglII site (for Csn3) and the KpnI site (for Zfp179) and targeted consecutively to each respective locus. The insertion of the vector creates a disruption of the Csn3 gene, while for Zfp179, a duplication of the gene is achieved. wt, wild-type. (D) Southern analysis of ES cells. The ES cells positive for the first targeting event (Zfp179) were recognized by the visualization of the expected 16-kb and 12.5-kb hybridizing SacI bands diagnostic of the targeting. These ES cell clones were then utilized for the second targeting (Csn3), and the expected 18-kb EcoRI band diagnostic of the targeting was identified.
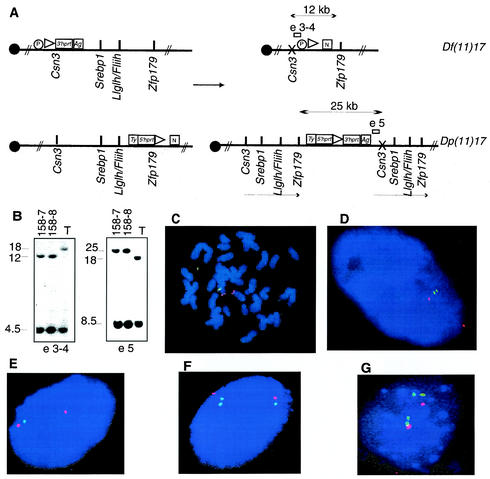
FIG. 2.
Genetically engineered chromosomes. (A) Double-targeted ES cell clones were subjected to transient expression of Cre recombinase. The predicted products of the recombination are schematized, with arrows indicating the expected fragment sizes when genomic DNA from rearranged chromosomes was digested with NdeI. Open rectangles represent the probes used in genomic Southern analysis of ES cells; probe e3-4 is predicted to detect a 12-kb band in the cell line with the deletion, whereas e5 is anticipated to detect a 25-kb band in the cell line with the duplication. The presence of an X over Csn3 denotes that a mutation of this gene is achieved by the insertion of the targeting vector. Ty, tyrosinase minigene; Ag, K14Agouti transgene; N, neomycin resistance gene; P, puromycin resistance gene. (B) Southern analysis of ES clones selected by using HAT media after the induction of Cre-loxP-mediated recombination. Two clones, 158-7 and 158-8, were analyzed sequentially with both probes, and the expected pattern for duplication and deletion of the predicted regions was identified in genomic DNA digested with NdeI. T, double-targeted ES cell not subjected to Cre recombinase. Independent confirmation of the predicted structure for rearranged chromosomes was obtained by FISH analysis with large insert BAC probes. (C) ES cells from clone 158-7 with deletion and duplication metaphase chromosomes were analyzed with probes containing Fliih (green) as well as D11Mit11 (red) as a reference probe that lies outside the rearranged interval. Note the absence of a green (Fliih) signal on one chromosome 11 homologue, signifying deletion. (D) Interphase nuclei from the same ES cell clone probed with Srebp1 (green) and Pmp22 (red) probes. The extended interphase chromosomes enable visualization of the submicroscopic duplicated interval (two nearby green dots). Identical results were obtained with ES cell clone 158-8. (E, F, and G) Depicted are results from FISH analysis of interphase nuclei of tail fibroblasts obtained from animals with the following genotypes: Df(11)17/+, Dp(11)17/+, and Dp(11)17/Dp(11)17, respectively. Srebp1 (green) and Pmp22 (red) were used as probes.
Chromosomal rearrangements.
Three ES cell clones double targeted at Zfp179 and Csn3 (clones 127, 158, and 168) were used to induce the rearrangements. Transient Cre recombinase expression and selection of recombination products by using HAT media were performed as described previously (15, 26). Twelve colonies of each clone were analyzed by mini-Southern blot analysis. Fluorescence in situ hybridization (FISH) was performed on chromosome-engineered ES cell spreads by utilizing BAC probes (from the RPCI-24 male [C57BL/6J] mouse bacterial artificial chromosome [BAC] library) following a standard protocol (2) in order to confirm the rearrangements. Two clones (158-7 and 158-8) carrying the deletion, termed del(11)(Csn3-Zfp179) and referred to as Df(11)17, of the region flanked by the two loxP sites in one chromosome and the duplication, termed dup(11)(Csn3-Zfp179) and referred to as Dp(11)17, of the same fragment in the other chromosome were injected into C57BL/6 Tyrc-Brd blastocysts. The resulting chimeric males were bred to C57BL/6 Tyrc-Brd wild-type females in order to obtain F1 mice carrying the Df(11)17 and the Dp(11)17 chromosomes.
Mouse strains and genotyping.
The phenotypes were analyzed in a mixed genetic background: C57BL/6 Tyrc-Brd × 129S5/SvEvBrd. The animals were genotyped by Southern blot analysis (utilizing the same probe as that used for the Csn3 targeting event) when the mice were 3 weeks of age and by FISH of tail fibroblast spreads. In addition, Dp(11)17/+ and Dp(11)17/Dp(11)17 mice can be distinguished from each other and their wild-type littermates by coat color since both coat color markers present in the vector are retained and functional in these animals, thus facilitating genotyping. Every four generations, randomly selected mice were tested for the stable transmission of rearranged chromosomes by FISH analysis of tail fibroblast spreads.
Histopathology and electrophysiology.
Animals were perfused with buffered 4% formaldehyde following inhalation of anesthesia. Tissues were paraffin embedded. Histological staining was performed with hematoxylin and eosin.
The caudal epididymal sperm from adult male mice was collected by incubating minced caudal epididymis in phosphate-buffered saline medium (250 μl) at 37°C for 10 min. The sperm number was calculated by using a hemocytometer for sperm at a 1:10 dilution. Sperm morphology was assessed by staining the sample smears with hematoxylin for 3 min.
Skeletal preparations were performed as described previously (18). Briefly, mice were skinned, eviscerated, and fixed in 95% ethanol. After cartilage was stained in a solution containing 0.05% alcian blue 8GX and 5% acetic acid, the specimens were refixed in 95% ethanol and treated in 2% KOH until mostly clear. The skeletons were then stained in a solution containing 0.015% alizarin red S and 1% KOH and cleared in a 1:1 mixture of 95% ethanol and glycerol. Both newborns and adult mice were processed by using the same procedures, with modifications in timing only. For statistical analysis, different landmarks were chosen in two-dimensional images of wild-type and Df(11)17/+ skulls captured with Image Pro Plus software (Media Cybernetics). Relative measurements (in centimeters) were taken by utilizing Adobe Photoshop version 5.5 (Adobe Systems, Incorporated).
Electroencephalogram (EEG) studies were executed by following standard procedures. Teflon-coated silver wire electrodes (0.005-in. diameter) soldered to a microminiature connector were implanted bilaterally into the subdural space over the frontal and parietal cortexes of anesthetized mice at least 24 h before recording (21). Cortical activity was recorded by using a digital video electroencephalograph (Stellate Harmonie) on animals moving freely in the test cages for prolonged periods of time. All video EEGs were reviewed for evidence of seizures and abnormal electrographic activity.
Statistical analysis.
Statistical analysis was performed by using Student's t test run on a Microsoft Excel (version 2002-SP1; Redford, Wash.) software program. A P value of <0.05 was considered significant.
RESULTS
Chromosome engineering for human CGS.
Human chromosome region 17p11.2 is highly syntenic to the 32-to-34-cM region of murine chromosome 11 (Fig. 1A) (3). To generate chromosomal rearrangements that include the common (syntenic) deletion region, Csn3 was selected as the proximal anchor point for one loxP site and Zfp179 was selected as the distal anchor point in murine chromosome 11. We sequentially inserted chromosome-engineering cassettes by gap repair recombination (39) into Zfp179 (Fig. 1C) and Csn3 (Fig. 1B), which are located approximately 3 Mb apart (3). Double-targeted ES cells were then subjected to Cre-mediated site-specific recombination, and the ES cells carrying rearrangements were selected in HAT media by virtue of the reconstituted Hprt gene. The recombination frequencies were 296 × 10−7, 12 × 10−7, and 176 × 10−7 per electroporated cells for clones 127, 158, and 168, respectively. HAT-resistant clones were also resistant to G418 and puromycin, consistent with a trans recombination event. For each of three different double-targeted ES clones (127, 158, and 168), 12 colonies were analyzed by mini- Southern analysis. Two clones, 158-7 and 158-8, demonstrated a pattern consistent with a deletion, termed del(11)(Csn3-Zfp179) and hereafter referred to as Df(11)17, and a reciprocal duplication, termed dup(11)(Csn3-Zfp179) and hereafter referred to as Dp(11)17, of the corresponding regions on the chromosome 11 homologues (Fig. 2A and B). To confirm these rearrangements, FISH was performed by utilizing different pairs of BAC clones as probes. For BAC probes outside the rearranged region, D11Mit11 (330P14) and Pmp22 (480F3) were utilized, whereas for probes inside the rearranged region, Fliih (278I21), Srebp1 (438J17), or Aldh3 (40J4) was used.
FISH results for compound heterozygous Df(11)17/Dp(11)17 ES cells are shown in Fig. 2C and D. When metaphase chromosomes were analyzed by utilizing D11Mit11 and Fliih probes, the D11Mit11 marker was present on both chromosomes but Fliih was present on only one homologue. As the size of the deletion or duplication region is ∼3 Mb, below the resolution of metaphase spreads, interphase nuclei were analyzed in order to confirm the presence of duplication. Pmp22, which maps outside the SMS syntenic region, was present on both chromosomes; Srebp1, which maps within the common region, was absent from one homologue but present in two copies (two signals) on the homologous chromosome, confirming the duplication of this region. The results with each different combination of probes indicate that the rearrangements were successfully obtained (data not shown).
Viability of mice harboring rearranged chromosomes.
Two independent ES clones containing both rearrangements were injected into blastocysts. Chimeric male mice were mated with wild-type C57BL/6 Tyrc-Brd females, and germ line transmission of both Df(11)17 and Dp(11)17 chromosomes was independently achieved. Heterozygous animals were analyzed by FISH of mouse tail fibroblasts (Fig. 2E and F). A ratio of approximately 1 to 1 for Df(11)17/+ to Dp(11)17/+ animals (n = 95) was obtained, indicating that both rearrangements were transmissible and viable in the heterozygous state. The life span of both Df(11)17/+ and Dp(11)17/+ animals appears to be normal; the oldest mice are currently 2 years of age.
To examine the viability of the rearrangements in the homozygous state after birth, heterozygous animals were mated. Df(11)17/+ × Df(11)17/+ matings yielded a wild type-to-heterozygote ratio of approximately 1 to 2, but no homozygous Df(11)17/Df(11)17 animals were found (n = 40), indicating that the deletion is lethal in the homozygous state. This outcome was expected since at least two genes included in this region, Top3α (12) and Fliih (4), have been independently targeted and null alleles are lethal in the homozygous state. However, Dp(11)17/+ × Dp(11)17/+ matings gave rise to homozygous Dp(11)17/Dp(11)17 animals (Fig. 2G).
Df(11)17/+ × Dp(11)17/+ matings were set up. This enabled us to obtain animals carrying both Df(11)17 and Dp(11)17 chromosome rearrangements, and the gene dosage in these animals is balanced. Df(11)17/Dp(11)17 animals were obtained at the expected Mendelian ratio and with no apparent phenotype, consistent with the absence of overt phenotypes secondary to position effects.
Phenotypic analysis reveals no obvious major organ system maldevelopment.
Clinical manifestations in SMS patients include craniofacial abnormalities (brachycephaly, prominent forehead, synophrys, epicanthic folds, broad nasal bridge, ear anomalies, and prognathism) in 52 to 93% of patients, brachydactyly (85%), self-injurious behaviors (74%), sleep abnormalities (69%), speech delay, and various degrees of mental retardation (100%). Less commonly reported are hearing impairment (63%), urinary tract and renal anomalies (35%), congenital heart defects (29%), and seizures (19%) (6, 8). For dup(17)(p11.2p11.2) patients, the clinical phenotype is still being established, given limited studies, but it usually appears to be less severe, with prominent features including borderline to mild mental retardation and behavioral difficulties. No prominent dysmorphic features or major organ developmental abnormalities were seen in the majority of these patients (23).
The chromosome-engineered mice were analyzed for specific phenotypic features to determine if indeed these animals are appropriate models to study these CGS. We performed analyses with mice at 3 to 4 months of age except when otherwise specified. Gross necropsy studies were done for all genotypes. No gross developmental defects were identified in the urinary tracts or in the hearts of any of the analyzed animals in this mixed C57BL/6 Tyrc-Brd × 129S5/SvEvBrd genetic background [Df(11)17/+, n = 30; Dp(11)17/+, n = 30]. This finding is consistent with the lack of gross developmental abnormalities observed in most duplication patients. Heart and renal defects are present in 28 and 29% of SMS patients, respectively (8). The lack of organ developmental abnormalities in the Df(11)17/+ mice may reflect the genetic background. Histological examinations were performed with brains, hearts, spleens, and kidneys, and no obvious defects were identified in any of these tissues for any of the genotypes analyzed [Dp(11)17/+ and Df(11)17/+; n = 20 for each phenotype]. Although ophthalmological abnormalities were observed in 85% of SMS patients, no iris abnormalities, microcorneas, or retinal detachments were identified in Df(11)17/+ (n = 12) or Dp(11)17/+ (n = 12) animals. However, we did observe multiple other phenotypes specific to the rearrangement genotypes, and these include craniofacial abnormalities, seizures and abnormal EEGs, growth (weight) differences, and reduced fertilities, most of which appear to result from gene dosage effects.
Craniofacial abnormalities.
One of the characteristic physical features of SMS patients is craniofacial abnormalities, including brachycephaly, midface hypoplasia, broad face, short and broad nose, down-turned lip, and prognathism (1, 8, 31). A study with detailed facial measurements from 55 SMS patients indicated that the facial dysmorphism evolves over age (1). Skeletal preparation was carried out with heterozygous Df(11)17/+ newborns and adult mice, with wild-type littermates as controls. While no apparent difference was observed between Df(11)17/+ newborns and the wild-type littermates (data not shown), obvious craniofacial abnormalities were found in all Df(11)17/+ adult animals studied (Fig. 3).
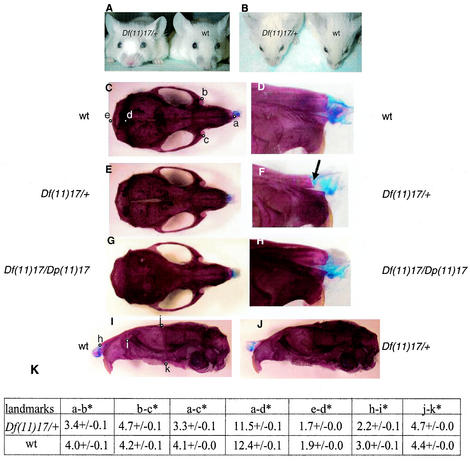
FIG. 3.
Craniofacial abnormalities. (A and B) Df(11)17/+ and wild-type (wt) animals are pictured together. (A) Note the positions of the snouts and the broad distance between the eyes (hypertelorism) of the Df(11)17/+ mouse compared with that between the eyes of the wild-type littermate. (B) The short distance between the eyes and the nose of the Df(11)17/+ mouse compared with the distance between those of the wild-type littermate can also be visualized. Skeletal preparations of wild-type (C, D, and I), Df(11)17/+ (E, F, and J), and Df(11)17/Dp(11)17 (G and H) skulls of 3-month-old male animals are shown for comparison. The different positions of the noses in the Df(11)17/+ mice can be explained by the shape of the nasal bone (F, arrow) compared with that of the wild type (D). (H) This phenotype is completely rescued with the addition of an extra copy of the genes that are deleted [Df(11)17/Dp(11)17 animals]. (K) The different landmarks pictured in panels C and I were used to objectively measure the distances between the landmarks. Cranial landmarks are as follows: a and h, nasal; b, c, and i, anterior notch on frontal process situated laterally in relation to the intraorbital fissure; d, intersection of parietal and intraparietal bones; e, intersection of interparietal and occipital bones at the midline; j, bregma; and k, intersection of maxilla and sphenoid on inferior alveolar ridge. The relative distances (in centimeters; see Materials and Methods) were used for the statistical analysis, and the averages of the distances are shown in the table. The asterisks denote that significant differences (P of <0.05) between the measurements for Df(11)17/+ (n = 3) and wild-type littermates (n = 3) were found.
Different landmarks of wild-type skulls (Fig. 3C and I) were established in order to perform objective measurements (27). Relative distances were measured for the wild type (Fig. 3C and I) and Df(11)17/+ (Fig. 3E and J) and analyzed (Fig. 3K). Df(11)17/+ mice had shorter overall skulls (distances from a to d and d to e) and broader and shorter snouts and nasal bones (distances from a to b, a to c, b to c, and h to i) than the wild type. A lateral view reveals that the skull is broader for the Df(11)17/+ heterozygous mouse than for the wild type (distances from j to k). Of note, the shapes of the nasal bones are very distinctive for Df(11)17/+ animals and their wild-type littermates (Fig. 3F and D). To determine whether the skeletal phenotype observed for Df(11)17/+ mice is due to haploinsufficiency of genes within the SMS deleted region, Df(11)17/Dp(11)17 animals were analyzed. Skeletons of 3-month-old animals exhibited a wild-type phenotype, demonstrating that the craniofacial phenotype was rescued by the addition of the Dp(11)17 chromosome (Fig. 3G and H). These results indicate that the craniofacial features in SMS patients are due to a gene dosage effect and are not the result of a position effect.
Seizures and abnormal EEGs in Df(11)17/+ mice.
About 19% of the patients with SMS manifest a seizure disorder at some point in their lives (8), but only a small proportion of patients with dup(17)(p11.2p11.2), one in seven, were reported to have seizures (23). We observed overt clinical seizures in Df(11)17/+ animals between the ages of 4 weeks and 6 months, but no seizures were observed in Dp(11)17/+ or Df(11)17/Dp(11)17 animals. The percentage of Df(11)17/+ animals with overt seizures was 22% (for all genotypes, n = 50). All seizures witnessed in Df(11)17/+ animals were generalized and tonic clonic in nature.
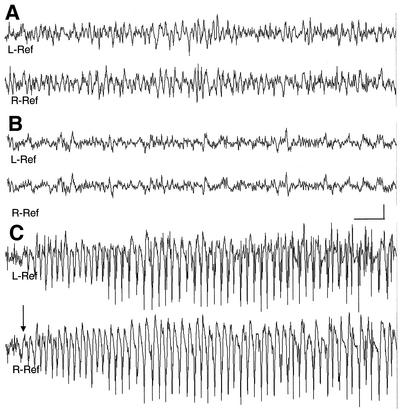
Prolonged video EEG monitoring was performed with freely moving mice with the following genotypes: Df(11)17/+ (n = 6), wild type (n = 3), Dp(11)17/+ (n = 4), and Df(11)17/Dp(11)17 (n = 4). No apparent abnormalities were seen in Dp(11)17/+ mice or wild-type littermates. In both, the waking background EEG activity consisted of low voltage activity with 4 to 5, 8 to 9, and 18 to 25 Hz being the most predominant pattern observed. During behavioral sleep, higher voltage delta (1 to 4 Hz) rhythms increased, and occasional sharp wave activity was noted. In contrast, in Df(11)17/+ mice, paroxysmal spikes and slow discharges were seen. Spike and wave activity occurred in singles and occasional doubles; rare polyspikes were observed. Bilaterally synchronous spike and slow wave activity occurred in five of six Df(11)17/+ animals analyzed. During behavioral sleep, spikes were seen for five of six animals. Of the mice recorded by using EEGs, generalized tonic clonic seizures were witnessed in two of six animals, and one of these animals had myoclonic jerks without a clear EEG signature being identified. In a recorded seizure, single spikes progressed to polyspike bursts of increasing duration which coincided with tail extension. Subsequently, a generalized seizure ensued, with continuous spike and wave activity lasting 23 s, followed by postictal depression (Fig. 4). No photically induced abnormalities were seen for any of the analyzed genotypes. Prolonged monitoring of Df(11)17/Dp(11)17 mice (n = 4) showed a single, milder electrographic seizure with behavioral arrest, suggesting that positional effects of genes within the deletion interval may potentially contribute to seizure susceptibility.
FIG. 4.
Chronic bilateral electrocorticograms reveal epileptic seizures in Df(11)17/+ mice. Background EEG rhythms from a freely behaving wild-type mouse (A) and a Df(11)17/+ mouse (B) are shown. (C) Onset of spontaneous seizure discharge (arrow) in Df(11)17/+ mouse. Bars, 1 s and 500 μV. L-Ref, left reference electrode; R-Ref, right reference electrode.
Histological analysis of brain sections from a Df(11)17/+ mouse in which seizures were witnessed, a Df(11)17/+ mouse without overt seizures but with abnormal EEG, and a wild-type littermate failed to demonstrate any obvious cytopathology in the brain hippocampal or cortical regions (data not shown). Thus, although behavioral seizures were witnessed and objective electrophysiological evidence was recorded, no specific anatomical features causative of the seizures could be identified.
Gene dosage effects on weight.
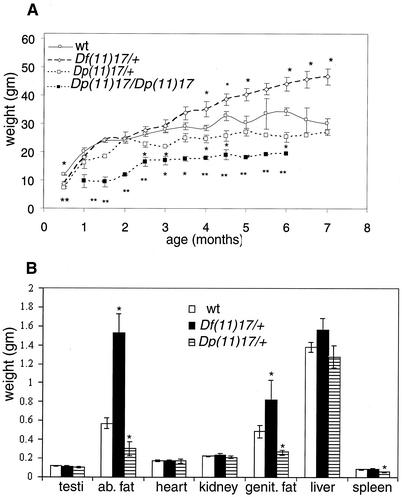
Marked differences have been observed in the weights of Df(11)17/+ and Dp(11)17/+ chromosome-engineered animals compared to those of their wild-type littermates. Weight measurements from males of the different genotypes are shown in Fig. 5A. Remarkably, both Df(11)17/+ and Dp(11)17/+ animals are significantly lighter than the wild type in the first month of life. Dp(11)17/+ animals remain underweight relative to wild-type siblings during their entire lives. This difference is even more significant for homozygous Dp(11)17/Dp(11)17 animals, and in these animals a size difference is overt. In contrast, Df(11)17/+ animals are significantly overweight compared to the wild type, beginning at 4 months of age. The body weights for the Df(11)17/+ animals at ∼8 months of age are greater than 60 g for both males and females.
FIG. 5.
(A) Weight curves for Df(11)17/+, Dp(11)17/+, Dp(11)17/Dp(11)17, and wild-type (wt) male mice are presented. Each point represents the average of 10 weights, with error bars indicated. (B) Different organs and body fat (ab. fat, abdominal fat; genit. fat, genital fat) from mice with the indicated phenotypes were weighed, and each bar represents the average of results for 10 samples. Error bars are indicated. *, P < 0.05; **, P < 0.01. Asterisks representing P values appear on top of or below each time point for which significant differences were found.
Several organs (hearts, spleens, kidneys, testes, and livers) and abdominal white fat from 6-month-old male animals (n = 10) with different genotypes were analyzed and weighed (Fig. 5B). There are no significant differences in organ weights, with the exception that the spleens of Dp(11)17/+ animals are smaller than those of the wild type (0.070 ± 0.004 g versus 0.056 ± 0.006 g; P = 0.03). However, abdominal fat contents are different, measuring 1.93 ± 0.20 g for Df(11)17/+ (P = 1.05 × 10−5), 0.30 ± 0.06 g for Dp(11)17/+ (P = 0.02), and 0.56 ± 0.05 g for wild-type animals. The abdominal fat comprises 4.5, 1.2, and 2% of the total body weight, respectively. The extent of correlation with the human phenotype needs to be examined further. Df(11)17/Dp(11)17 animals show a wild-type weight curve (data not shown), indicating that the phenotype can be rescued if the dosage of the genes included in this region is restored to wild-type levels.
Reduced fertility of male Df(11)17/+ mice.
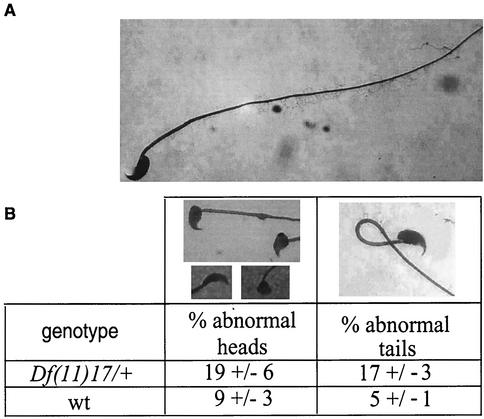
Df(11)17/+ × Df(11)17/+ matings produced fewer litters than matings of wild-type mice. To determine whether the apparent reduction in fertility was sex limited, matings between Df(11)17/+ and wild-type animals were carried out. From the matings of seven Df(11)17/+ males with wild-type females, only seven litters were obtained in a period of 11 months. During the same time period, seven Df(11)17/+ females mated with wild-type males yielded more than 55 litters. Histology of the testes revealed no abnormalities in Df(11)17/+ males, indicating that initial spermatogenesis is apparently not affected. Sperm samples were examined, and they revealed a significant reduction in sperm counts [36 × 106 ± 7 × 106 cells/ml (n = 8) for the wild type compared to 13 × 106 ± 4 × 106 cells/ml for Df(11)17/+ mice (n = 8; P = 0.035)]. The morphologies of the sperm were analyzed, and the results are shown in Fig. 6. There is a significantly higher percentage of abnormal sperm tails (17 versus 5%) and heads (19 versus 9%) in the Df(11)17/+ sperm samples than in wild-type sperm samples (P = 0.016).
FIG. 6.
Fertility testing for Df(11)17/+, Dp(11)17/+, and Df(11)17/Dp(11)17 animals. Sperm smears from Df(11)17/+ (n = 11) and wild-type (n = 9) males were analyzed. Two hundred sperm cells were scored independently for heads or tails. (A) Normal sperm cell. (B) The percentage of abnormalities found for each genotype is shown in the table.
DISCUSSION
SMS and dup(17)(p11.2p11.2) are CGS both resulting from DNA rearrangements due to NAHR via LCRs present in the 17p11.2 genomic interval. Taking advantage of the conserved synteny between human chromosome 17p11.2 and the 32-to-34-cM region of murine chromosome 11, we engineered mice carrying either a deletion [Df(11)17/+] or the reciprocal duplication [Dp(11)17/+]. We were able to reproduce several SMS phenotypic features in the Df(11)17/+ mice, thus constructing a partial model for this human syndrome.
Craniofacial dysmorphism is a consistent feature of many malformation syndromes. Recognition of dysmorphic patterns and specific facial features can be essential for establishing a clinical diagnosis of such genetic conditions (11). However, the molecular basis for facial dysmorphism remains largely unknown. We hypothesize that there are developmental parallels (i.e., homologous genes and expression profiles [3]) responsible for the craniofacial dysmorphism observed in SMS patients and Df(11)17/+ mice. Genetic dissection of this deleted region in mice should enable the definition of the genetic basis of craniofacial abnormalities of SMS.
The presence of seizures and abnormal EEG patterns in Df(11)17/+ mice suggests a direct effect of the deletion on neuronal excitability. Identifying the exact genes responsible will require further analysis, since comparison of the human and mouse genomic regions shows an inversion that leads to the inclusion inside the mouse rearranged region of at least one gene (Kcnj12) that is excluded in the human patients (3). Kcnj12 codes for Kir2.2, the alpha subunit of an inward rectifying potassium channel (Kir) (34). The physiological role of this specific subunit is unknown. However, the phenotype present in the mouse seems very similar to that observed in humans, suggesting that a gene other than Kcnj12 is responsible for this phenotype in SMS patients. In a recent study reviewing EEG recordings of SMS patients, background activity was almost uniformly within normal limits, with paroxysmal abnormalities, most of which were generalized in nature, in nearly half the recordings (unpublished information). Parents reported overt seizures of various types in 18.3% of SMS patients.
The correspondence of the weight differences found for the different genotypes with the phenotypes found in patients needs to be addressed, as diet and social behavior can mask this phenotype in SMS and dup(17)(p11.2p11.2) patients. One candidate potentially responsible for this phenotype was Srebp1, a gene that encodes sterol regulatory element binding protein 1. However, targeted disruption of this gene produced no differences in body weights or epididymal fat contents (29), suggesting that another gene in this genomic region is responsible for this phenotype.
Which gene(s) is responsible for SMS? We have learned from the analysis of the Df(11)17/+ animals that several phenotypic features observed in humans can be reproduced in mice. These observations suggest that at least the gene(s) responsible for these phenotypes should be shared in both organisms. Disruptions of the genes Srebp1 (29), Pemt (35), Myo 15 (24), Tnfrsf13b (Taci) (36), Top3α (12), and Fliih (4) have been reported and no phenotypes have been described in the heterozygous animals, indicating that these genes are not responsible for the phenotypes observed in Df(11)17/+ animals. Patients harboring the common deletion region are haploinsufficient for CSN3. By the methodology utilized in this study, a disruption of Csn3 has been achieved, making this particular gene a potential candidate for involvement in SMS. However, Csn3−/+ mice are normal (unpublished observations) and display none of the phenotypes observed in the Df(11)17/+ animals, thus suggesting that Csn3 haploinsufficiency is not involved in any of the described phenotypes. On the other hand, Zfp179 was duplicated as a result of the insertion of the targeting vector, leading to the presence of two copies of the gene in the Df(11)17/+ mice, so this excludes the gene from contributing to the phenotypes found in our mouse model. In addition, Mfap4 can be excluded from responsibility for any of the phenotypes found in Df(11)17/+ mice since it is not included in the mouse deleted region. The common deleted region is covered by six mouse large insert BAC genomic clones (3) that are currently being used for BAC complementation analysis in order to address this question. Considering that more than 80 to 90% of SMS patients have a common deletion region, it is very difficult to perform a phenotype-genotype correlation in humans to establish the genes manifesting haploinsufficiency effects. Our mouse model is an invaluable tool for addressing which gene(s) is responsible for the SMS phenotype.
The Dp(11)17/+ animals should enable better understanding of the dup(17)(p11.2p11.2) syndrome. To date, only a few patients with this duplication have been described, so this animal model should help to distinguish not only which genes are important or dosage sensitive but also what phenotype is expected for these patients. Homozygous duplication animals, which contain four copies of a candidate gene potentially involved in regulating fat content, have a more profound and severe phenotype than heterozygous animals with three copies. This finding should prompt careful reevaluation of dup(17)(p11.2p11.2) patients.
Most of the phenotypes found in our engineered animal models appear to be due to gene dosage effects secondary to haploinsufficiency or segmental trisomy for a gene(s) present in this region. Craniofacial abnormalities, reduced fertility, and weight differences can be prevented when the normal two copies of the genes are present [Df(11)17/Dp(11)17 animals]. Our mouse models will be important not only for finding the genes responsible for each phenotype but also for studying the phenotypic consequences of gene dosage in this region. The approach of chromosome engineering of the mouse genome will likely be of great utility for delineating the consequences of gene dosage effects in humans and correlating specific phenotypic features to genes within defined genomic intervals.
Acknowledgments
We appreciate the critical reviews of P. Stankiewicz, B. Lee, M. Justice, L. Potocki, and J. I. Young and the technical assistance of A. Clark with the sperm analysis and D. Lewis with the ophthalmological evaluation of the mice.
This work was supported in part by the National Cancer Institute, NIH (PO1CA75719).
REFERENCES
- 1.Allanson, J. E., F. Greenberg, and A. C. M. Smith. 1999. The face of Smith-Magenis syndrome: a subjective and objective study. J. Med. Genet. 36:394-397. [PMC free article] [PubMed] [Google Scholar]
- 2.Baldini, A., and E. A. Lindsay. 1994. Mapping human YAC clones by fluorescence in situ hybridization using Alu-PCR from single yeast colonies. Methods Mol. Biol. 33:75-84. [DOI] [PubMed]
- 3.Bi, W., J. Yan, P. Stankiewicz, S. S. Park, K. Walz, C. F. Boerkoel, L. Potocki, L. G. Shaffer, K. Devriendt, M. J. Nowaczyk, K. Inoue, and J. R. Lupski. 2002. Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res. 12:713-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, H. D., S. Fountain, I. S. McLennan, L. A. Berven, M. F. Crouch, D. A. Davy, J. A. Hooper, K. Waterford, K.-S. Chen, J. R. Lupski, B. Ledermann, I. G. Young, and K. I. Matthaei. 2002. Fliih, a gelsolin-related cytoskeletal regulator essential for early mammalian embryonic development. Mol. Cell. Biol. 22:3518-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, K.-S., P. Manian, T. Koeuth, L. Potocki, Q. Zhao, A. C. Chinault, C. C. Lee, and J. R. Lupski. 1997. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat. Genet. 17:154-163. [DOI] [PubMed] [Google Scholar]
- 6.Chen, K.-S., L. Potocki, and J. R. Lupski. 1996. The Smith-Magenis Syndrome [del(17)(p11.2)]: clinical review and molecular advances. Ment. Retard. Dev. Disabil. Res. Rev. 2:122-129. [Google Scholar]
- 7.Greenberg, F., V. Guzzetta, R. Montes de Oca-Luna, R. E. Magenis, A. C. Smith, S. F. Richter, I. Kondo, W. B. Dobyns, P. I. Patel, and J. R. Lupski. 1991. Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2). Am. J. Hum. Genet. 49:1207-1218. [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg, F., R. A. Lewis, L. Potocki, D. Glaze, J. Parke, J. Killian, M. A. Murphy, D. Williamson, F. Brown, R. Dutton, C. McCluggage, E. Friedman, M. Sulek, and J. R. Lupski. 1996. Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2). Am. J. Med. Genet. 62:247-254. [DOI] [PubMed] [Google Scholar]
- 9.Inoue, K., and J. R. Lupski. 2002. Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet. 3:199-242. [DOI] [PubMed] [Google Scholar]
- 10.Jerome, L. A., and V. E. Papaioannou. 2001. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 27:286-291. [DOI] [PubMed] [Google Scholar]
- 11.Jones, K. L. 1997. Smith's recognizable patterns of human malformation, 5th ed. W. B. Saunders Co., Philadelphia, Pa.
- 12.Li, W., and J. C. Wang. 1998. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc. Natl. Acad. Sci. USA 95:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsay, E. A., A. Botta, V. Jurecic, S. Carattini-Rivera, Y. C. Cheah, H. M. Rosenblatt, A. Bradley, and A. Baldini. 1999. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 401:379-383. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay, E. A., F. Vitelli, H. Su, M. Morishima, T. Huynh, T. Pramparo, V. Jurecic, G. Ogunrinu, H. F. Sutherland, P. J. Scambler, A. Bradley, and A. Baldini. 2001. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410:97-101. [DOI] [PubMed] [Google Scholar]
- 15.Liu, P., H. Zhang, A. McLellan, H. Vogel, and A. Bradley. 1998. Embryonic lethality and tumorigenesis caused by segmental aneuploidy on mouse chromosome 11. Genetics 150:1155-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupski, J. R. 1998. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14:417-422. [DOI] [PubMed] [Google Scholar]
- 17.Matzuk, M. M., M. J. Finegold, J.-G. J. Su, A. J. W. Hsueh, and A. Bradley. 1992. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature 360:313-319. [DOI] [PubMed] [Google Scholar]
- 18.McLeod, M. J. 1980. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22:299-301. [DOI] [PubMed] [Google Scholar]
- 19.Merscher, S., B. Funke, J. A. Epstein, J. Heyer, A. Puech, M. M. Lu, R. J. Xavier, M. B. Demay, R. G. Russell, S. Factor, K. Tokooya, B. S. Jore, M. Lopez, R. K. Pandita, M. Lia, D. Carrion, H. Xu, H. Schorle, J. B. Kobler, P. Scambler, A. Wynshaw-Boris, A. I. Skoultchi, B. E. Morrow, and R. Kucherlapati. 2001. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619-629. [DOI] [PubMed] [Google Scholar]
- 20.Mills, A. A., and A. Bradley. 2001. From mouse to man: generating megabase chromosome rearrangements. Trends Genet. 17:331-339. [DOI] [PubMed] [Google Scholar]
- 21.Noebels, J. L., and R. L. Sidman. 1979. Inherited epilepsy: spike-wave and focal motor seizures in the mutant mouse tottering. Science 204:1334-1336. [DOI] [PubMed] [Google Scholar]
- 22.Park, S.-S., P. Stankiewicz, W. Bi, C. Shaw, J. Lehoczky, K. Dewar, B. Birren, and J. R. Lupski. 2002. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 12:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potocki, L., K.-S. Chen, S. S. Park, D. E. Osterholm, M. A. Withers, V. Kimonis, A. M. Summers, W. S. Meschino, K. Anyane-Yeboa, C. D. Kashork, L. G. Shaffer, and J. R. Lupski. 2000. Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 24:84-87. [DOI] [PubMed] [Google Scholar]
- 24.Probst, F. J., R. A. Fridell, Y. Raphael, T. L. Saunders, A. Wang, Y. Liang, R. J. Morell, J. W. Touchman, R. H. Lyons, K. Noben-Trauth, T. B. Friedman, and S. A. Camper. 1998. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science 280:1444-1447. [DOI] [PubMed] [Google Scholar]
- 25.Ramirez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855-878. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Solis, R., P. Liu, and A. Bradley. 1995. Chromosome engineering in mice. Nature 378:720-724. [DOI] [PubMed] [Google Scholar]
- 27.Richtsmeier, J. T., L. L. Baxter, and R. H. Reeves. 2002. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev. Dyn. 217:137-145. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer, L. G., D. H. Ledbetter, and J. R. Lupski. 2001. The metabolic and molecular bases of inherited disease, 8th ed., vol. I, p. 1291-1327. The McGraw-Hill Companies, Inc., New York, N.Y.
- 29.Shimano, H., I. Shimomura, R. E. Hammer, J. Herz, J. L. Goldstein, M. S. Brown, and J. D. Horton. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Investig. 100:2115-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, A. C. M., A. L. Gropman, J. E. Bailey-Wilson, O. Goker-Alpan, S. H. Elsea, J. Blancato, J. R. Lupski, and L. Potocki. 2002. Hypercholesterolemia in children with Smith-Magenis syndrome: del (17) (p11.2p11. 2). Genet. Med. 4:118-125. [DOI] [PubMed] [Google Scholar]
- 31.Smith, A. C. M., L. McGavran, J. Robinson, G. Waldstein, J. Macfarlane, J. Zonona, J. Reiss, M. Lahr, L. Allen, and E. Magenis. 1986. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am. J. Med. Genet. 24:393-414. [DOI] [PubMed] [Google Scholar]
- 32.Stankiewicz, P., and J. R. Lupski. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18:74-82. [DOI] [PubMed] [Google Scholar]
- 33.Stankiewicz, P., and J. R. Lupski. 2002. Molecular-evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 1:312-319. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, N., K. I. Morishige, A. Jahangir, M. Yamada, I. Findlay, H. Koyama, and Y. Kurachi. 1994. Molecular cloning and functional expression of cDNA encoding a second class of inward rectifier potassium channels in the mouse brain. J. Biol. Chem. 269:23274-23279. [PubMed] [Google Scholar]
- 35.Walkey, C. J., L. R. Donohue, R. Bronson, L. B. Agellon, and D. E. Vance. 1997. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 94:12880-12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, M., H. Wang, B. Chan, M. Roose-Girma, S. Erickson, T. Baker, D. Tumas, I. S. Grewal, and V. M. Dixit. 2001. Activation and accumulation of B cells in TACI-deficient mice. Nat. Immunol. 2:638-643. [DOI] [PubMed] [Google Scholar]
- 37.Yu, Y., and A. Bradley. 2001. Engineering chromosomal rearrangements in mice. Nat. Rev. Genet. 10:780-790. [DOI] [PubMed] [Google Scholar]
- 38.Zhao, Q., K.-S. Chen, B. A. Bejjani, and J. R. Lupski. 1998. Cloning, genomic structure and expression of mouse ringer finger protein gene Znf179. Genomics 49:394-400. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, B., A. A. Mills, and A. Bradley. 1999. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 27:2354-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]