Abstract
A variety of stresses on the heart initiate a number of subcellular signaling pathways, which finally reach the nuclei of cardiac myocytes and cause myocyte hypertrophy with heart failure. However, common nuclear pathways that lead to this state are unknown. A zinc finger protein, GATA-4, is one of the transcription factors that mediate changes in gene expression during myocardial-cell hypertrophy. p300 not only acts as a transcriptional coactivator of GATA-4, but also possesses an intrinsic histone acetyltransferase activity. In primary cardiac myocytes derived from neonatal rats, we show that stimulation with phenylephrine increased an acetylated form of GATA-4 and its DNA-binding activity, as well as expression of p300. A dominant-negative mutant of p300 suppressed phenylephrine-induced nuclear acetylation, activation of GATA-4-dependent endothelin-1 promoters, and hypertrophic responses, such as increase in cell size and sarcomere organization. In sharp contrast to the activation of cardiac MEK-1, which phosphorylates GATA-4 and causes compensated hypertrophy in vivo, p300-mediated acetylation of mouse cardiac nuclear proteins, including GATA-4, results in marked eccentric dilatation and systolic dysfunction. These findings suggest that p300-mediated nuclear acetylation plays a critical role in the development of myocyte hypertrophy and represents a pathway that leads to decompensated heart failure.
Heart failure arises from a number of diverse primary cardiovascular disorders and is associated with significant morbidity and mortality. Therefore, elucidating the mechanisms of this disease is of clinical importance. Previous studies have demonstrated that a variety of stresses on the heart activate neuronal and hormonal factors, such as the renin-angiotensin system and factors regulating the sympathetic nervous systems. These factors initiate a number of subcellular signaling pathways, which finally reach the nuclei of cardiac myocytes and change the pattern of gene expression associated with hypertrophy (reviewed in references 14 and 53). In order to establish appropriate therapy for heart failure, it is critical to identify a common nuclear pathway which can be targeted by pharmacological agents in the future.
We have been interested in transcription factors that mediate changes in gene expression during myocardial-cell hypertrophy. A zinc finger protein, GATA-4, is one such factor and is required for transcriptional activation of cardiac genes whose expression is upregulated during myocardial-cell hypertrophy (22, 25; reviewed in reference 46). While overexpression of GATA-4 in cardiac myocytes causes hypertrophy, expression of a dominant-negative form of GATA-4 inhibits Gq protein-coupled receptor agonist-induced hypertrophy (37). During myocardial-cell hypertrophy, GATA-4 is phosphorylated at a serine residue and shows increased DNA-binding ability (38, 47). Phosphorylation of cardiac GATA-4 requires activation of MEK1/extracellular signal-regulated kinase (ERK) 1/2. On the other hand, activation of MEK1 in cardiac myocytes results in phosphorylation of GATA-4 and in concentric left-ventricular hypertrophy in vivo (9). Other transcription factors that control myocardial-cell hypertrophy include myocyte-enhancing factor 2 (MEF2) (34), serum response factor (49), TEF-1 (32), and AP-1 (25). MEF2 is a downstream target for calmodulin-dependent protein kinase IV, the activation of which is sufficient to induce cardiac hypertrophy in vivo (50). The involvement of multiple transcription factors in hypertrophic responses suggests that these factors are activated through some common pathway during myocardial-cell hypertrophy. Regarding this hypothesis, Molkentin et al. have reported that the calcineurin-NF-AT pathway is one of the common pathways that transduces signals for cardiac hypertrophy (45). However, several studies have demonstrated that blockade of calcineurin fails to inhibit the development of heart failure in pathophysiological settings, suggesting the existence of redundant networks in cardiac nuclear signaling (63; reviewed in reference 44).
p300 is an adenovirus E1A-associated protein that acts as a transcriptional coactivator that mediates the interaction of DNA-binding transactivators with the basal transcriptional complex (17). Consistent with this function, p300 protein interacts directly with components of the basal transcriptional apparatus (62) and diverse enhancer-binding proteins, modulating many examples of enhancer-mediated transcription. Several lines of evidence suggest that p300 plays a critical role in the physiological growth and differentiation of cardiac myocytes during development. Mice lacking a functional p300 gene die between days 9 and 11.5 of gestation, exhibiting defects of cardiac muscle differentiation and trabeculation (61). The expression of myocardial contractile proteins is clearly reduced in the mutants compared with the wild type (WT). Furthermore, p300 is required for maintenance of the G1 phase of the cell cycle in differentiated cardiac myocytes (5, 39). p300 protein also serves as an adaptor for hypertrophy-responsive transcription factors, such as GATA-4, MEF2, serum response factor, and AP-1, and these interactions are required for the full transcriptional activities of these factors (16, 31, 56). However, the direct roles of p300 in cardiac myocyte hypertrophy have not been clarified.
In addition to its “bridging function,” p300 protein possesses an intrinsic histone acetyltransferase (HAT) activity (48) which promotes a transcriptionally active chromatin configuration. The HAT activity of p300 is required for the functions of diverse proteins in transcriptional activation (42, 52, 54). The PHD finger of p300 has been reported to be dispensable for its HAT activity and its transactivating function (7). p300 protein can also acetylate certain nonhistone transcription-related proteins, including transcriptional activators (20), coactivators (12), and basal transcription factors (28). Acetylation is emerging as a posttranslational modification of nuclear proteins that is essential for the regulation of transcription and that modifies transcription factor affinity for binding sites on DNA, stability, and/or nuclear localization. However, it is unknown whether p300 is able to acetylate hypertrophy-responsive transcription factors such as GATA-4 and, if so, whether p300-mediated acetylation plays a role in myocardial-cell hypertrophy.
For all of these reasons, we were interested in clarifying the role of the intrinsic HAT activity of p300 in myocardial-cell hypertrophy. We show here that p300 acetylates GATA-4 and increases its DNA-binding ability, as well as causing nuclear hyperacetylation of cardiac myocytes. In addition, this process is closely associated with the development of myocardial-cell hypertrophy. In sharp contrast to MEK1 transgenic (TG) mice (which exhibit concentric hypertrophy with normal systolic function), acetylation of cardiac nuclear proteins, including GATA-4, by overexpression of p300 in mice results in marked left-ventricular dilatation and systolic dysfunction. Thus, p300-mediated acetylation may be involved in the development of decompensated heart failure and represents a pharmacological target of heart failure therapy in humans.
MATERIALS AND METHODS
Plasmid constructs.
The expression vectors pcDNAG4, pCMVβ-gal, pCMVwtp300, and pCMV1514-1922p300 contain the cytomegalovirus promoter-enhancer fused to murine GATA-4 cDNA (4, 29), β-galactosidase cDNA, a full-length human p300 cDNA (17), and a portion of p300 cDNA encoding amino acid residues 1514 to 1922 (62), respectively. Plasmid pwtE1A is an expression vector for WT 12S E1A. pwtE1A and pCMVwtp300 were gifts from Richard Eckner and David M. Livingston (Harvard Medical School, Boston, Mass.), and pCMV1514-1922p300 was provided by Antonio Giordano (Thomas Jefferson University, Philadelphia, Pa.). pRSVCAT and pRSVluc contain chloramphenicol acetyltransferase (CAT) and Luc genes, respectively, driven by Rous sarcoma virus long terminal repeat sequences (22, 23). The plasmid construct pETCAT contains the transcription start site-proximal 204 bp of the WT rat endothelin-1 (ET-1) promoter fused to a bacterial CAT gene (33) and was a gift of Thomas Quertermous (Stanford University, Palo Alto, Calif.).
Transfection and luciferase-CAT assays.
COS7 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. The cells were washed twice with serum-free medium and then transfected with a total of <12 μg of DNA in 100-mm-diameter plates using Lipofectamine (Life Technologies, Inc.) according to the manufacturer's recommendation. After a 5-h incubation with DNA-Lipofectamine complex, the cells were washed twice with serum-free medium and further incubated in the medium with 10% fetal bovine serum for 48 h. The cells were then washed twice with ice-cold phosphate-buffered saline and lysed as described previously (31, 47). Luc and CAT activities were determined in the same cell lysate as described previously (31). The relative CAT activity was calculated from the ratio of CAT background to Luc background and was expressed as the mean ± standard error.
Primary neonatal rat ventricular cardiac myocytes were prepared as previously described (23, 31, 47). Cardiac myocytes were cotransfected with the appropriate amounts of DNA using Lipofectamine Plus (Life Technologies, Inc.) according to the manufacturer's recommendation. After a 2-h incubation with DNA-Lipofectamine complex, the cells were washed twice with serum-free medium and further incubated for 48 h in serum-free medium in the presence of 10−5 M phenylephrine (PE) or saline as a control. The cells were then washed twice with ice-cold phosphate-buffered saline, lysed with lysis buffer, and subjected to assays for luciferase and CAT activities as described previously (47).
Analysis of the acetylation state of GATA-4 and Western blotting.
In analysis of the acetylation state of GATA-4, 50 mM NaF and 1 mM Na3VO4 were added to all buffers. For pulse-labeling, COS7 cells (108) transfected with GATA-4 were resuspended in 2.5 ml of medium containing 0.05 mCi of [1-14C]acetic acid sodium salt/ml (200 μCi/ml; Amersham Pharmacia Biotech) and incubated for 3 h. Ethanol, the solvent of [1-14C]acetic acid sodium salt, was evaporated to minimize the cytotoxic effect of the ethanol in the culture medium. Nuclear extracts from these cells were immunoprecipitated using anti-GATA-4 antibody (Santa Cruz Biotechnology, Inc.) or normal goat immunoglobulin G (IgG) in low-stringency buffer (50 mM Tris, pH 7.4, 0.15 M NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 10 mg each of aprotinin and leupeptin/ml, and 0.5 mM phenylmethylsulfonyl fluoride; all from Sigma) for 16 h at 4°C and incubated with protein G beads for 1 h at 4°C. The precipitate was washed four times in the same buffer, resuspended in 50 ml of sodium dodecyl sulfate (SDS) lysis buffer (20 mM Tris, pH 7.5, 50 mM NaCl, 0.5% SDS, 1 mM dithiothreitol), heated to 95°C for 2 min, electrophoresed on an SDS-polyacrylamide gel (10% acrylamide), fixed, and autoradiographed using a bioimaging analyzer (BAS 2000; FUJIX, Tokyo, Japan).
For immunoprecipitation and Western blotting, nuclear extracts were immunoprecipitated with anti-GATA-4 antibody (Santa Cruz Biotechnology, Inc.), which was captured by adding protein G beads. The precipitate was washed four times in the low-stringency buffer, resolved by SDS-polyacrylamide gel electrophoresis, transferred to Immobilon membranes, and reacted with anti-acetylated lysine antibody (New England Biolabs, Inc.), which was subsequently detected using horseradish peroxidase-conjugated anti-rabbit IgG. Signals were detected using the ECL Western blotting detection system (Amersham Pharmacia Biotech) according to the manufacturer's instructions. To normalize for protein loading after immunoprecipitation, the blots were stripped by incubation in 62.5 mM Tris-HCl, pH 6.8, 100 mM β-mercaptoethanol, and 2% SDS for 30 min at 50°C, washed twice with phosphate-buffered saline and 0.05% Tween, and then probed with anti-GATA-4 antibody. The primary antibody for p300/CBP associated factor (PCAF) was obtained from Santa Cruz Biotechnology, Inc. Western blotting for GATA-4, p300, E1A, and β-actin was performed as described previously (23, 31, 47).
Electrophoretic mobility shift assays (EMSAs).
Double-stranded oligonucleotides were designed to contain GATA motifs from the rat ET-1 promoter. The sequences of the sense strands of these oligonucleotides were as follows: WT, 5′-CCTCTAGAGCCGGGTCTTATCTCCGGCTGCACGTTGC-3′, and mut-GATA, 5′-CCTCTAGAGCCGGGTCTGCACTCCGGCTGCACGTTGC-3′ (mutated sequences are in italics).
We also used a double-stranded oligonucleotide (purchased from Santa Cruz Biotechnology, Inc.) that contained the Sp-1 binding site as a control probe. EMSAs were carried out at 4°C for 20 min in 15-μl reaction mixtures containing 10 μg of nuclear extract, 0.25 ng (>20,000 cpm) of radiolabeled double-stranded oligonucleotide, 500 ng of poly(dI-dC), 5 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.5 mM dithiothreitol, 37.5 mM KCl, and 4% Ficoll 400. For cold competition experiments, a 100 M excess of unlabeled competitor oligonucleotide was included in the binding reaction mixture. Protein-DNA complexes were separated by electrophoresis on 4% nondenaturing polyacrylamide gels in 0.25× TBE (1× TBE is 100 mM Tris, 100 mM boric acid, and 2 mM EDTA) at 4°C.
Immunocytochemistry and measurement of cell diameter.
The cardiac myocytes were grown in flask style chambers with glass slides (Nalgen Nunc) and stained for β-myosin heavy chain (MHC) using the indirect immunofluorescence method, as previously described (60). Immunocytochemical staining for acetylated lysine was performed using the indirect immuno-alkaline phosphatase method. Cells were incubated with anti-acetylated lysine polyclonal antibody (New England Biolabs, Inc.) at a dilution of 1:30. Signals of acetylated lysine were detected using anti-rabbit alkaline phosphatase-conjugated secondary antibody (Sigma) at a dilution of 1:250 for 45 min.
Measurement of cell diameter was performed in cardiac myocytes stained by anti-β-MHC antibody as previously described (60).
Creation of TG mice and Southern analysis.
The α-MHC- p300 DNA plasmid was constructed by subcloning a NotI-HindIII fragment of pCMVwtp300 (kindly donated by Richard Eckner, Zurich, Switzerland) into the SalI-HindIII site of clone 26, a 5.5-kb mouse α-MHC promoter-containing construct (kindly donated by Jeffery Robbins, Cincinnati, Ohio). The α-MHC- p300 DNA fragment was gel purified and injected into newly fertilized C57BL/6 oocytes, which were transferred to the oviducts of pseudopregnant C57BL/6 recipients. TG founders were identified by Southern blotting using the 32P-labeled 450-bp α-MHC- p300 sequence region as a probe.
RNA analysis.
Detection of mRNA for β-MHC, atrial natriuretic factor (ANF), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed by Northern blot analysis as previously described (22, 23, 31).
To detect prepro-ET-1 mRNA, reverse transcriptase (RT) PCR was carried out as described previously (30). The PCR primers were designed on the basis of the published cDNA sequence for mouse ET-1 and GAPDH as follows: sense for ET-1, 5′-CATCATCTGGGTCAACACTC-3′; antisense for ET-1, 5′-CATGGGGCTCGCACTATATA-3′; sense for GAPDH, 5′-TTGCCATCAACGACCCCTTC-3′; and antisense for GAPDH, 5′-TTGTCATGGATGACCTTGGC-3′. To define the optimal amplification conditions, a series of pilot studies were performed by the use of various amounts of RT products and 20 to 45 cycles of PCR amplification as described previously (30). On the basis of these initial experiments, the linear portion of the amplification was determined for the ET-1 and GAPDH genes.
Transthoracic echocardiography.
The cardiac functions of WT and TG mice were evaluated noninvasively by echocardiography. The animals were anesthetized with ketamine (50 μg/g of body weight) and xylazine (2.5 μg/g). Transthoracic echocardiography was performed with a cardiac ultrasound recorder (Toshiba Power Vision, Tokyo, Japan), using a 7.5-MHz transducer. After the acquisition of high-quality two-dimensional images, M mode images of the left ventricle were recorded. Measurements of left-ventricular end diastolic (LVDD) and end systolic (LVESD) internal dimensions were performed according to the leading edge-to-leading edge convention adopted by the American Society of Echocardiography. Percent fractional shortening (%FS) was calculated as follows: %FS = [(LVDD − LVESD)/LVDD] × 100. At least three independent M mode measurements per animal were obtained by an examiner blinded to the genotype of the animal.
Statistical analysis.
Data are presented as means ± standard errors. Statistical comparisons were performed using unpaired two-tailed Student's t tests or analysis of variance with Scheffe's test where appropriate, with a probability value of <0.05 taken to indicate significance.
RESULTS
p300 protein acetylates lysine residues of GATA-4, enhances its DNA-binding activity, and participates in GATA-4-dependent ET-1 transcription.
To give positive proof of GATA-4 acetylation, expression plasmids encoding GATA-4 (pcDNAG4) and p300 (pCMVwtp300) were transfected into COS7 cells, which lack all GATA factors (31, 47). These cells were pulse-labeled with the [14C]acetic acid, sodium salt and subjected to immunoprecipitation with antiserum against GATA-4 or with normal goat IgG as a negative control. As shown in Fig. 1, GATA-4 protein incorporated sodium [14C]acetate, indicating acetylation of GATA-4.
FIG. 1.
In vitro acetylation of GATA-4 in COS7 cells. COS7 cells were transfected with 2 μg of pcDNAG4 and 9 μg of pCMVwtp300 and were pulse-labeled with [14C]acetic acid sodium salt for 3 h. The nuclear extracts were immunoprecipitated with anti-GATA-4 antibody or with normal goat IgG, resolved by SDS-polyacrylamide gel electrophoresis, fixed, and autoradiographed using a bioimaging analyzer.
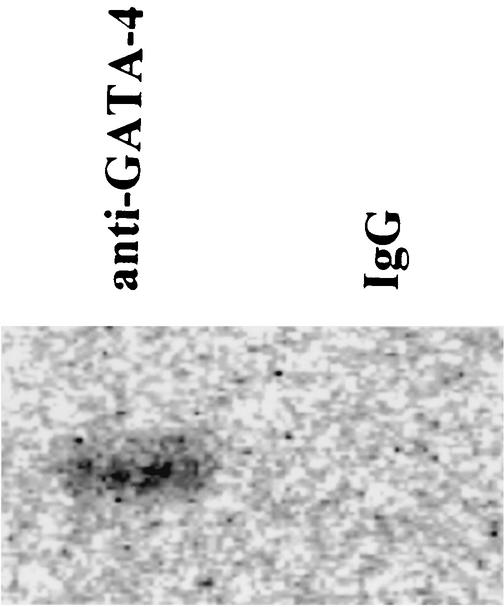
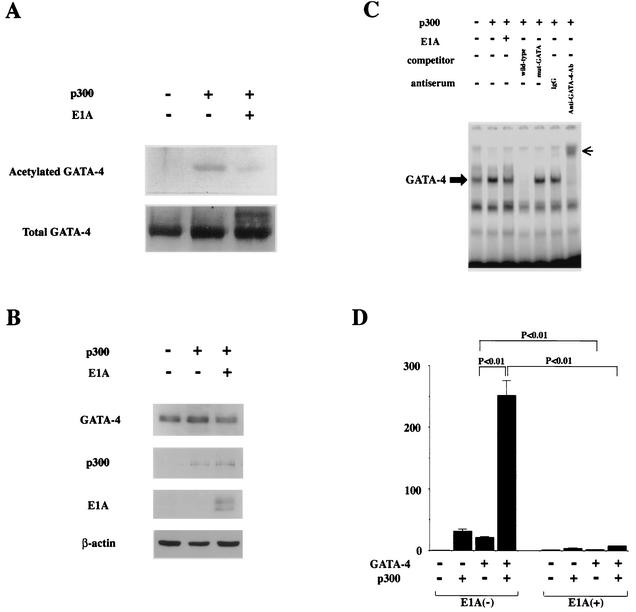
To determine whether GATA-4 is acetylated by p300, expression plasmids encoding GATA-4 (pcDNAG4), p300 (pCMVwtp300), and E1A (pwtE1A) were transfected (Fig. 2) into COS7 cells. Nuclear extracts from these cells were subjected to immunoprecipitation with an anti-GATA-4 antibody, followed by Western blotting using anti-acetylated lysine antibody. As shown in Fig. 2A, the forced expression of p300 induced acetylation of GATA-4. However, when E1A, which disrupts p300 function, was expressed in addition to p300, it almost completely inhibited this acetylation. Expression of p300 or E1A did not influence the amounts of GATA-4 and β-actin produced by transfecting pcDNAG4 (Fig. 2B). To determine whether acetylation of GATA-4 by p300 modulates the DNA-binding activity of GATA-4, EMSAs were performed. The same nuclear extracts used to detect acetylation were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site of the rat ET-1 promoter. As shown in Fig. 2C, competition EMSAs demonstrated that the retarded band represented specific binding, as evidenced by the fact that it was competed out by an excess of unlabeled ET-1 GATA oligonucleotide (fourth lane from left) but not by the same amount of an oligonucleotide containing the ET-1 GATA site with a mutation (fifth lane). To further confirm that the retarded band represents an interaction of the probe with GATA-4, we performed supershift experiments. The retarded band was supershifted by anti-GATA-4 antibody (seventh lane) but not by control goat IgG (sixth lane). The amount of specific complex formed with GATA-4 markedly increased in nuclear extracts from p300-expressing cells (second lane) compared with those from β-galactosidase (β-Gal)-expressing cells (first lane). Coexpression of E1A, which inhibited GATA-4 acetylation, blocked the p300-mediated increase in GATA-4- DNA binding (third lane).
FIG. 2.
p300 acetylates lysine residues of GATA-4, enhances its DNA-binding activity, and is involved in GATA-4-dependent ET-1 transcription. (A) COS7 cells were transfected with 2 μg of pcDNAG4 or with (+) 9 μg of pCMVwtp300 (p300) and/or 1 μg of pwtE1A (E1A) as indicated. The total amount of DNA was kept constant by cotransfecting pCMVβ-gal. Nuclear extracts (300 μg of protein) from these cells were immunoprecipitated with anti-GATA-4 antibody, followed by sequential Western blotting with anti-acetylated lysine antibody and with anti-GATA-4 antibody. (B) The nuclear extracts used for panel A before immunoprecipitation were subjected to Western blotting using the anti-GATA-4 antibody, anti-p300 antibody, anti-E1A antibody, or anti-β-αctin antibody. (C) The same nuclear extracts were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site in the ET-1 promoter. Ab, antibody; small arrow, supershifted band of GATA-4. (D) COS7 cells were transfected with 2.0 μg of pETCAT, 0.1 μg of pRSVluc, 0.5 μg of pcDNAG4 or pCMVβ-gal, 2.5 μg of pCMVwtp300 or pCMVβ-gal, and 0.3 μg of pwtE1A or pCMVβ-gal. The results are expressed as n-fold activation of the normalized CAT activity (CAT/luc) relative to that resulting from transfection with 3.3 μg of pCMVβ-gal without pcDNAG4, pCMVwtp300, or pwtE1A. The data shown are the means and standard errors of the mean from three independent experiments.
To examine whether p300-mediated acetylation is involved in GATA-4-dependent ET-1 transcription, we performed transient-transfection experiments. As seen in Fig. 2D, the expression of GATA-4 resulted in a 22-fold activation of the 204-bp rat ET-1 promoter (pETCAT), and coexpression of E1A almost completely blocked this increase. Consistent with the acetylation of GATA-4 by p300, expression of p300 dramatically potentiated GATA-4-mediated transactivation of the ET-1 promoter (252-fold). This potentiation was also repressed by coexpression of E1A, which inhibits p300-mediated acetylation.
PE induces acetylation and DNA-binding of GATA-4, as well as p300 expression in cardiac myocytes.
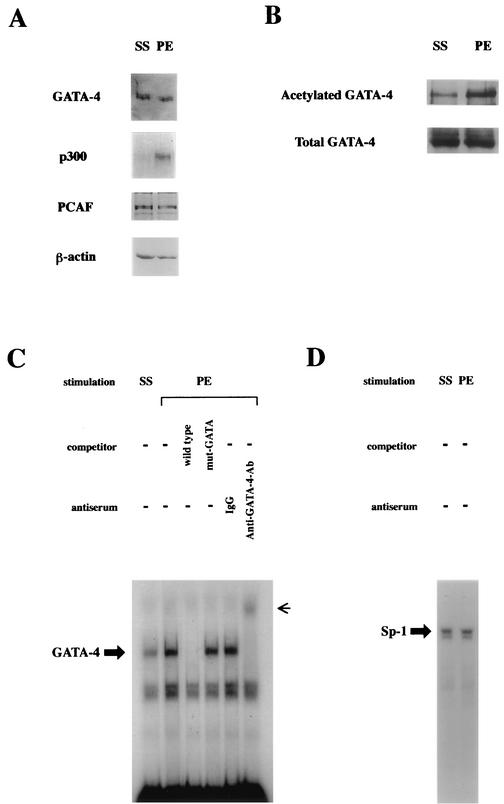
To examine whether acetylation of GATA-4 actually occurs during myocardial-cell hypertrophy, cultured ventricular myocytes prepared from neonatal rats were incubated with an α1-adrenergic agonist, PE, at 10−5 M or with saline as a control for 48 h. As shown in Fig. 3A, the expression levels of GATA-4, PCAF, and β-actin were similar in saline- and PE-stimulated cells. In contrast, p300 levels were markedly increased by PE. Nuclear extracts from cardiac myocytes were immunoprecipitated with anti-GATA-4 antibody, followed by Western blotting with anti-acetylated lysine antibody. As shown in Fig. 3B, PE treatment resulted in a marked increase in the level of the acetylated form of GATA-4. To correct for differences in protein loading after immunoprecipitation, the same membrane was reblotted with anti-GATA-4 antibody. The amounts of total GATA-4 in lysates after immunoprecipitation were similar in saline- and PE-stimulated cells. Therefore, PE markedly increased the ratio of the acetylated form of GATA-4 relative to the total GATA-4 in cardiac myocytes.
FIG. 3.
PE induces p300 expression, acetylation, and DNA binding of GATA-4 in cardiac myocytes. (A) Primary cardiac myocytes from neonatal rats were stimulated with saline (SS) or PE (10−5 M) for 48 h. Nuclear extracts from these cells were subjected to Western blotting with anti-GATA-4 antibody, anti-p300 antibody, anti-PCAF antibody, or anti-β-actin antibody. (B) The same nuclear extracts (100 μg of protein) were immunoprecipitated with anti-GATA-4 antibody and sequentially subjected to Western blotting with anti-acetylated lysine antibody and with anti-GATA-4 antibody. (C and D) The same nuclear extracts were probed with a radiolabeled double-stranded oligonucleotide containing the ET-1 GATA site (C) and with one containing the Sp-1 site (D). −, absent.
To determine whether PE stimulation influences the DNA-binding activity of GATA-4 in cardiac myocytes, EMSAs were performed with a radiolabeled double-stranded oligonucleotide containing the ET-1 GATA site. As shown in Fig. 3C, a retarded band indicated sequence-specific binding (third and fourth lanes from left), and it was supershifted by anti-GATA-4 antibody (sixth lane) but not by control goat IgG (fifth lane). These findings demonstrate that this band represents a specific GATA-4 complex. The intensity of this band was increased in nuclear extracts from PE-stimulated cardiac myocytes (second lane) compared with those from saline-stimulated cells (first lane). However, the DNA-binding activities of Sp-1 in cells stimulated with saline (Fig. 3D, left lane) and those stimulated with PE (Fig. 3D, right lane) did not differ.
p300(1514-1922) inhibits p300-induced acetylation and DNA binding of GATA-4.
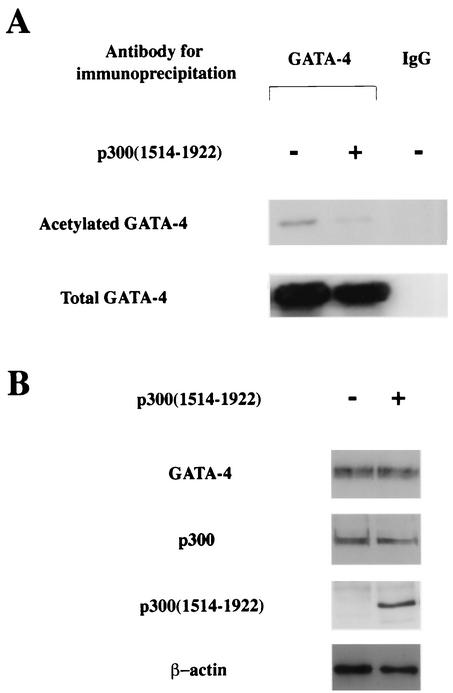
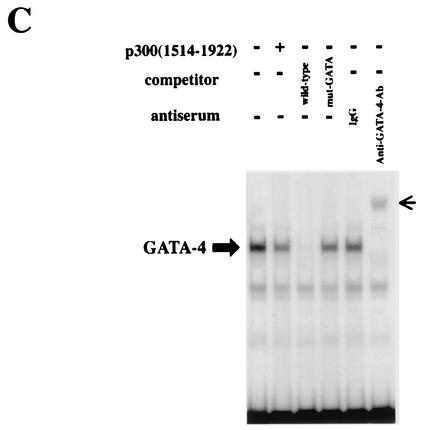
Yuan et al. reported that the portion of p300 containing the affinity to MyoD was detected in fragment 1514 to 1922, covering the third cysteine-histidine-rich conserved domain (62). Dai and Markham showed that this cysteine-histidine-rich region of p300 interacts with N and C zinc finger domains of GATA-4 (16). However, this small fragment of p300(1514-1922) lacks the HAT activity, suggesting that this fragment may act as a dominant-negative mutant and impair the function of endogenous p300. To determine whether p300(1514-1922) inhibits p300-induced acetylation of GATA-4, expression plasmids encoding GATA-4 (pcDNAG4), p300 (pCMVwtp300), and p300(1514-1922) (pCMV1514-1922p300) were transfected into COS7 cells. Nuclear extracts from these cells were subjected to immunoprecipitation with an anti-GATA-4 antibody (first two lanes on the left) or with normal goat IgG as a negative control (third lane), followed by Western blotting using anti-acetylated lysine antibody. As shown in Fig. 4A, the extent of GATA-4 acetylation decreased when p300(1514-1922) was expressed in addition to p300. Expression of p300 or p300(1514-1922) did not influence the amount of GATA-4 produced by transfecting pcDNAG4 (Fig. 4B). To determine whether p300(1514-1922) modulates the DNA-binding activity of GATA-4, EMSAs were performed (Fig. 4C). The same nuclear extracts used to detect acetylation were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site of the rat ET-1 promoter. The specific band indicating GATA-4 binding was determined by competition EMSAs and by supershift experiments (third to sixth lanes from the left). Coexpression of p300(1514-1922), which inhibited GATA-4 acetylation, also decreased GATA-4- DNA binding (compare the first and second lanes).
FIG. 4.
p300(1514-1922) inhibits p300-induced acetylation and DNA binding of GATA-4. (A) COS7 cells were transfected with (+) 2 μg of pcDNAG4, 9 μg of pCMVwtp300, and 1 μg of pCMV1514-1922p300 as indicated. The total amount of DNA was kept constant by cotransfecting pCMVβ-gal. Nuclear extracts (300 μg of protein) from these cells were immunoprecipitated with anti-GATA-4 antibody or normal goat IgG, followed by sequential Western blotting with anti-acetylated lysine antibody and with anti-GATA-4 antibody. (B) The nuclear extracts used for panel B before immunoprecipitation were subjected to Western blotting using the anti-GATA-4 antibody, anti-p300 antibody, or anti-β-actin antibody. (C) The same nuclear extracts were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site in the ET-1 promoter. Small arrow, supershifted band of GATA-4.
p300 protein is involved in PE-induced nuclear acetylation of cardiac myocytes.
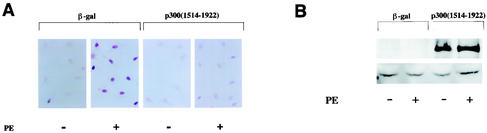
We next evaluated whether nuclear hyperacetylation of cardiac myocytes occurs during hypertrophy and, if so, whether p300 is involved in the process. Cardiac myocytes were transfected with a plasmid encoding p300(1514-1922), or β-Gal as a control, and stimulated with saline or 10−5 M PE for 48 h. By Western blotting, the specific band indicating p300(1514-1922) was intensely detected in cardiac myocytes transfected with pCMV1514-1922p300 but not in myocytes transfected with pCMVβ-gal (Fig. 5B). Then, these cells were immunostained with an anti-acetylated lysine antibody. As shown in Fig. 5A, in β-Gal-transfected cardiac myocytes, PE stimulation markedly induced nuclear staining, indicating hyperacetylation. This change in β-Gal-expressing cells was compared with that in p300(1514-1922)-expressing cells. PE-induced nuclear hyperacetylation was almost completely blocked by transfection of an expression plasmid encoding p300(1514-1922).
FIG. 5.
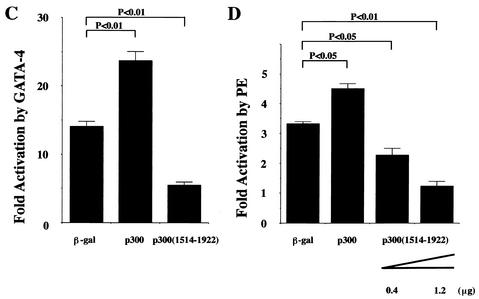
p300(1514-1922) inhibits PE-induced nuclear hyperacetylation and GATA-4-dependent and PE-induced ET-1 transcription in cardiac myocytes. (A) Cardiac myocytes were transfected with 0.7 μg of pCMV1514-1922p300 or pCMVβ-gal (β-gal). Then, these cells were stimulated with saline or PE (10−5 M) for 48 h and subjected to immunocytochemical staining with antibody against acetylated lysine. +, present. (B) Nuclear extracts from these cells were subjected to Western blotting using the anti-p300 and anti-β-actin antibodies. (C) Cardiac myocytes were cotransfected with 1 μg of pETCAT, 0.05 μg of pRSVluc, 0.25 μg of pcDNAG4 or pCMVβ-gal, and 1.25 μg of pCMVwtp300, pCMV1514-1922p300, or pCMVβ-gal. The results are expressed as n-fold activation by GATA-4 of the normalized CAT activity (CAT/luc). The data shown are the means and standard errors of the mean of two independent experiments, each carried out in duplicate. (D) Cardiac myocytes were cotransfected with 2 μg of pETCAT, 0.1 μg of pRSVluc, and 0.4 μg of pCMVwtp300 or 0.4 or 1.2 μg of pCMV1514-1922p300. The total DNA content was equalized in each sample with pCMVβ-gal. The results are expressed as n-fold activation by PE of the normalized CAT activity (CAT/luc). The data shown are the means and standard errors of the mean of two independent experiments, each carried out in duplicate.
p300 protein is involved in PE-induced ET-1 transcription and hypertrophy in cardiac myocytes.
To test whether p300 is involved in GATA-4- and PE-induced ET-1 transcription in cardiac myocytes, we transfected an ET-1 reporter plasmid into cardiac myocytes. As shown in Fig. 5C and D, coexpression of WT p300 further activated GATA-4- and PE-induced ET-1 promoter activities. However, coexpression of p300(1514-1922), which blocks PE-induced nuclear acetylation, inhibited GATA-4- and PE-induced ET-1 transcription.
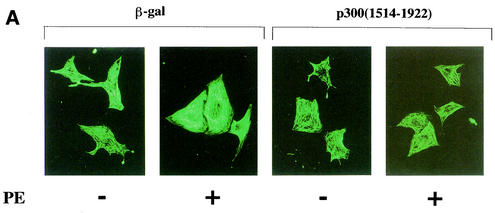
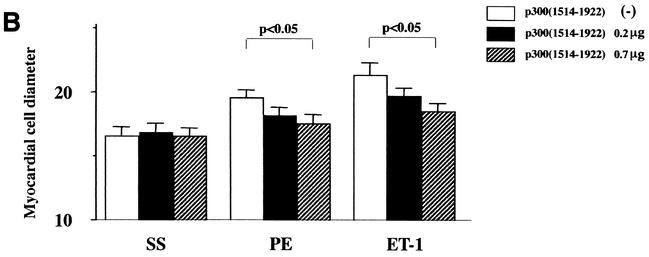
To investigate whether p300 is involved in hypertrophic responses, such as increase in cell size and sarcomere organization, cardiac myocytes were subjected to immunofluorescence with an antibody against β-MHC. As shown in Fig. 6A, in β-Gal-expressing cardiac myocytes, PE stimulation caused increases in cell size and in the thickness and density of striated sarcomeric actin fibers compared with saline stimulation. Expression of p300(1514-1922) markedly inhibited PE-induced changes of cell size and myofibrillar organization. As shown in Fig. 6B, p300(1514-1922) dose-dependently inhibited the increase of cell diameter induced by PE or ET-1. However, p300(1514-1922) alone did not affect the cell diameter in saline-stimulated cardiac myocytes. These data demonstrate that p300(1514-1922) selectively suppresses hypertrophic responses.
FIG. 6.
p300(1514-1922) blocks hypertrophic responses in cardiac myocytes. (A) Cardiac myocytes were transfected with a total of 0.7 μg of pCMV1514-1922p300 or pCMVβ-gal, stimulated with saline (SS) or PE (10−5 M) for 48 h, and subjected to immunofluorescent staining with antibody to β-MHC. −, absent; +, present. (B) Measurement of cell diameter (in micrometers). The values are means and standard errors of the mean. The data are from 50 cells in each group.
Cardiac overexpression of p300 in vivo results in increased level of acetylation and DNA binding of GATA-4.
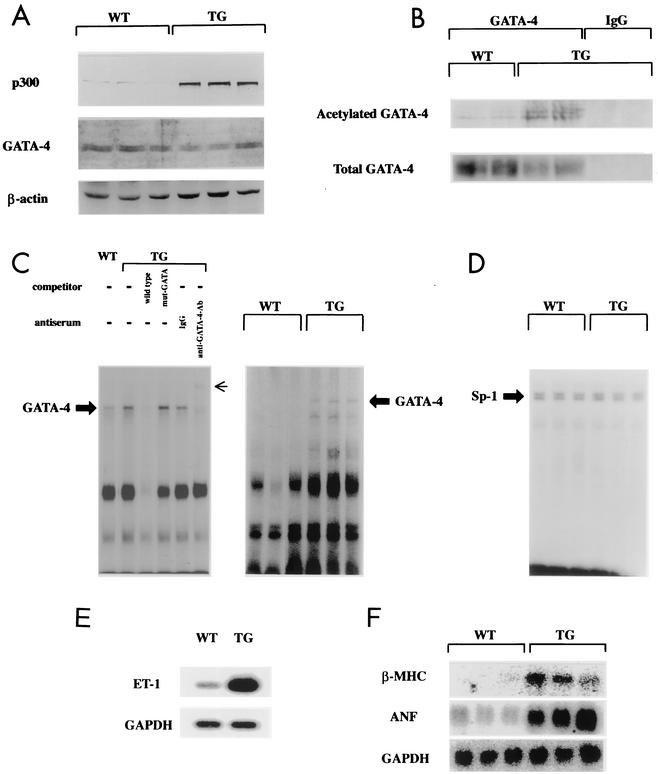
To further investigate the role of p300 in cardiac hypertrophy and heart failure in vivo, we generated TG mice expressing p300 in the heart. The injected construct consisted of the full-length human p300 cDNA driven by a cardiomyocyte-specific 5.5-kb mouse α-MHC promoter. Three p300 TG founders were identified by Southern blotting analysis and bred with C57BL/6 mice to generate F1 heterozygotes. To examine the expression of the p300 transgene in different tissues of TG mice, we performed RT-PCR using primers that specifically recognize the p300 transgene but not the endogenous p300 gene. The expression of the p300 transgene in all lines of TG mice was detected in the heart but not in the lung, liver, or kidney. These findings are consistent with prior observations that the α-MHC promoter specifies cardiac-restricted transgene expression (51).
Myocardial expression levels of p300 protein were determined by Western blot analysis. Cardiac nuclear extracts isolated from 8-week-old TG and WT mice were subjected to Western blotting with anti-p300 antibody, which recognizes both the transgene and endogenous p300. TG mouse hearts showed an eightfold increase in total p300 content compared with WT mouse hearts, whereas the total amounts of cardiac GATA-4 and β-actin in WT and TG mice did not differ (Fig. 7A). Next, we investigated whether cardiac p300 overexpression results in acetylation of GATA-4 in mice. Nuclear extracts isolated from TG and WT mouse hearts were subjected to immunoprecipitation with anti-GATA-4 antibody or with normal goat IgG as a negative control, followed by sequential Western blotting with anti-acetylated lysine antibody and anti-GATA-4 antibody. As shown in Fig. 7B, the ratio of the acetylated form of GATA-4 to total GATA-4 was markedly increased in TG mouse hearts compared with that in WT mouse hearts, indicating hyperacetylation of GATA-4 by p300 expression.
FIG. 7.
Cardiac overexpression of p300 results in acetylation and increased DNA-binding activity of GATA-4. (A) Nuclear extracts (10 μg of protein) from WT or p300 TG mouse hearts were subjected to Western blotting with anti-p300 antibody, anti-GATA-4 antibody, or anti-β-actin antibody. (B) Nuclear extracts (400 μg of protein) from WT or TG mouse hearts were immunoprecipitated with anti-GATA-4 antibody and control goat IgG and sequentially subjected to Western blotting with anti-acetylated lysine antibody and anti-GATA-4 antibody. (C and D) Nuclear extracts from WT and TG mouse hearts were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site in the ET-1 promoter (C) and with one containing the Sp-1 site (D). Small arrow, supershifted band of GATA-4. (E) Analysis of ET-1 mRNA levels in WT and TG mouse hearts was performed by RT-PCR. (F) Northern blotting of total RNA from WT and TG mouse hearts for β-MHC, ANF, and GAPDH.
To determine whether GATA-4 acetylation results in increased DNA-binding activity of cardiac GATA-4 in p300 TG mouse hearts, EMSAs were performed with cardiac nuclear extracts from 8-week-old WT and TG mice. Nuclear extracts were probed with a radiolabeled double-stranded oligonucleotide containing the GATA-4 site of the ET-1 promoter. Competition and supershift experiments demonstrated that the band represents an interaction of the probe with cardiac GATA-4 (Fig. 7C, left gel, third to sixth lanes from left). As shown in Fig. 7C, the intensity of the specific band indicating GATA-4 binding was increased in cardiac nuclear extracts from TG mice (second lane in the left gel and fourth to sixth lanes in the right gel) compared with those from WT mice (first lane in the left gel and first to third lanes in the right gel). In contrast, Sp-1 binding activities did not differ in WT and TG mouse hearts (Fig. 7D).
Cardiac overexpression of p300 results in increased expression of GATA-4-dependent hypertrophy-responsive genes.
To determine if cardiac p300 overexpression alters the expression of GATA-4-dependent hypertrophy-responsive genes, RNA isolated from mouse ventricles was subjected to RT-PCR for the detection of prepro-ET-1 mRNA and to Northern blotting for detection of ANF and β-MHC mRNAs. TG ventricles showed markedly increased levels of ET-1 (Fig. 7E), ANF, and β-MHC (Fig. 7F) mRNAs compared with WT ventricles. However, the levels of ventricular control GAPDH mRNA in WT and TG mice did not differ.
Cardiac overexpression of p300 results in increased mortality and left-ventricular myocyte hypertrophy, dilatation, and dysfunction.
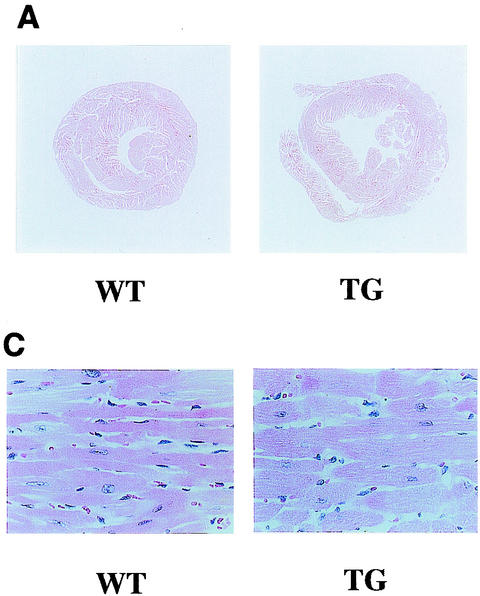
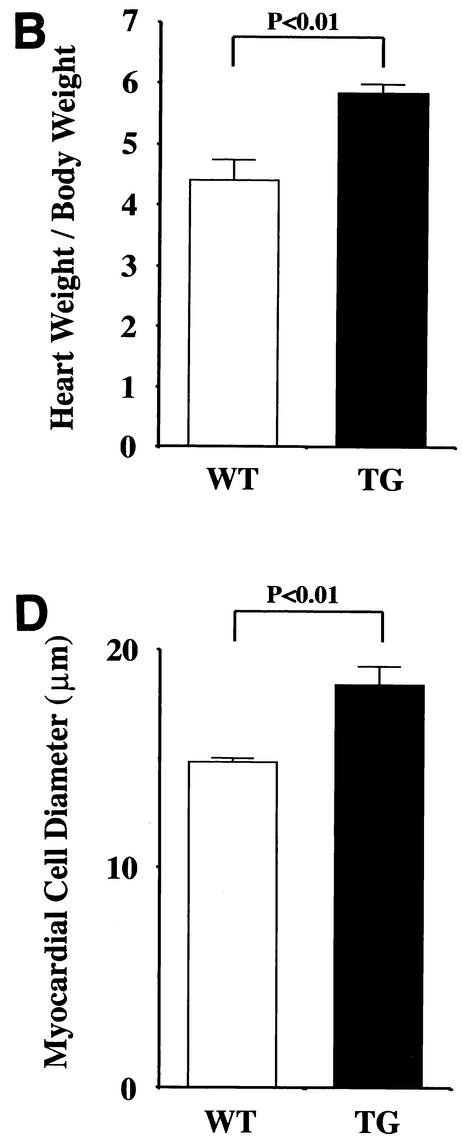
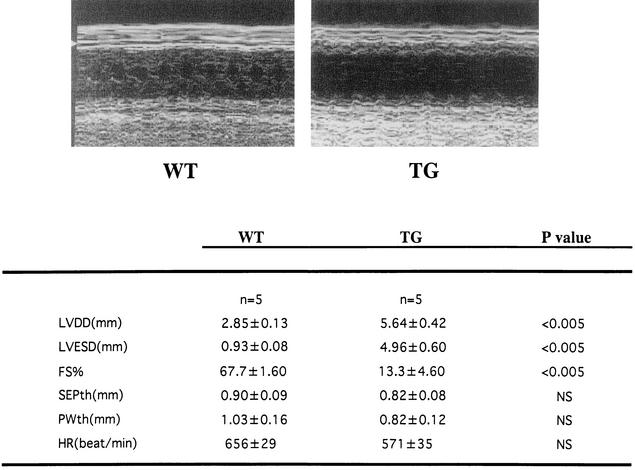
Finally, we evaluated whether TG mice exhibit representative symptoms of heart failure, such as increased mortality, reduced systolic function, and dilatation of the left ventricles. TG mice were prone to premature death after 20 weeks of age and displayed significantly lower survival rates than WT mice at 42 weeks of age (WT [n = 45], 100%; TG [n = 45], 76%; P < 0.0001). TG mice sacrificed at the age of 24 weeks demonstrated dilatation of the left ventricles without a significant increase in wall thickness (Fig. 8A). Hearts from TG mice showed a significant (P < 0.01) increase in the heart weight-to-body weight ratio (Fig. 8B). Histological analysis demonstrated that TG mouse hearts showed obvious hypertrophy of individual myocytes but no evidence of an increase in fibrosis, myofibrillar disarray, or inflammatory changes compared with WT mouse hearts (Fig. 8C). The cross-sectional myocardial-cell diameter was significantly (P < 0.01) increased in the TG mice compared with the WT mice (Fig. 8D). To determine the effects of cardiac p300 overexpression on ventricular function, we performed echocardiography. As shown in Fig. 9, TG mice at the age of 24 weeks revealed markedly depressed fractional shortening and increased cavity diameter of the left ventricles. However, the left-ventricular wall thicknesses and heart rates in WT and TG mice did not differ. These changes were observed in all three lines of p300 TG mice.
FIG. 8.
Histological sections of hearts from WT and p300 TG mice. (A) Gross view of histological sections of WT and TG mice at 24 weeks of age. Both sections were cut at the midsagittal level and parallel to the base. (B) Heart weight/body weight ratio (1,000) of WT and TG mice at 24 weeks of age (n = 5 for each group). (C) Histological sections at a magnification of ×200. (D) Cell diameter was measured as described in Materials and Methods. The values are means and standard errors of the mean. The data are from 50 cells in each group.
FIG. 9.
Echocardiographic parameters of WT and p300 TG mice. TG mice or WT littermates at the age of 24 weeks were subjected to transthoracic echocardiography. SEPth, septal wall thickness; PWth, left-ventricular posterior-wall thickness; HR, heart rate; FS, fractional shortening, which was calculated as [(LVDD − LVESD)/LVDD] × 100.
DISCUSSION
GATA-4 acetylation as a new mode of posttranslational modification during myocardial-cell hypertrophy.
GATA-4 is a member of the GATA family of zinc finger transcription factors and plays a critical role in heart development (57). GATA-4 also mediates hypertrophic responses of cardiac myocytes (22, 25; reviewed in reference 46). p300 protein serves as a transcriptional coactivator of GATA-4 and provides a bridge between GATA-4 and the basal transcriptional machinery (16, 31). In addition to its bridging function, p300 exhibits HAT activity and is able to acetylate DNA-binding transcription factors, as well as histones (20). p300 protein also frequently forms complexes with other HATs, including PCAF (48), SRC-1 (58), and P/CIP/ACTR/AIBI (13). Protein acetylation often facilitates protein-protein and protein-DNA interactions. The present study has demonstrated that p300 is sufficient to induce GATA-4 acetylation, which results in an increase in the DNA-binding activity of GATA-4. Adenovirus E1A oncoprotein, which inhibited p300-mediated acetylation, also perturbed p300/GATA-4 dependent transcription. These results suggest that p300-mediated acetylation of GATA-4 is required for its full activity as a transcriptional regulator. The lysine residues acetylated by p300/CBP in GATA-1/3 have been mapped (8, 59). Although these residues are not conserved in GATA-4, several lysine residues are possible acetylation sites around the DNA-binding domain of GATA-4. Dissecting these sites remains a major theme for future study.
The present study demonstrates that E1A inhibits p300-mediated acetylation of GATA-4. Our data are compatible with the finding that E1A interferes in the association of p300/CBP with DNA-binding transcription factors, such as p53 and GATA-1, and inhibits acetylation of these factors (10, 26). However, there are reports indicating that E1A does not interfere with the activity of p300 to acetylate histones (3, 6, 19). The reasons for this discrepancy are unclear at present. It is possible that E1A weakens the acetylation of transcription factors by disrupting the association of p300 with these factors rather than affecting HAT activity itself. Alternatively, E1A might have some other direct or indirect effects on histones to reinforce the HAT activity of p300/CBP. Further studies of how E1A regulates the HAT activity of p300/CBP are needed.
During myocardial-cell hypertrophy, the DNA binding of GATA-4 markedly increases while the level of expression of GATA-4 remains unchanged. These findings suggest that posttranslational mechanisms are involved in the activation of cardiac GATA-4 during the process of hypertrophy. One such mechanism is phosphorylation of GATA-4, which requires activation of MEK1/ERKs by Gq protein-coupled receptor agonists (38, 47). The present study demonstrated that during myocardial-cell hypertrophy, an acetylated form of GATA-4 markedly increases, which results in an increase in the DNA-binding activity of cardiac GATA-4. These findings provide evidence that the acetylation of GATA-4 is a novel mode of posttranslational modification during hypertrophic responses. However, the time courses of phosphorylation and acetylation are clearly distinct. The acetylation of cardiac GATA-4 occurs 48 h after agonist stimulation. In contrast, the phosphorylation occurs at much earlier stages (3 and 12 h after stimulation) and decreases at later stages (47). These findings suggest that these two modes of posttranslational modification are regulated through independent mechanisms.
Role of p300 in nuclear hyperacetylation of cardiac myocytes during hypertrophy.
Histone acetylation is now recognized as one of the hallmark properties of transcriptionally active chromatin (24), and it appears to influence cell cycle progression (35), chromosome dynamics (18), and DNA recombination (55), as well as DNA repair and apoptosis (27). The present study provides the first evidence that nuclear hyperacetylation of cardiac myocytes occurs during hypertrophic responses. Acetylation is regulated not only by intrinsic HATs, such as p300, but also by histone deacetylases. Activation of calcium-calmodulin-dependent protein kinase signaling, which generally occurs during myocardial-cell hypertrophy (50), releases MEF2 from histone deacetylases 4 and 5 (40, 41, 43). This results in unmasking of MEF2 transcriptional activity and in nuclear export of histone deacetylases. It is also possible that the exit of histone deacetylases from the nucleus contributes to nuclear hyperacetylation of hypertrophied cardiac myocytes. The present study demonstrated that expression of p300 in cardiac myocytes is markedly increased during hypertrophy and that a dominant-negative form of p300 blocks the acetylation caused by a hypertrophic agonist. These findings suggest that p300 is required for nuclear hyperacetylation of hypertrophied myocytes. However, they do not exclude the possibility that other HATs, such as CBP and PCAF, play a role in acetylation of GATA-4 in cardiac myocytes.
The question arises as to how the HAT activity of p300 is regulated during myocardial-cell hypertrophy. Increased levels of p300 protein may contribute to the nuclear hyperacetylation of hypertrophied myocytes. In addition, phosphorylation of p300 has been reported to influence the HAT activity of p300/CBP. For example, the HAT activity of p300/CBP increases in association with cyclin E/CDK-mediated phosphorylation of p300/CBP at the G1/S boundary of the mammalian cell cycle (1). It has been proposed that activation of p44 MAP kinase leads to enhanced phosphorylation of p300/CBP in the C-terminal region and thereby enhances its HAT activity (2). Protein kinase A (15), calcium-calmodulin-dependent kinase N (11), and MAP kinase (2) are known to phosphorylate p300/CBP, thereby enhancing p300/CBP-mediated transcriptional activation. In contrast, the transcriptional regulator Twist appears to reduce p300/CBP HAT activity (21). Further studies are needed to clarify how the HAT activity of p300 is regulated during myocardial-cell hypertrophy.
Nuclear acetylation by p300 causes decompensated heart failure in mice.
Phosphorylation of GATA-4 is one mechanism by which its DNA-binding activity is increased during myocardial-cell hypertrophy (38, 47). Activation of MEK1/ERK1/ERK2 either by agonist stimulation or by expression of a constitutively active form of MEK1 results in phosphorylation of GATA-4 at serine residue 105 (38). TG mice overexpressing a constitutively active form of MEK1 in the heart exhibit concentric hypertrophy associated with hyperdynamic systolic function (9). To test whether acetylation of cardiac GATA-4 by p300 in vivo results in a cardiac phenotype similar to or distinct from that induced by phosphorylation of GATA-4, we have generated TG mice overexpressing p300 under the control of the cardiac-specific α-MHC promoter. In contrast to mice with cardiac MEK-1 activation, mice overexpressing p300 in the heart exhibit increased mortality and marked eccentric dilatation and systolic dysfunction of the left ventricles. In these mice, acetylation and the increased DNA-binding activity of cardiac GATA-4 occurred as early as 8 weeks of age and preceded the development of left-ventricular dilatation and dysfunction. These findings raise the hypothesis that two modes of posttranslational modification, phosphorylation and acetylation, are involved in distinct cardiac phenotypes: concentric hypertrophy with normal systolic function and eccentric dilatation with systolic dysfunction, respectively. However, the present study provides insufficient evidence that p300-mediated acetylation and the resultant increase in the DNA-binding activity of cardiac GATA-4 are the primary causes of heart failure in these mice. To confirm this hypothesis, it is necessary to perform further studies using TG mice expressing equivalent amounts of p300 mutant lacking the HAT activity.
The cardiac phenotype of p300 TG mice also significantly differs from that of GATA-4 TG mice (37). These two kinds of TG mice exhibit similar increases (30 to 40%) in the heart weight/body weight ratio at 6 to 8 months of age. At that stage, the left ventricles of p300 TG mice show a 98% increase in the end diastolic dimension and an 80% decrease in fractional shortening compared with those of WT mice. In contrast, GATA-4 TG mice exhibit an increase in the end diastolic dimension of only 11% and a 42% decrease in fractional shortening. Thus, overexpression of p300 induces similar levels of cardiac growth but much more severe left-ventricular dilatation and dysfunction than overexpression of GATA-4. While the mechanisms that account for this difference are unclear at present, p300 may activate (acetylate) not only GATA-4 but also other transcription factors, including MEF2, serum response factor, and AP-1. Activation of multiple transcription factors may be involved in the more severe dilatation and systolic dysfunction of the left ventricles than those caused by increased levels of GATA-4 protein alone.
The present study demonstrated that a dominant-negative form of p300 inhibits not only nuclear hyperacetylation but also all of the characteristics of hypertrophic responses examined. These findings suggest that acetylation, which results in transcriptionally active chromatin, plays a key role in the development of hypertrophy, as well as changes in cardiac gene expression during this process. In addition, p300-mediated acetylation of cardiac nuclear proteins, including GATA-4, in mice results in pathophysiological changes that mimic those associated with heart failure in humans. These findings suggest that acetylation by p300 provides a pharmacological target for the treatment of heart failure. Recently, Lau et al. reported that Lys-coenzyme A is a selective inhibitor of p300 HAT activity (36). The selectivity of this compound for p300 is at least 400 times higher than that for PCAF. The search for cardiac-specific HAT inhibitors will be of considerable importance in establishing a novel therapy modality for heart failure in humans.
Acknowledgments
This work was supported in part by the Advanced and Innovational Research program in Life Science and by grants to T.K. and K.H. from the Ministry of Education, Science and Culture of Japan.
We thank N. Sowa for his excellent technical assistance.
Three authors, T. Yanazume, T. Morimoto, and T. Kawamura, equally contributed to this work.
REFERENCES
- 1.Ait-Si-Ali, S., A. Polesskaya, S. Filleur, R. Ferreira, A. Duquet, P. Robin, A. Vervish, D. Trouche, F. Cabon, and A. Harel-Bellan. 2000. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene 19:2430-2437. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., D. Carlisi, S. Ramirez, L. C. Upegui-Gonzalez, A. Duquet, P. Robin, B. Rudkin, A. Harel-Bellan, and D. Trouche. 1999. Phosphorylation by p44 MAP kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 262:157-162. [DOI] [PubMed] [Google Scholar]
- 3.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. [DOI] [PubMed] [Google Scholar]
- 4.Arceci, R. J., A. A. King, M. C. Simon, S. H. Orkin, and D. B. Wilson. 1993. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol. Cell. Biol. 13:2235-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 6.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 7.Bordoli, L., S. Husser, U. Luthi, M. Netsch, H. Osmani, and R. Eckner. 2001. Functional analysis of the p300 acetyltransferase domain: the PHD finger of p300 but not of CBP is dispensable for enzymatic activity. Nucleic Acids Res. 29:4462-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 9.Bueno, O. F., L. J. De Windt, K. M. Tymitz, S. A. Witt, T. R. Kimball, R. Klevitsky, T. E. Hewett, S. P. Jones, D. J. Lefer, C. F. Peng, R. N. Kitsis, and J. D. Molkentin. 2000. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 11.Chawla, S., G. E. Hardingham, D. R. Quinn, and H. Bading. 1998. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science 281:1505-1509. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 13.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 14.Chien, K. R., H. Zhu, K. U. Knowlton, W. Miller-Hance, M. van Bilsen, T. X. O'Brien, and S. M. Evans. 1993. Transcriptional regulation during cardiac growth and development. Annu. Rev. Physiol. 55:77-95. [DOI] [PubMed] [Google Scholar]
- 15.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H.Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 16.Dai, Y. S., and B. E. Markham. 2001. p300 functions as a coactivator of transcription factor GATA-4. J. Biol. Chem. 276:37178-37185. [DOI] [PubMed] [Google Scholar]
- 17.Eckner, R., M. E. Ewen, D. Newsome, M. Gerdes, J. A. DeCaprio, J. B. Lawrence, and D. M. Livingston. 1994. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 8:869-884. [DOI] [PubMed] [Google Scholar]
- 18.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 19.Fax, P., O. Lehmkuhler, C. Kuhn, H. Esche, and D. Brockmann. 2000. E1A12S-mediated activation of the adenovirus type 12 E2 promoter depends on the histone acetyltransferase activity of p300/CBP. J. Biol. Chem. 275:40554-40560. [DOI] [PubMed] [Google Scholar]
- 20.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 21.Hamamori, Y., V. Sartorelli, V. Ogryzko, P. L. Puri, H. Y. Wu, J. Y. Wang, Y. Nakatani, and L. Kedes. 1999. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96:405-413. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa, K., S. J. Lee, S. M. Jobe, B. E. Markham, and R. N. Kitsis. 1997. cis-Acting sequences that mediate induction of beta-myosin heavy chain gene expression during left ventricular hypertrophy due to aortic constriction. Circulation 96:3943-3953. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa, K., M. B. Meyers, and R. N. Kitsis. 1997. Transcriptional coactivator p300 stimulates cell type-specific gene expression in cardiac myocytes. J. Biol. Chem. 272:20049-20054. [DOI] [PubMed] [Google Scholar]
- 24.Hebbes, T. R., A. W. Thorne, and C. Crane-Robinson. 1988. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 7:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herzig, T. C., S. M. Jobe, H. Aoki, J. D. Molkentin, A. W. Cowley, Jr., S. Izumo, and B. E. Markham. 1997. Angiotensin II type 1a receptor gene expression in the heart: AP-1 and GATA-4 participate in the response to pressure overload. Proc. Natl. Acad. Sci. USA 94:7543-7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung, H. L., J. Lau, A. Y. Kim, M. J. Weiss, and G. A. Blobel. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 28.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 29.Ip, H. S., D. B. Wilson, M. Heikinheimo, Z. Tang, C. N. Ting, M. C. Simon, J. M. Leiden, and M. S. Parmacek. 1994. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol. Cell. Biol. 14:7517-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaburagi, S., K. Hasegawa, T. Morimoto, M. Araki, T. Sawamura, T. Masaki, and S. Sasayama. 1999. The role of endothelin-converting enzyme-1 in the development of α1-adrenergic-stimulated hypertrophy in cultured neonatal rat cardiac myocytes. Circulation 99:292-298. [DOI] [PubMed] [Google Scholar]
- 31.Kakita, T., K. Hasegawa, T. Morimoto, S. Kaburagi, H. Wada, and S. Sasayama. 1999. p300 protein as a coactivator of GATA-5 in the transcription of cardiac-restricted atrial natriuretic factor gene. J. Biol. Chem. 274:34096-34102. [DOI] [PubMed] [Google Scholar]
- 32.Kariya, K., I. K. Farrance, and P. C. Simpson. 1993. Transcriptional enhancer factor-1 in cardiac myocytes interacts with an alpha 1-adrenergic- and beta-protein kinase C-inducible element in the rat beta-myosin heavy chain promoter. J. Biol. Chem. 268:26658-26662. [PubMed] [Google Scholar]
- 33.Kawana, M., M. E. Lee, E. E. Quertermous, and T. Quertermous. 1995. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol. Cell. Biol. 15:4225-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolodziejczyk, S. M., L. Wang, K. Balazsi, Y. DeRepentigny, R. Kothary, and L. A. Megeney. 1999. MEF2 is upregulated during cardiac hypertrophy and is required for normal post-natal growth of the myocardium. Curr. Biol. 9:1203-1206. [DOI] [PubMed] [Google Scholar]
- 35.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 36.Lau, O. D., T. K. Kundu, R. E. Soccio, S. Ait-Si-Ali, E. M. Khalil, A. Vassilev, A. P. Wolffe, Y. Nakatani, R. G. Roeder, and P. A. Cole. 2000. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5:589-595. [DOI] [PubMed] [Google Scholar]
- 37.Liang, Q., L. J. De Windt, S. A. Witt, T. R. Kimball, B. E. Markham, and J. D. Molkentin. 2001. The transcription factors GATA-4 and GATA-6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276:30245-30253. [DOI] [PubMed] [Google Scholar]
- 38.Liang, Q., R. J. Wiese, O. F. Bueno, Y. S. Dai, B. E. Markham, and J. D. Molkentin. 2001. The transcription factor GATA-4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lill, N. L., S. R. Grossman, D. Ginsberg, J. DeCaprio, and D. M. Livingston. 1997. Binding and modulation of p53 by p300/CBP coactivators. Nature 387:823-827. [DOI] [PubMed] [Google Scholar]
- 40.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 41.Lu, J., T. A. McKinsey, R. L. Nicol, and E. N. Olson. 2000. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc. Natl. Acad. Sci. USA 97:4070-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molkentin, J. D. 2000. Calcineurin and beyond: cardiac hypertrophic signaling. Circ. Res. 87:731-738. [DOI] [PubMed] [Google Scholar]
- 45.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molkentin, J. D., and E. N. Olson. 1997. GATA4: a novel transcriptional regulator of cardiac hypertrophy? Circulation 96:3833-3835. [PubMed] [Google Scholar]
- 47.Morimoto, T., K. Hasegawa, S. Kaburagi, T. Kakita, H. Wada, T. Yanazume, and S. Sasayama. 2000. Phosphorylation of GATA-4 is involved in α1-adrenergic agonist-responsive transcription of the endothelin-1 gene in cardiac myocytes. J. Biol. Chem. 275:13721-13726. [DOI] [PubMed] [Google Scholar]
- 48.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 49.Paradis, P., W. R. MacLellan, N. S. Belaguli, R. J. Schwartz, and M. D. Schneider. 1996. Serum response factor mediates AP-1-dependent induction of the skeletal alpha-actin promoter in ventricular myocytes. J. Biol. Chem. 271:10827-10833. [DOI] [PubMed] [Google Scholar]
- 50.Passier, R., H. Zeng, N. Frey, F. J. Naya, R. L. Nicol, T. A. McKinsey, P. Overbeek, J. A. Richardson, S. R. Grant, and E. N. Olson. 2000. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Investig. 105:1395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robbins, J., J. Palermo, and H. Rindt. 1995. In vivo definition of a cardiac specific promoter and its potential utility in remodeling the heart. Ann. N. Y. Acad. Sci. 752:492-505. [DOI] [PubMed] [Google Scholar]
- 52.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 53.Sadoshima, J., and S. Izumo. 1997. The cellular and molecular response of cardiac myocytes to mechanical stress. Annu. Rev. Physiol. 59:551-571. [DOI] [PubMed] [Google Scholar]
- 54.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlissel, M. S. 2000. Perspectives: transcription. A tail of histone acetylation and DNA recombination. Science 287:438-440. [DOI] [PubMed] [Google Scholar]
- 56.Slepak, T. I., K. A. Webster, J. Zang, H. Prentice, A. O'Dowd, M. N. Hicks, and N. H. Bishopric. 2001. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J. Biol. Chem. 276:7575-7585. [DOI] [PubMed] [Google Scholar]
- 57.Soudais, C., M. Bielinska, M. Heikinheimo, C. A. MacArthur, N. Narita, J. E. Saffitz, M. C. Simon, J. M. Leiden, and D. B. Wilson. 1995. Targeted mutagenesis of the transcription factor GATA-4 gene in mouse embryonic stem cells disrupts visceral endoderm differentiation in vitro. Development 121:3877-3888. [DOI] [PubMed] [Google Scholar]
- 58.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 59.Yamagata, T., K. Mitani, H. Oda, T. Suzuki, H. Honda, T. Asai, K. Maki, T. Nakamoto, and H. Hirai. 2000. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO J. 19:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yanazume, T., K. Hasegawa, H. Wada, T. Morimoto, M. Abe, T. Kawamura, and S. Sasayama. 2002. Rho/ROCK pathway contributes to the activation of extracellular signal-regulated kinase/GATA-4 during myocardial cell hypertrophy. J. Biol. Chem. 277:8618-8625. [DOI] [PubMed] [Google Scholar]
- 61.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 62.Yuan, W., G. Condorelli, M. Caruso, A. Felsani, and A. Giordano. 1996. Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem. 271:9009-9013. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, W., R. C. Kowal, F. Rusnak, R. A. Sikkink, E. N. Olson, and R. G. Victor. 1999. Failure of calcineurin inhibitors to prevent pressure-overload left-ventricular hypertrophy in rats. Circ. Res. 84:722-728. [DOI] [PubMed] [Google Scholar]