Abstract
Friend leukemia integration 1 (Fli-1) is a member of the Ets family of transcriptional activators that has been shown to be an important regulator during megakaryocytic differentiation. We undertook a two-hybrid screen of a K562 cDNA library to identify transcription factors that interacted with Fli-1 and were potential regulators of megakaryocyte development. Here we report the physical interaction of Fli-1 with GATA-1, a well-characterized, zinc finger transcription factor critical for both erythroid and megakaryocytic differentiation. We map the minimal domains required for the interaction and show that the zinc fingers of GATA-1 interact with the Ets domain of Fli-1. GATA-1 has previously been shown to interact with the Ets domain of the Fli-1-related protein PU.1, and the two proteins appear to inhibit each other's activity. In contrast, we demonstrate that GATA-1 and Fli-1 synergistically activate the megakaryocyte-specific promoters GPIX and GPIbα in transient transfections. Quantitative electrophoretic mobility shift assays using oligonucleotides derived from the GPIX promoter containing Ets and GATA binding motifs reveal that Fli-1 and GATA-1 exhibit cooperative DNA binding in which the binding of GATA-1 to DNA is increased approximately 26-fold in the presence of Fli-1 (from 4.2 to 0.16 nM), providing a mechanism for the observed transcriptional synergy. To test the effect on endogenous genes, we stably overexpressed Fli-1 in K562 cells, a line rich in GATA-1. Overexpression of Fli-1 induced the expression of the endogenous GPIX and GPIbα genes as measured by Northern blot and fluorescence-activated cell sorter analysis. This work suggests that Fli-1 and GATA-1 work together to activate the expression of genes associated with the terminal differentiation of megakaryocytes.
The successive activation of tissue-specific genes during cellular differentiation is orchestrated by the formation of different transcriptional complexes consisting of cell-specific and ubiquitous transcription factors (24, 30). This process is arguably best exemplified in the hematopoietic system, where different transcriptional complexes control the production of distinct cellular lineages from a common hematopoietic stem cell precursor. Among the rarest of the mature hematopoietic cells are megakaryocytes, large polyploid cells that reside in the bone marrow and whose cytoplasmic fragments are extruded into the bloodstream to form platelets.
The key transcription factors involved in megakaryocyte differentiation are coming to light (for a review, see Shivdasani [28]). One of these, Friend leukemia integration 1 (Fli-1), is a member of the Ets family of transcription factors. Ets factors encompass a family of over 40 members that are characterized by an 85-amino-acid region of homology termed the Ets domain, which mediates binding to the core Ets recognition element 5′-GGA(A/T)-3′ (36; F. D. Karim, L. D. Urness, C. S. Thummel, M. J. Klemsz, S. R. McKercher, A. Celada, C. Van Beveren, R. A. Maki, C. V. Gunther, J. A. Nye, et al., Letter, Genes Dev. 4:1451-1453, 1999). Fli-1 was originally discovered as a gene that was commonly activated as a result of proviral insertion of the Friend leukemia virus in mice (5).
Several pieces of evidence suggest that Fli-1 plays an important role during the normal development of megakaryocytes. Early experiments showed that Fli-1 overexpression in K562 cells caused these cells to acquire a megakaryocytic phenotype similar to that observed when the cells were treated with phorbol esters (1). Fli-1 protein expression has been demonstrated in primary megakaryocytes and platelets (3). The same investigators demonstrated Fli-1 also binds and transactivates the promoters of a number of megakaryocyte-specific genes in transient transfection experiments. However, perhaps the most convincing evidence of Fli-1's role in megakaryocyte differentiation comes from knockout models (14, 31). Inactivation of the Fli-1 gene in mice is embryonic lethal at day E11.5, with death resulting from brain hemorrhage and endothelial cell dysfunction. Fli-1 knockout mice produce small, undifferentiated megakaryocytic progenitors with abnormal ultrastructural features such as reduced α-granule numbers and disrupted demarcation membrane systems. Levels of megakaryocyte-specific genes normally expressed late during differentiation, such as GPIX (for glycoprotein IX), are also markedly reduced (14). Moreover, Fli-1−/− embryonic stem cells are unable to produce megakaryocytic colonies or multilineage colonies containing megakaryocytes in colony formation assays (16).
In order to identify transcription factors that physically interact and potentially cooperate with Fli-1 to promote megakaryocyte differentiation, we conducted a yeast two-hybrid screen of a K562 cDNA library by using Fli-1 as bait. Here we identify GATA-1, a well-characterized zinc finger transcription factor crucial for both erythroid and megakaryocytic differentiation as a partner of Fli-1. In contrast to the antagonistic interaction between the Ets family protein PU.1 and GATA-1 described previously (22, 27, 38, 39), we demonstrate that the interaction between Fli-1 and GATA-1 results in synergistic activation of megakaryocyte-specific genes through cooperative DNA binding. One of the hallmark features of the promoter regions of almost all megakaryocyte-specific genes studied to date is the presence of multiple binding sites for GATA and Ets family members (17). We provide evidence that both Fli-1 and GATA-1 are key determinants for the high-level expression of Mk-specific genes and for the normal differentiation of megakaryocytes.
MATERIALS AND METHODS
DNA constructs.
The pGex-Fli-1(76-452) construct was provided by D. Hickstein (Fred Hutchinson Cancer Research Center). To generate the pGBKT7/Fli-1(238-452) construct used in the two-hybrid screen, the region of Fli-1 containing amino acids 237 to 452 was amplified by PCR with 5′-GGG CAA TCA TAT GAA TTC TGG CCT C-3′ and 5′-TGC AGG ATC CGC TTC AGC TAG AAG-3′ as primers. The NdeI/BamHI-digested fragment was subcloned into the pGBKT7 plasmid (Clontech) predigested with the same enzymes. To generate pGex/GST-Fli-1(238-452), the corresponding region of Fli-1 was digested from pGBKT7/Fli-1Δ237 with EcoRI/HindIII, passaged through pBluescript II KS+, digested with EcoRV/HindIII, and cloned into the SmaI/HindIII sites of pGexJT (pGex2T with a modified multiple cloning site). pGexJT/Fli-1(237-364) and pGexJT/Fli-1(362-452) were constructed by amplifying the corresponding regions of Fli-1 with the following primer pairs for the fragments from amino acids 237 to 364 and 362 to 452, respectively: 5′-GGC AAT AAC ATG GAA TTC GGC CTC AAG-3′ and 5′-GCC TGG TCG ACG CCG TGG AAG TC-3′ and 5′-TTG AAT TCC ACG GCA TTG CCC AG-3′ and 5′-GCT AGA AGT CGA CTG ATG AGT AAG C-3′. The inserts were digested with EcoRI and SalI and subcloned into the same sites of pGexJT. The GST-mGATA-1 and pRcCMV-mGATA-1 constructs used have been described previously (11, 23). The GPIX-567 luciferase reporter construct has also been published previously (9). The GPIbα reporter construct pGPIb-253 was a kind gift from J. Ware (Scripps Institute). The pIRES2-Fli-1 EGFP plasmid was constructed by amplifying full-length Fli-1 cDNA with the following primer pairs: 5′-CTA TAG GGA GAT CTA AGC TTC CGC-3′ and 5′-TCC AAT GCA TGG AGT AAG TGT GC-3′ out of pcDNA-Fli-1 FLAG. The insert was digested with BglII/NsiI and cloned into the BglII/PstI sites of pIRES2-EGFP (Clontech). pcDNA-FLAG-Fli-1 was generated by subcloning the BamHI/XhoI Fli-1 fragment from pGex-Fli-1(76-452) into pcDNA3-FLAG, amplifying the first 75 amino acids from Dami cell cDNA with the primer pair 5′-CTG CAG ATC TGG CCA AAT GGA CG-3′ and 5′-CCT GGA TCC ATT CAT GTG GTC A-3′, and cloning this fragment into the HindIII/BamHI site.
Tissue culture.
HeLa cells were purchased from the American Type Culture Collection and cultured in Dulbecco's modified Eagle medium supplemented with 3.7 g of sodium bicarbonate/liter and 10% fetal bovine serum (FBS) (Invitrogen) in a humidified incubator at 37°C with 5% CO2. K562 cells were cultured in RPMI 1640 supplemented with 2.0 g of sodium bicarbonate per liter and 10% FBS (Invitrogen) under the same conditions.
Yeast two-hybrid screen.
The Ets and carboxy-terminus activation domains of human Fli-1 (amino acids 238 to 452) were fused in frame to the GAL4 DNA-binding domain (GAL4DBD) in the pGBKT7 vector (Clontech) and used to screen a K562 cDNA library inserted into the EcoRI sites of the pGAD10 vector (Clontech) in Saccharomyces cerevisiae strain AH109 as described in the Matchmaker GAL4 Two-Hybrid System 3 protocol (Clontech).
Coimmunoprecipitation.
Total cell extracts from HeLa cells expressing Fli-1 and/or GATA-1 (see below) were precleared by incubating with 20 μl of protein G agarose beads for 30 min at 4oC. Subsequently, 900 μl of the precleared sample was immunoprecipitated for 3 h at 4oC with 20 μl of protein G agarose beads that had been previously cross-linked to anti-GATA-1 monoclonal antibody N6 (catalog no. sc-265; Santa Cruz Biotechnology) with dimethyl-pimelimidate (Sigma). In addition, 90 μl of sample number 5 (Fig. 1D; corresponding to 10% input Fli-1) was incubated with 10 μl of protein A agarose beads that had been previously cross-linked to anti-Fli-1 polyclonal antibody (catalog no. SC-356X, Santa Cruz). This sample was used as a control for immunoprecipitation of Fli-1. Following immunoprecipitation, samples were washed four times with NP-40 buffer (0.5% NP-40/Igepal, 150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1 μg of leupeptin/ml, 1 μg of aprotonin/ml, and 1 mM phenylmethylsulfonyl fluoride), and beads were resuspended in 20 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye and subject to immunoblotting with either Fli-1 or GATA-1 monoclonal antibody as described below.
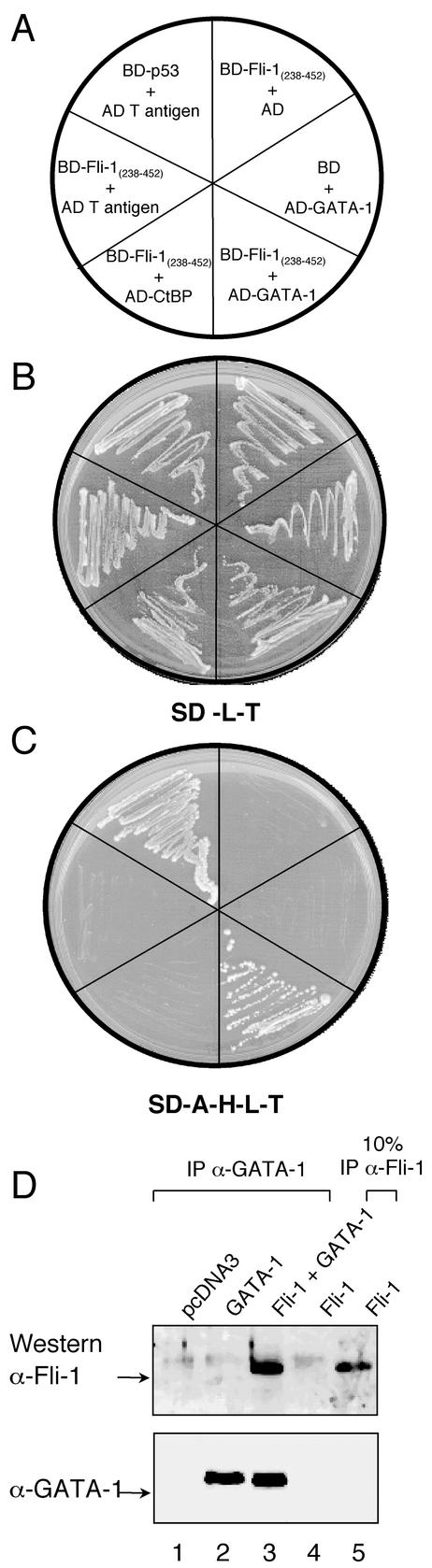
FIG. 1.
Fli-1 interacts with GATA-1. (A) Yeast strain AH109 was transformed with the indicated plasmids as shown and plated onto the corresponding sectors shown in panels B and C. (B and C) Growth of AH109 yeast harboring the plasmids as shown in panel A after 5 days of incubation at 30°C on minimal medium lacking Leu and Trp (B) and minimal medium lacking Ade, His, Leu, and Trp (C). (D) Coimmunoprecipitation of Fli-1 with GATA-1. HeLa cells were transfected with cDNAs encoding for pcDNA3 backbone (lane 1), GATA-1 (lane 2), Fli-1 and GATA-1 (lane 3), or Fli-1 alone (lanes 4 and 5) and were immunoprecipitated with anti-GATA-1 monoclonal antibody (lanes 1 to 4) or anti-Fli-1 monoclonal antibody as control (lane 5). Immunoprecipitates were then subjected to immunoblotting with anti-GATA-1 or anti-Fli-1 monoclonal antibody, as outlined in Materials and Methods.
Immunoblotting.
Samples were subject to SDS-PAGE on 10% Tris-glycine gels and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) at 100 V for 1 h. Following transfer, membranes were blocked overnight in 5% skim milk-PBS-0.05% Tween 20 and incubated with either anti-Fli-1 monoclonal antibody (1:10,000 dilution; catalog no. 554267; BD Pharmingen) or anti-GATA-1 monoclonal antibody N6 (1: 2,000 dilution; Santa Cruz) for 1 h. Membranes were washed four times in PBS-Tween 20 and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (1:2,500 dilution; Dako) for a further hour, washed four times with PBS-Tween 20, and subjected to chemiluminescence detection (Perkin-Elmer Life Sciences).
GST pulldown studies.
The various Fli-1 and GATA-1 constructs were obtained by PCR and subcloned in frame into the pGex2T vector (Pharmacia) and pGexJT vector. Glutathione S-transferase (GST) and GST fusion proteins were expressed in Escherichia coli strain BL-21 according to standard protocol. 35S-labeled proteins were prepared by in vitro translation by using the TNT T7 coupled reticulocyte lysate system (Promega). GST interaction assays were performed as previously described (15).
Transient transfection and reporter assays.
Transient transfection of HeLa cells was carried out in six-well plates by using Lipofectamine (Invitrogen) reagent according to the manufacturer's instructions. HeLa cells were transfected with 200 to 800 ng of the reporter plasmid and expression plasmids encoding either Fli-1 or GATA-1, as indicated. The total amount of DNA in each transfection was kept constant at 1.2 μg by adding the pcDNA3 backbone. Cells were harvested 48 h after transfection, and 10 μl of lysate was assayed for luciferase activity by using the Promega luciferase assay system kit.
For Western blotting experiments, 30 (Fig. 5B) or 50 μg (Fig. 5D) of total cell lysate was subjected to immunoblotting as described above. The assay for human growth hormone was performed using the human growth hormone radioisotopic assay kit as indicated by the manufacturer's instructions (Nichols Institute Diagnostics). Results were not normalized to an internal control plasmid due to activation of the internal control by Fli-1 or GATA-1 similar to that observed with other activators by previous investigators (4, 10). Rather, the results shown represent average fold increase in firefly luciferase or growth hormone activity ± standard deviation of the mean from three experiments performed in triplicate.
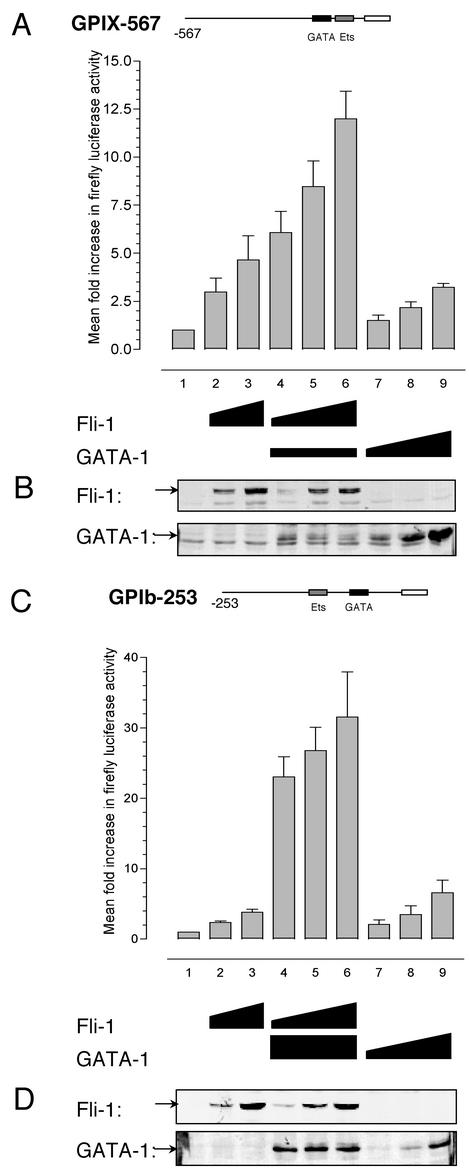
FIG. 5.
The Fli-1-GATA-1 combination results in synergistic activation of the megakaryocyte-specific promoters GPIX and GPIbα in HeLa cells. (A) HeLa cells were transfected with 200 ng of the GPIX-567 luciferase reporter plasmid alone (bar 1) or together with increasing amounts of expression plasmids for Fli-1 (bars 2 and 3, respectively, 200 and 400 ng) or were transfected with 200 ng of GATA-1 expression plasmid together with increasing amounts of Fli-1 (bars 4 to 6, respectively, 100, 200, and 400 ng) or increasing amounts of GATA-1 expression plasmid alone (bars 7 to 9, respectively, 200, 400, and 800 ng). Cells were harvested 48 h posttransfection in passive lysis buffer, and 10 μl was used in the luciferase assay. Values are expressed as mean increases ± standard deviations relative to a value of 1 for each reporter. Results shown are means from three experiments performed in triplicate. (B) Western blotting was performed on 30 μg of total cell extract from a single representative experiment described in panel A and probed for Fli-1 or GATA-1 expression to rule out possible effects of GATA-1 expression on Fli-1 levels and vice versa. Fli-1 and GATA-1 are indicated by the arrows. (C) HeLa cells were transfected with 800 ng of the GPIbα-253 luciferase reporter plasmid alone (bar 1) or together with increasing amounts of expression plasmids for Fli-1 (bars 2 and 3, respectively, 100 and 200 ng); 50 ng of GATA-1 expression plasmid together with increasing amounts of Fli-1 (bars 4 to 6, respectively, 50, 100, and 200 ng); or increasing amounts of GATA-1 expression plasmid alone (bars 7 to 9, respectively, 10, 20, and 50 ng). Cells were harvested and assayed as described for panel A. (D) Western blotting, as described for panel B, on 50-μg total cell extracts from a single representative experiment from panel C.
For coimmunoprecipitation experiments, HeLa cells were transiently transfected in 100-mm-diameter dishes with 1.75 μg of the indicated GATA-1 and/or Fli-1 expression plasmids in conjunction with 500 ng of GPIX-567 luciferase reporter plasmid to confirm functional expression and synergy. Total DNA was kept constant by adding pcDNA backbone to a total of 4 μg. Total cell extracts were washed once in 1× PBS, harvested in 1× passive lysis buffer, and subjected to coimmunoprecipitation as described above.
Creation of stably transfected K562 cells overexpressing Fli-1.
K562 cells (1.0 × 107) were transfected by electroporation in 0.6 ml of RPMI 1640 in a 0.4-cm-gap cuvette at 280 V and 960 μF by using a Bio-Rad gene pulser II with 20 μg of the indicated plasmid that had been linearized previously with VspI. The day following transfection, cells were diluted 1 in 10 and 1 in 100 into 24-well plates in RPMI 1640-10% FBS containing 1 mg of G418/ml (Amresco). After 14 days, G418-resistant colonies were picked and expanded. Cells were later analyzed for green fluorescent protein (GFP) expression by fluorescence-activated cell sorting (FACS) and for Fli-1 expression by Northern analysis. Only cells exhibiting GFP levels greater than 102 log units were analyzed.
RNA isolation and Northern blotting.
Total RNA was harvested by using TRIzol RNA isolation reagent (Life Technologies) according to the manufacturer's instructions. Poly (A)+ RNA was purified from 1 mg of total RNA by using the poly ATtract mRNA isolation kit (Promega) according to the manufacturer's instructions. Three micrograms of poly (A)+ RNA was subject to electrophoresis on a 1% agarose gel containing formaldehyde in 1× morpholinepropanesulfonic acid (MOPS) and transferred to Hybond N+ nylon membrane (Amersham). Membranes were prehybridized for 5 h at 42°C. Hybridization was performed overnight at 42°C with the described cDNA probes that had been radiolabeled with 20 μCi of [α-32P]dCTP (GeneWorks) by using nick translation and separated from unincorporated label by using Sephadex G-50 quick-spin columns (Boehringer Mannheim). Membranes were washed in 2× SSC-0.5% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.2× SSC-0.5% SDS at 42°C and then exposed to a Kodak (Rochester, N.Y.) phosphor screen overnight and developed on a Personal FX phosphorimager and analyzed by using Quantity One software (Bio-Rad).
Flow cytometry.
Flow cytometry was performed on a FACStar Plus cytometer (Becton Dickinson) as previously described (6). Cells in suspension were removed and pelleted by centrifugation at 180 × g. The adherent cells were detached from culture dishes with 0.5 mmol of EDTA/liter in PBS and combined with any corresponding suspension cells from the same flask. Cells were then washed three times in PBS-1% bovine serum albumin (BSA). The cells (2.0 × 105/reaction) were incubated with monoclonal anti-GPIX antibody GR-P (Serotec) or monoclonal anti-GPIbα antibody anti-CD42b (Dako) specific for the GPIX and GPIbα subunits, respectively. Additionally, an irrelevant isotype-matched mouse immunoglobulin G control antibody (against keyhole limpet hemocyanin) (Becton Dickinson) was used as a control. Cells were incubated for 10 min at room temperature, washed three times in 1% BSA-PBS, and then incubated at room temperature with the relevant phycoerythrin-conjugated secondary antibody (goat anti-mouse immunoglobulin G at a 1-in-200 dilution). Cells were washed three times and were kept in the dark until analysis.
EMSAs.
Analysis by electrophoretic mobility shift assays (EMSAs) was performed by using GST-purified recombinant Fli-1 [GST-Fli-1(237-364)] corresponding to the minimal interacting Ets domain and maltose-binding protein-purified mGATA-1 [MBP-GATA-1(200-318)] bearing the interacting zinc finger region. The oligonucleotides used in EMSA analysis were as follows: GATA-1 consensus oligonucleotide, 5′-GAT CTC CGG CAA CTG ATA AGG ATT CCC TG-3′; GPIX GATA-Ets oligonucleotide, 5′-CAC TGG GGG GAT AAG CCA GGC TAT TTT CAT CAC TTC CTT CCG CCC G-3′; and their complements. Probes were annealed and end labeled with polynucleotide kinase and [γ-32P]ATP followed by purification on MicroSpin G-25 columns according to the manufacturer's instructions (Amersham Pharmacia Biotech Inc). Binding reactions were performed on ice for 30 min in 25 mM Tris Cl (pH 5.7), 10% glycerol, 6 mM MgCl2, 0.5 mM EDTA, 60 mM KCl, 0.5 mM DTT, and 200 μg of BSA/ml. After binding, the reaction was subject to PAGE on 5% polyacrylamide gels in 0.5× Tris-borate-EDTA at 4°C. Gels were dried and exposed to a Kodak phosphor screen overnight and developed on a Personal FX phosphorimager, and relative radioactivity was quantified as the adjusted volume (volume minus background) of individual bands by using Quantity One software (Bio-Rad). As described previously (13), for all experiments, the DNA concentration used (0.2 fmol per 20-μl reaction) was well below the expected KD for GATA-1 binding (≤10−1 M) to ensure that the total MBP-GATA-1(200-318) concentration ([GT]) was an accurate estimate of the free MBP-GATA-1(200-318) concentration ([G]). For protein titrations of MBP-GATA-1(200-318) or GST-Fli-1(237-364), a range of protein concentrations were used to result in 0 to >90% binding. For the experiments that measured cooperative DNA binding of MBP-GATA-1 (200-318) in the presence of saturating amounts of GST-Fli-1(237-364), the concentration of GST-Fli-1(237-364) chosen (1.25 × 10−9 M) ensured greater than 90% DNA binding of GST-Fli-1(237-364) to its specific site on the probe.
Quantitative EMSA analysis.
Apparent DNA binding affinities for MBP-GATA-1 (200-318) in the absence and presence of GST-Fli-1(237-364) (KGD and KFD,G, respectively) and the binding affinity for GST-Fli-1(237-364) alone (KFD) for the GPIX-GATA-Ets oligonucleotide were calculated by quantitative gel shift assays similar to that described by Goetz et al. (13). For the single GATA-1 binding species (labeled G.D in Fig. 8A), the fraction of free DNA (D/Dt) was measured by calculating the free DNA signal at each protein concentration (D) and dividing it by the total DNA signal in a control lane containing no protein (Dt). The fraction of DNA in a complex with GATA-1 protein (G-D) can then be calculated from the equation G-D/Dt = 1 − D/Dt. The fraction of DNA in a complex with Fli-1 protein (F-D/Dt) (Fig. 8B) was measured in a similar manner. To measure the potential cooperative binding between Fli-1 and GATA-1 on the GPIX-GATA-Ets probe, the experiment was repeated in the presence of a fixed, saturating amount of Fli-1 which ensured >90% binding to the specific site on the probe. As previously described (13), several assumptions were made in this calculation: the signal from Fli-1-DNA binary complex (labeled F.D in Fig. 8C) was used as the value for D in lanes containing GATA-1 (instead of the free DNA used in single binary species reaction), and Dt was defined as the signal of the Fli-1-DNA binary complex in a control lane lacking GATA-1 protein. The fraction of DNA then bound in the ternary complex was defined as G-D/Dt = 1 − D/Dt, as above. Each experiment was performed four times to provide means for each data point.
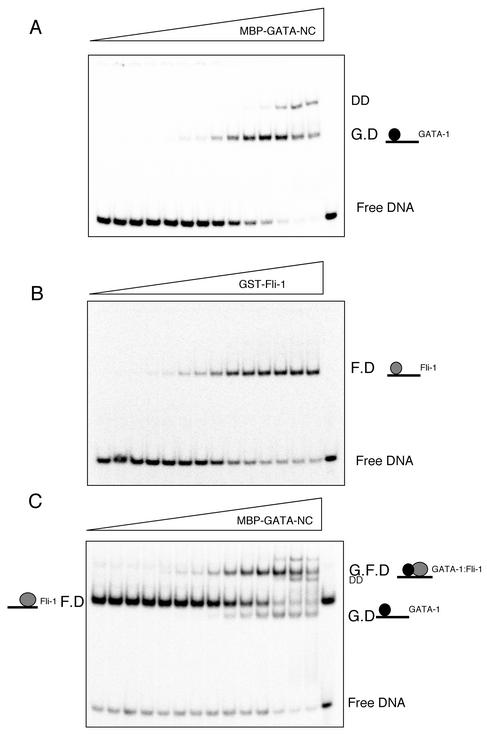
FIG. 8.
Fli-1 and GATA-1 exhibit cooperative DNA binding. (A) EMSA of equilibrium binding studies of increasing amounts of MBP-GATA-NC titrated onto the 32P-labeled GPIX-GATA-Ets oligonucleotide (0.2 fmol/20-μl reaction). The wedge indicates increasing concentrations of MBP-GATA-NC (2 × 10−13 M to 2 × 10−8 M). G.D corresponds to GATA-1-DNA binary complex. DD corresponds to GATA-1 dimer. (B) EMSA, as described for panel A, with increasing amounts of GST-Fli-1 Ets (5 × 10−13 M to 5 × 10−7 M). F.D corresponds to Fli-1-DNA binary complex. (C) EMSA, as described for panel A, with identical amounts of MBP-GATA-NC and probe in the presence of a saturating amount of GST-Fli-1. F.D corresponds to Fli-1-DNA complex, G.D corresponds to GATA-1-DNA complex, and G.F.D corresponds to GATA-1-Fli-1-DNA ternary complex.
Calculation of KD values for binary and ternary complex formation.
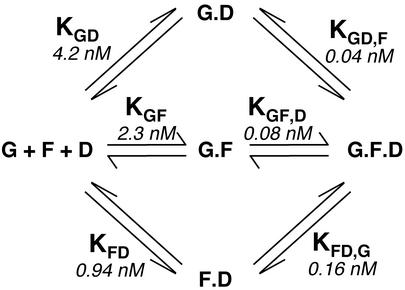
Figure 10 describes the formation of the three possible binary and the final ternary complexes in equilibrium mixtures of GATA-1, Fli-1, and DNA. In the model, GATA-1 interacts with DNA with dissociation constant KGD, GATA-1 interacts with Fli-1 with dissociation constant KGF, and Fli-1 interacts with DNA with dissociation constant KFD. Fli-1-DNA binary complex interacts with GATA-1 to form the ternary GATA-1-Fli-1-DNA complex with dissociation constant KFD,G. The formation of the ternary complex can also occur through interaction of GATA-1-DNA or GATA-1-Fli-1 binary complexes with Fli-1 or DNA, respectively, with dissociation constants KGD,F or KGF,D.
FIG. 10.
Schematic representation of the possible pathways for GATA-1-Fli-1-DNA ternary complex formation. Ternary complex formation between GATA-1 (G), Fli-1 (F), and GPIX-GATA-Ets DNA oligonucleotide (D) can occur through three possible pathways via the intermediary binary complexes GATA-1-DNA, GATA-1-Fli-1, or Fli-1-DNA. The calculated dissociation constants are also shown.
The equilibrium binding model, where [G-D] = {[G] · [D]}/KGD and [G-F] = {[G] · [F]}/KGF and [F-D] = {[F] · [D]}/KFD, is described by the equation [G-F-D] = {[F] · [D] · [G]}/{KFD · KFD,G}, where, [GT] = [G] + [G-D] + [G-F] + [G-F-D], [FT] = [F] + [G-F] + [F-D] + [G-F-D], and [DT] = [D] + [G-D] + [F-D] + [G-F-D], where the subscript T denotes total concentration of reactant.
The dissociation constants for formation of the binary GATA-1-DNA and Fli-1-DNA complexes were calculated by fitting of the individual binding isotherms to a single rectangular hyperbola by least-squares minimization. The isotherm for binding of GATA-1 to DNA in the presence of a fixed concentration of Fli-1 was fit by numerical integration of the equilibrium binding model equation using least-squares minimization (Scientist software; Micromath, Salt Lake City, Utah) with KGF and KFD,G being the unknown parameters. KGD and KFD were fixed at their separately determined values.
RESULTS
The Ets family member Fli-1 physically interacts with GATA-1.
A GAL4DBD-Fli-1 fusion was constructed and used as a bait to screen 2.0 × 106 transformants of a K562 cDNA expression library in the yeast strain AH109. K562 cells were chosen since they are a human erythroleukemic cell line which exhibits some early Mk features. The screen yielded 27 potential positive clones. The sequence from one of the clones identified (represented twice) corresponded to full-length human GATA-1, a zinc finger transcription factor important for the terminal differentiation of both erythroid cells and megakaryocytes.
Yeast strain AH109 was transformed with a series of plasmids containing Fli-1 and GATA-1 fusions and controls, as depicted in Fig. 1A. All yeast strains harboring both GAL4DBD fusions and GAL4AD fusions were able to grow on minimal medium lacking Leu and Trp, indicating the presence of both plasmids (Fig. 1B). However, apart from the p53/T antigen interaction which served as a positive control, only the expression of both GAL4DBD-Fli-1(238-452) and GAL4AD-GATA-1 in AH109 allowed growth on minimal medium lacking Ade, His, Leu, and Trp, indicating a physical interaction between the Fli-1 and GATA-1 domains within these two protein fusions (Fig. 1C).
To confirm that Fli-1 and GATA-1 associate in vivo, HeLa cells were transfected with expression vectors encoding for Fli-1 and/or GATA-1 cDNAs or vector backbone alone as control. Whole-cell extracts were then subjected to immunoprecipitation with anti-GATA-1 antibody, followed by immunoblotting with anti-Fli-1 antibody or anti-GATA-1 antibody as control. Ten-percent input of extracts from Fli-1-transfected cells subject to immunoprecipitation with an anti-Fli-1 antibody served as a positive control for Fli-1 detection (Fig. 1D, lane 5). As shown in the top panel of Fig. 1D (lane 3), Fli-1 coimmunoprecipitated with GATA-1 only in those cells transfected with both GATA-1 and Fli-1 expression constructs. No Fli-1 was detected in cells transfected with the vector backbone (Fig. 1D, lane 1), GATA-1 expression construct (Fig. 1D, lane 2) or Fli-1 expression construct (Fig. 1D, lane 4) alone. The membrane was then stripped and reprobed with anti-GATA-1 antibody. As expected, GATA-1 was detected in only those cells transfected with GATA-1 cDNA (Fig. 1D, lanes 2 and 3), demonstrating that the GATA-1 immunoprecipitation was successful. Taken together these results show that Fli-1 can physically interact with GATA-1 both in yeast and in mammalian cells.
GATA-1 binds the Fli-1 Ets domain through its zinc fingers.
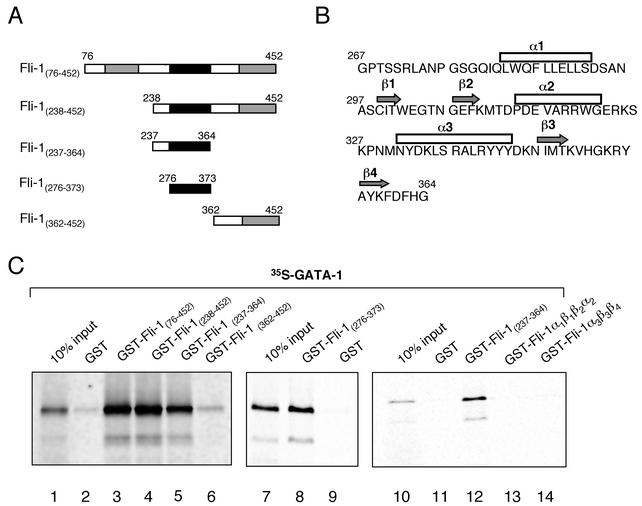
In order to map the domains on each protein required for the interaction, we constructed a number of truncated and mutant Fli-1 and GATA-1 fusion constructs (Fig. 2A and 3A) and performed GST pulldown assays. Both GST-Fli-1(76-452) and GST-Fli-1(238-452) sequestered 35S-labeled GATA-1 equally well (Fig. 2C, lanes 3 and 4), whereas deletion of the Fli-1 Ets domain (amino acids 237 to 364) abolished the interaction (Fig. 2C, lane 6). Additionally, the Fli-1 Ets domain was sufficient to retain 35S-labeled GATA-1 (Fig. 2C, lane 5), indicating that the interaction between Fli-1 and GATA-1 occurs through the Ets domain.
FIG. 2.
Fli-1 domains required for the Fli-1-GATA-1 interaction. (A) Schematic representations of the Fli-1 domains used in GST-pulldown assays. (B) Amino acid sequence of the Fli-1 Ets domain showing α-helical and β-sheet regions. (C) GST pulldowns demonstrating the Fli-1 domains required for the interaction with GATA-1. The indicated GST and GST-Fli-1 fusion constructs were expressed in E. coli strain BL-21, purified using GST-agarose beads, and incubated in the presence of 35S-labeled mGATA-1. After extensive washing, the GST- or GST-Fli-1-coated beads were boiled in loading buffer and subjected to electrophoresis, after which retained GATA-1 was visualized by phosphorimaging. Lane 1 contains 10% of the input in vitro-translated 35S-labeled GATA-1 protein. Other lanes contain the GST-Fli-1 deletions as shown.
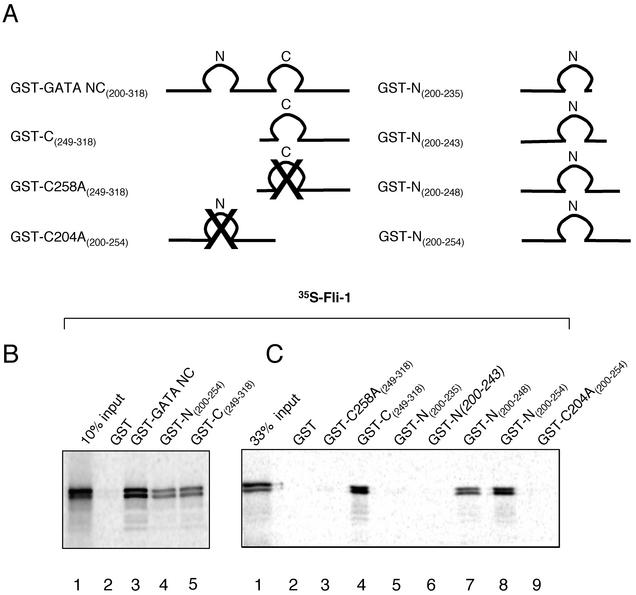
FIG. 3.
GATA domains required for the Fli-1-GATA-1 interaction. (A) Schematic representations of the GST-GATA-1 constructs used in GST pulldown assays. (B) GST pulldown assays using the GST-GATA-1 deletions and mutations shown schematically above. Lane 1, 10% of the input in vitro-translated 35S-labeled Fli-1 protein; lane 2, GST-coated beads; lanes 3 to 5, GST-GATA constructs, as indicated. (C) Finer mapping of the domains of GATA-1 required for the interaction with Fli-1. Lane 1, 33% of the input in vitro-translated 35S-labeled Fli-1 protein; lane 2, GST; lane 3, GATA-C finger with a mutated cysteine C258A; lane 4, wild-type C finger; lanes 5 to 9, GATA-N finger constructs, as shown. Samples were incubated as described in the legend for Fig. 2.
To map the interaction site(s) further, we constructed progressively smaller deletions of the Fli-1 Ets domain. A region corresponding to the minimal Ets domain (amino acids 276 to 373) was still able to interact with Fli-1 (Fig. 2C, lane 8). However, further deletion of the Ets domain into two subdomain regions, GST-Fli-1α1β1β2α2 (amino acids 278 to 330) and GST-Fli-1α3β3β4 (amino acids 327 to 364) corresponding to the first two alpha helices and two beta-pleated sheets and to the third alpha helix and third and fourth beta-pleated sheets, respectively, resulted in complete loss of 35S-GATA-1 binding activity (Fig. 2C, lanes 13 and 14). These results suggest that the entire Fli-1 Ets domain must be expressed as a complete functional unit to fold properly and retain its capacity to interact with GATA-1.
We next turned to mapping the domains of GATA-1 that were required for the interaction. GATA-1 contains two zinc fingers that conform to a CX2CX17CX2C consensus. GST pulldown experiments using truncated and mutated GATA-1 constructs (Fig. 3A) demonstrated that the central region of GATA-1 that contains the two zinc fingers GST-GATA-NC (amino acids 200 to 318) is sufficient to mediate the interaction with Fli-1 (Fig. 3B, lane 3). Both the N-terminal finger (amino acids 200 to 254) and C finger (amino acids 249 to 318) constructs were able to interact with Fli-1 equally well (Fig. 3B, lanes 4 and 5); however, binding was reduced by about 50% in comparison to that of the GST-GATA-NC construct. Finer mapping of the N-finger domain of GATA-1 implicated residues 243 to 248 in the tail of the N finger as being essential for the interaction (that is, the region following the fourth cysteine residue). C-terminal deletion of the N finger beyond amino acid 243 abolished the interaction with Fli-1 (Fig. 3C, lanes 5 and 6). Moreover, disruption of the zinc finger structure by mutation of critical zinc-chelating cysteines to alanine in either finger (amino acids C204A and C258A in the N and C fingers, respectively) abolished the interaction with Fli-1 (Fig. 3C, lane 9 and 3). Equivalent amounts of GST-fusion proteins were used in each lane, as assessed by Coomassie gels (data not shown).
The Fli-1 and GATA-1 interaction is different from the PU.1-GATA-1 interaction.
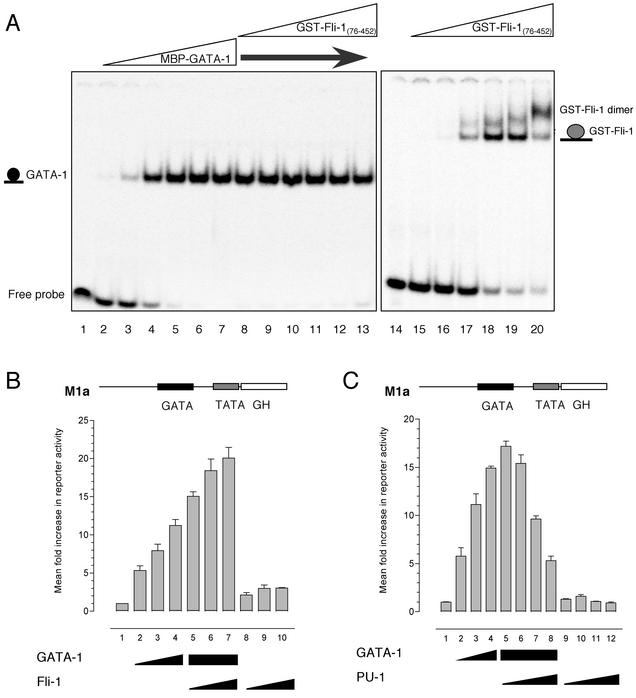
As noted above, the Ets family member PU.1 has been shown to physically interact with GATA-1. PU.1 is able to abolish GATA-dependent transactivation by binding the GATA-1 zinc finger (i.e., DNA binding) domain and preventing GATA-1 from binding to DNA (39). In order to test whether Fli-1 also acted in an antagonistic manner and interfered with DNA binding by GATA-1, we conducted EMSAs. An oligonucleotide probe containing a consensus GATA binding site was end labeled with [γ-32P]ATP and incubated in the presence of increasing amounts (0.0004 to 40 nM) of recombinant maltose-binding protein-tagged GATA-1 (MBP-GATA-1(200-318) prior to being subjected to EMSA) (Fig. 4A, lanes 2 to 7). Incubation of the probe in the presence of GATA-1 resulted in the formation of a strong nucleoprotein complex, which effectively saturated the probe at higher concentrations (Fig. 4A, lanes 5 to 7). To test whether Fli-1 could prevent GATA-1-DNA binding, increasing amounts of recombinant Fli-1(76-452) (0.008 to 800 nM) containing the homologous N-terminal region to PU.1 were added in the maintained presence of GATA-1 (4 nM). Titration of up to a 200-fold molar excess of Fli-1 had no effect upon the ability of GATA-1 to bind the GATA-1 oligonucleotide (Fig. 4A, lanes 8 to 13). This same amount of Fli-1 was sufficient to saturate an oligonucleotide from the GPIX promoter bearing only the Fli-1 binding site (Fig. 4A, lanes 15 to 20). The appearance of a higher-molecular-weight complex may be due to dimerization of the GST tag, as Fli-1 has been reported to bind DNA as a monomer (18).
FIG. 4.
Fli-1 does not antagonize GATA-1 DNA binding or transcriptional activity. (A) EMSA was performed by titrating increasing amounts of purified recombinant MBP-tagged GATA-NC with 0.2 fmol of the 32P-labeled GATA consensus oligonucleotide 5′-GATCTCCGGCAACTGATAAGGATTCCCTG-3′ (sense strand; Crossley et al. [8]) (the GATA site is shown in bold). The first lane contains probe alone. Lanes 2 to 17, increasing concentrations of GATA-1 protein (4 × 10−13 M to 4 × 10−8 M). Lanes 8 to 13, 4 × 10−9 M GATA-1 with increasing amounts of GST-Fli-1 protein (8 × 10−12 M to 8 × 10−7 M). Lane 14, GPIX-Ets probe alone (5′-ATTTTCATCACTTCCTTCCGCCCGCTCCC-3′, sense strand; Eisbacher et al. [9]). Lanes 15 to 20, increasing amounts of GST-Fli-1 protein (8 × 10−12 M to 8 × 10−7 M) in the presence of GPIX-Ets probe. (B) HeLa cells were transfected with 400 ng of M1α reporter alone (bar 1) or together with expression plasmids for GATA-1 alone (bars 2 to 4, respectively, 100, 200, and 400 ng); 400 ng of GATA-1 in combination with increasing amounts of Fli-1 expression plasmid (bars 5 to 7, respectively, 100, 200, and 400 ng); or increasing amounts of Fli-1 expression plasmid alone (bars 8 to 10, respectively, 100, 200, and 400 ng). Growth hormone levels were assayed after 48 h. Values are expressed as mean fold increase ± standard deviation relative to a value of 1 for each reporter. Results shown are means from three experiments performed in triplicate. (C) HeLa cells were transfected as described for panel B, except that Fli-1 was replaced with PU.1. Bars correspond to 400 ng of M1α reporter alone (bar 1); expression plasmids for GATA-1 alone (bars 2 to 4, respectively, 100, 200, and 400 ng); GATA-1 in combination with increasing amounts of PU.1 expression plasmid (bars 5 to 8, respectively, 100, 200, 400, and 800 ng); or increasing amounts of PU.1 expression plasmid alone (bars 9 to 12, respectively, 100, 200, 400, and 800 ng).
The inability of Fli-1 to prevent GATA-1 from binding DNA was investigated functionally by conducting transactivation assays in HeLa cells with the reporter construct M1α-GH (19), in which a GATA binding site upstream of a minimal TATA box drives expression of the growth hormone gene (Fig. 4B and C). Consistent with previously published findings (39), cotransfection of a GATA-1 expression plasmid resulted in a dose-dependent transactivation of the reporter (Fig. 4B and C, bars 2 to 4). Moreover, in agreement with our EMSA studies, cotransfection of increasing amounts of a Fli-1 expression plasmid with GATA-1 had no inhibitory effect whatsoever on GATA-dependent transactivation of the M1α promoter. In fact, Fli-1 strongly enhanced GATA-1's transactivating activity (Fig. 4B, bars 5, 6, and 7). The cotransfection of equivalent amounts of Fli-1 in the absence of GATA-1 did not result in significant activation (Fig. 4B, bars 8, 9, and 10). As a control, we repeated the experiment but replaced the Fli-1 expression plasmid with one expressing PU.1. Similar to previously published findings, coexpression of PU.1 with GATA-1 resulted in repression of GATA-dependent activation of the M1α promoter (Fig. 4C, bars 5 to 8). Thus, the interaction between Fli-1 and GATA-1 is different from that between PU.1 and GATA-1, as Fli-1 appears to physically interact with and enhance the activity of GATA-1 rather than inhibiting its activity.
The physical interaction between Fli-1 and GATA-1 results in synergistic activation of the megakaryocyte-specific promoters GPIX and GPIbα.
In order to determine the functional consequences of the physical interaction between Fli-1 and GATA-1 further, we conducted transactivation assays with luciferase reporter constructs driven by Mk-specific promoters in HeLa cells. Fli-1 has previously been demonstrated to transactivate the GPIX and GPIbα promoters in other nonhematopoietic cells (3). Consistent with these observations, addition of increasing amounts of Fli-1 expression plasmid resulted in dose-dependent transactivation of both GPIX and GPIbα-dependent reporter activity of up to fivefold (Fig. 5A and C, bars 2 and 3). Similarly, increasing amounts of GATA-1 expression plasmid were also able to modestly transactivate both constructs in the absence of Fli-1 (Fig. 5A and C, bars 7, 8, and 9). However, addition of increasing amounts of Fli-1 plasmid in the presence of a constant amount of GATA-1 resulted in a marked increase in reporter activity of both reporters by up to 30-fold (Fig. 5A and C, bars 4, 5, and 6). These data demonstrate that the transactivation observed with the Fli-1-GATA-1 combination is much greater than the expected activation of both factors acting independently of one another. That is, the effect of the Fli-1-GATA-1 interaction upon activation of the GPIbα and GPIX genes is not only additive but synergistic.
In order to rule out possible effects of Fli-1 on GATA-1 expression levels, Western blotting was performed on whole-cell extracts from transfected cells (Fig. 5B and D). Immunoblotting with anti-GATA-1 and anti-Fli-1 monoclonal antibodies demonstrated no discernible change in levels of GATA-1 expression in the presence of Fli-1 (and vice versa).
A Fli-1 activation domain in conjunction with the Fli-1 Ets domain is required for functional synergy between Fli-1 and GATA-1.
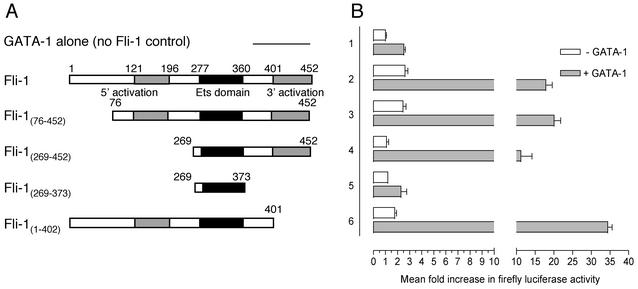
Fli-1 has two domains with helix-loop-helix and turn-loop-turn structures (the 5′ and 3′ activation domains, respectively) that have previously been implicated in transcriptional activation (26). Having observed the transcriptional synergy between Fli-1 and GATA-1 in activation of the GPIX and GPIbα reporters, we next turned to mapping the Fli-1 domains required for functional activity with GATA-1. Fli-1 expression plasmids, encoding deletions of Fli-1 cDNA (shown schematically in Fig. 6A), were cotransfected with backbone or GATA-1 expression plasmid and the GPIbα-253 luciferase reporter in HeLa cells.
FIG. 6.
A Fli-1 activation domain in conjunction with the Fli-1 Ets domain is required to retain transcriptional synergy with GATA-1. (A) Schematic representations of Fli-1 constructs used alone or in combination with GATA-1 to activate the GPIbα-253 luciferase reporter. (B) HeLa cells were cotransfected with 800 ng of GPIbα-253 luciferase reporter and either 200 ng of pcDNA3 backbone (lane 1) or 200 ng of the indicated Fli-1 constructs (lanes 2 to 6), either without (open bars) or with (filled bars) 50 ng of GATA-1 expression plasmid. Total DNA was kept constant in each sample by addition of pcDNA3 backbone. Luciferase activity was measured as described for Fig. 5. Results represent mean increases of reporter activity ± standard deviations relative to a value of 1 for GPIbα-253 reporter activity alone (lane 1, open bar). Results shown are the means from three experiments performed in triplicate. Each experiment was performed at least three times with similar results.
Consistent with the previous experiments, Fli-1 and GATA-1 synergistically activated the reporter to levels much higher than those anticipated if both factors were acting independently (Fig. 6, lanes 1 and 2). Deletion of the region encoding the first 75 amino acids of Fli-1 had no effect upon Fli-1's ability to activate the reporter in the absence of GATA-1 or its ability to synergize with GATA-1. In contrast, cotransfection of the Fli-1(269-452) construct, which lacks the 5′ activation domain of Fli-1 (amino acids 121 to 196), essentially abolished transactivation of the GPIbα-253 reporter in the absence of GATA-1 (Fig. 6, compare the open bars in lane 4 with those in lanes 2 and 3). Surprisingly, despite not being able to activate the GPIbα-253 reporter independently, the Fli-1(269-452) construct retained the capacity to synergize with GATA-1 functionally, albeit to a reduced extent (Fig. 6, compare the filled in bars in lane 4 with those in lanes 2 and 3). Deletion of the region encoding the 3′ activation domain of Fli-1 (amino acids 401 to 452) had no effect upon the ability of Fli-1 to activate the GPIbα-253 reporter in isolation (Fig. 6, lane 6, open bars). Interestingly, deletion of this region consistently potentiated functional synergy with GATA-1. In contrast, expression of the Fli-1 Ets domain in isolation [construct Fli-1(269-373)] resulted in complete loss of both independent activation by Fli-1 and functional synergy in combination with GATA-1 (Fig. 6, lane 5).
These results suggest that the presence of either the 5′ activation domain or the 3′ activation domain in combination with the Ets domain of Fli-1 is sufficient to impart functional synergy with GATA-1. Moreover, Fli-1 is able to retain its capacity to synergize with GATA-1 despite loss of the 5′ activation domain that is essential for Fli-1-dependent activation of GPIbα-253 in the absence of GATA-1.
Overexpression of Fli-1 in K562 cells results in expression of GPIbα and GPIX mRNA and protein.
Previous investigators have demonstrated that infection of K562 cells with a retroviral construct expressing Fli-1 resulted in a megakaryocytic phenotype with expression of early markers such as GPIIb (1). In order to determine whether Fli-1 could result in the expression of late-appearing Mk markers such as GPIbα and GPIX, we used a similar system. Above, we have shown that Fli-1 and GATA-1 could synergistically activate the GPIbα and GPIX promoters in transient transfection assays. We now wished to investigate whether the presence of GATA-1 and Fli-1 could induce expression of the endogenous genes. K562 cells, which are known to express abundant GATA-1, were transfected with a Fli-1 expression plasmid, pIRES2-EGFP/Fli-1 (or the backbone control), allowing both Fli-1 and GFP to be translated from the same bicistronic mRNA. Stably transfected cell clones were selected in G418 and analyzed for high levels of GFP expression by FACS (>102 log scale). Cells overexpressing Fli-1 acquired an adherent phenotype similar to that described previously (1), whereas control cells remained largely in suspension with no visible changes (data not shown). Both cell lines chosen for further study exhibited and retained similarly high levels of GFP expression (see below).
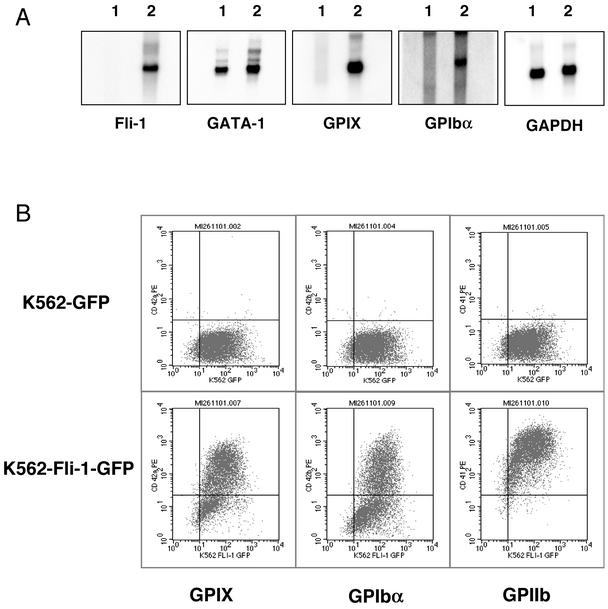
Fli-1 expression was confirmed by Northern blotting, which demonstrated the presence of Fli-1 mRNA only in the cells transfected with pIRES2-EGFP/Fli-1 (Fig. 7A, panel 1, lane 2). In order to determine whether Fli-1 overexpression resulted in any modulation of GATA-1 levels, the membrane was stripped and probed for GATA-1 mRNA. In contrast to previous reports (2), in our lines, overexpression of Fli-1 did not reduce GATA-1 expression levels (Fig. 7A, panel 2). To ascertain whether Fli-1 expression resulted in the expression of GPIX and GPIbα, the blot was then stripped and reprobed for both GPIX and GPIbα mRNAs. Those cells exhibiting Fli-1 expression also demonstrated high-level expression of both GPIbα and GPIX mRNAs (Fig. 7A, panels 3 and 4, lane 2). In contrast, control cells failed to express either of these mRNAs (Fig. 7A, panels 3 and 4, lane 1). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was similar in both lines (Fig. 7A, panel 5).
FIG. 7.
Fli-1 overexpression in K562 cells in the maintained presence of endogenous GATA-1. (A) Northern blot analysis of K562 cell lines stably transfected with control plasmid pIRES2-EGFP (lane 1) or Fli-1 expression plasmid pIRES2-Fli-1-EGFP (lane 2). Each lane contains 3 μg of poly(A+) RNA. The membrane was hybridized with the indicated 32P-labeled cDNA probes in the order shown. Membranes were stripped prior to each hybridization in the presence of boiling 0.1% SDS. (B) Flow cytometric analysis of K562-GFP and K562-Fli-1-GFP cell lines for the surface expression of markers associated with terminal differentiation of megakaryocytes. The indicated cell lines were incubated in the presence of anti-GPIX, anti-GPIbα, or anti-GPIIb primary antibodies prior to being labeled with phycoerythrin-conjugated secondary antibody, as shown.
We next investigated whether Fli-1 overexpression resulted in expression of GPIb-IX protein on the cell surface by using flow cytometry. Both control and Fli-1-expressing populations were analyzed with the monoclonal antibodies specific for the GPIX and GPIbα subunits with a phycoerythrin-conjugated goat anti-mouse secondary antibody. All cells expressed GFP, as demonstrated by a mean relative fluorescence (rFL) of 102 units along the x axis (Fig. 7B). In comparison, nontransfected cells exhibited a mean rFL of less than 101 units (data not shown). Those cells stably transfected with Fli-1 expression plasmid demonstrated markedly increased levels of GPIbα and GPIX on the cell surface, as reflected by a shift in the cell population from the lower right to the top right quadrant (Fig. 7B). In contrast, the control cells transfected with the pIRES2-EGFP backbone failed to express either GPIX or GPIbα, remaining in the lower right quadrant (Fig. 7B). As described previously, Fli-1-overexpressing cells also demonstrated markedly increased levels of the earlier marker, GPIIb (Fig. 7B). These results demonstrate that Fli-1 overexpression in K562 cells in the maintained presence of GATA-1 also results in high-level surface expression of late megakaryocytic markers such as GPIbα and GPIX.
Fli-1 and GATA-1 form a ternary complex and exhibit cooperative DNA binding on the GPIX promoter.
One mechanism used by transcription factors to mediate synergistic gene expression is through cooperative DNA binding (7). Considering the observed synergy in transcriptional activation of Mk-specific promoters by Fli-1 and GATA-1, we performed quantitative EMSA analysis in order to determine whether Fli-1 and GATA-1 exhibit cooperative DNA binding on the GPIX promoter.
Oligonucleotides corresponding to the region containing the GATA and Ets binding sites were end labeled with 32P, and 0.2 fmol of labeled probe was incubated with increasing concentrations of purified GATA-1. The appearance of the GATA-DNA complex (labeled G.D in Fig. 8A) and disappearance of the free DNA indicate 0 to >90% binding. Consistent with previous reports of GATA-1 self-association, a GATA-DNA dimer (labeled DD in Fig. 8A) was also observed at higher GATA-1 concentrations (8). Similar results were observed when increasing concentrations of Fli-1 were incubated with the probe (Fig. 8B); however, Fli-1 consistently bound to the GPIX-GATA-Ets oligonucleotide at lower concentrations than GATA-1, suggesting a higher affinity for the probe (see below). In order to test for cooperativity between GATA-1 and Fli-1, GATA-DNA binding was concurrently measured in the presence of a fixed, saturating amount of Fli-1 (1.25 × 10−9 M) (Fig. 8C). This concentration of Fli-1 ensured greater than 90% DNA occupancy (labeled F.D in Fig. 8C). Titration of increasing amounts of GATA-1 identical to those used in Fig. 8A resulted in the additional formation of a GATA-1-Fli-1-DNA ternary complex (labeled G.F.D in Fig. 8C) as well as a GATA-1-DNA binary complex of weaker intensity (labeled G.D in Fig. 8C). Moreover, this ternary complex consistently formed at a lower GATA concentration than that of the GATA-DNA binary complex (labeled G.D in Fig. 8A.). This result is evidence that GATA-1 loading has been facilitated by the binding of Fli-1; i.e., cooperative binding is occurring. Quantification of the cooperativity is described below.
Calculation of KD values for the binary GATA-DNA and Fli-1-DNA and ternary GATA-1-Fli-1-DNA complexes.
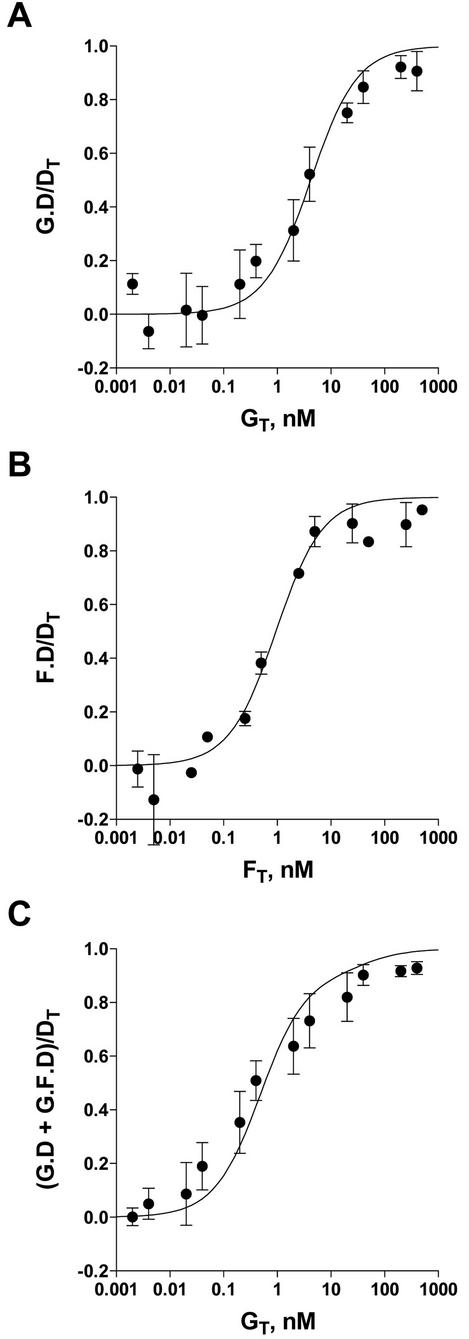
Quantitation of the fraction of free DNA in lanes containing protein relative to a control lane containing no protein from Fig. 8, as described in the Materials and Methods, allowed the calculation of binding isotherms for the GATA-1-DNA and Fli-1-DNA binary complexes shown in Fig. 9A and B.8, Fitting of this data by least-squares regression to a single rectangular hyperbola gave dissociation constants of 4.2 ± 0.7 nM (1 standard deviation) for KGD and 0.94 ± 0.18 nM for KFD. The isotherm for binding of GATA-1 to DNA in the presence of a fixed concentration of Fli-1 is shown in Fig. 9C. Fitting of this data to the scheme in Fig. 1 and the equilibrium binding model equation by numerical integration with KGD and KFD fixed at 4.2 and 0.94 nM, respectively, gave dissociation constants of 2.3 ± 3.9 nM for KGF and 0.16 ± 0.09 nM for KFD,G.
FIG.9.
Binding of GATA-1 (indicated as G) or Fli-1 (indicated as F) to DNA (indicated as D) and formation of the ternary GATA-1-Fli-1-DNA complex. Shown are the isotherms for binding of GATA-1 (A) or Fli-1 (B) to DNA and for binding of G to D in the presence of a fixed concentration of F (C). The solid lines in panels A and B represent the best fit of the data to a single rectangular hyperbola with dissociation constants of 4.2 (A) and 0.94 (B) nM. In panel C, the solid line represents the best fit of the data to the equilibrium binding model equation with dissociation constants of 2.3 and 0.16 nM for KGF and KFD,G, respectively. KGD and KFD were fixed at 4.2 and 0.94 nM, respectively.
Because of the linked equilibria shown in Fig. 10, the equality KGD · KGD,F = KGF · KGF,D = KFD · KFD,G applies.
Values of 0.04 and 0.08 nM for KGD,F and KGF,D ternary dissociation constants are therefore calculated. In summary, the dissociation constant for GATA-1 binding to DNA is increased from 4.2 nM in the absence of Fli-1 to 0.16 nM in its presence, demonstrating significant cooperative binding.
DISCUSSION
In this paper, we report and characterize in detail a protein-protein interaction between Fli-1 and GATA-1, two transcriptional regulators implicated in the differentiation of megakaryocytes. We have mapped the interacting domains and demonstrated that GATA-1 binds the Fli-1 Ets domain through its zinc fingers. Additionally, amino acids in the finger tails, such as amino acids 243 to 248 in the N finger of GATA-1, appear to be crucial for binding to Fli-1. We also investigated the functional consequences of the interaction between these two transcription factors in EMSAs and transient transfection assays and show that it is markedly different from the previously characterized PU.1-GATA-1 interaction. Our findings revealed the synergistic nature of the Fli-1-GATA-1 interaction, which results in the marked up-regulation of the Mk-specific promoters GPIbα and GPIX. Moreover, we showed in a cell line model that overexpression of Fli-1, in the maintained presence of endogenously expressed GATA-1, results in the transcriptional activation of Mk-specific genes, such as GPIbα and GPIX, which are associated with the terminal differentiation of megakaryocytes. Quantitative gel shift analysis with sequences derived from the GPIX promoter reveals that Fli-1 and GATA-1 exhibit significant DNA-binding cooperativity. The affinity of GATA-1 for the GPIX-GATA-Ets probe is increased approximately 26-fold in the presence of Fli-1 (from 4.2 to 0.16 nM). These quantitative data provide a mechanistic explanation for the observed transcriptional synergy between Fli-1 and GATA-1.
The GATA family of transcription factors bind the DNA motif 5′-T/AGATAA/G-3′ through conserved zinc fingers (for a review, see Weiss and Orkin [37]). The founding member, GATA-1, is abundantly expressed in the erythroid, mast cell, and megakaryocytic lineages (20). Early work demonstrated its importance in the regulation of globin genes and the terminal differentiation of erythroid cells (25). However, studies involving selective knockout of GATA-1 expression in the megakaryocytes of mice (29) and others involving overexpression of GATA-1 in cell line models (34) also revealed a key role for GATA-1 in the differentiation of megakaryocytes. GATA-1 contains two zinc finger domains, with the C-terminal finger mediating binding to DNA and the N-finger stabilizing binding (19). The zinc finger regions also mediate protein-protein interactions (21). Most notably, the N finger of GATA-1 was used to isolate a cofactor, Friend of GATA-1 (FOG), which subsequently has also been demonstrated to be essential for normal megakaryocytic differentiation (32, 33).
The present study demonstrates that the functional effects of the protein-protein interaction between GATA-1 and Fli-1 are markedly different from those of the previously described interaction with the Ets-related protein PU.1 (22, 27). The GATA-1-PU.1 interaction is an antagonistic one, with PU.1 inhibiting GATA-dependent activation and vice versa (38, 39). PU.1 prevents GATA-dependent activation by blocking GATA-1 from binding to DNA (39). In contrast, our EMSA analysis demonstrated that Fli-1 had no inhibitory effect upon GATA-1's ability to bind DNA. Moreover, further experiments, using an artificial GATA-dependent promoter (M1α) similar to that described previously, demonstrated that unlike PU.1, Fli-1 does not inhibit GATA-1-dependent transactivation. In fact, coexpression of Fli-1 consistently potentiated GATA-1's transactivating activity. Our findings that Fli-1 alone does not activate the M1α promoter despite containing an adjacent inverted Ets binding site (5′-AGCTTCCTCG-3′) are in agreement of those of Wang et al. (35), who showed by EMSA that Fli-1 was unable to bind this site. The potentiation of GATA-1 activation by Fli-1 in our assay may be explained by the effects of the Fli-1-GATA-1 protein-protein interaction, which might be expected to stabilize Fli-1 binding to the Ets site, despite differences in the nucleotides flanking the core.
GATA-1 interacts with two separate domains on PU.1, the N-terminal 70 amino acids and the β3-β4 region of the Ets domain. It is the N-terminal region of PU.1 that is responsible for mediating the inhibitory effect upon GATA-1 DNA binding (39). We found that the protein-protein interaction between GATA-1 and Fli-1 requires only the Fli-1 Ets domain. PU.1 is the most divergent of all Ets family members (38). Moreover, the homology between PU.1 and Fli-1 outside the Ets domain is very low. The divergence between the N-terminal regions of PU.1 and Fli-1 may explain the different functions of these two Ets family members with respect to their effect upon GATA-1 DNA binding and GATA-1-dependent activation. It is also interesting that two members from the same family of transcription factors can have such opposite effects on the same protein partner.
Further dissection of the subdomains within the Ets domain of Fli-1 required for the interaction with GATA-1 failed to identify specifically which region was involved. This was possibly due to failure of the individual subdomains to fold properly. This finding suggests that the entire Ets domain must be expressed to remain functional in terms of its interaction with GATA-1. This is supported in part by other experiments in which mutation of the β3 and/or β4 regions of the Fli-1 Ets domain prevents not only the interaction with GATA-1 but also Fli-1's ability to bind DNA (data not shown). As such, it may prove difficult to separate the DNA-binding activity of Fli-1 from its ability to interact with GATA-1 by creating mutations in the Ets domain.
Our in vitro experiments demonstrated that both zinc fingers of GATA-1 were able to bind Fli-1. The residues in the tail of the N finger appear to be crucial for its interaction with Fli-1. These residues differ from those required for the interaction with GATA-1's cofactor FOG, which is mediated through the residues within the core of the N finger itself and not the tail (11). Previous studies (12) have suggested that FOG might tether to a number of Ets factors (including Fli-1, Ets-1, and PU.1) to mediate the expression of the GPIIb gene. Thus, it may be possible that Fli-1, GATA-1, and FOG are able to form a trimolecular complex which mediates the high-level expression of Mk-specific genes.
Indeed, recent findings of Wang et al. (35) add further support to the GATA-Fli-1-FOG model and underscore the biological relevance of the Fli-1-GATA-1 interaction described here. Wang et al. (35) demonstrated that the type of Ets binding site found adjacent to a GATA-1 binding site plays an important role in determining whether FOG stimulates or inhibits GATA-1 activity. For example, mutation of sequences flanking the Ets 5′-GGAA-3′ core to enable Fli-1 binding converts FOG from being an inhibitor of GATA-1 function to a coactivator (35). This conversion of FOG activity is specific to Fli-1, as other experiments in the same study using engineered GATA-GAL4 reporter and GAL4DBD-Ets fusion proteins demonstrated that GATA-FOG activation was observed only in the presence of a GAL4-Fli-1 fusion and not with GAL4-PU.1. The residues of Fli-1 required for the effect upon FOG in that study differ from those implicated in the Fli-1-GATA-1 interaction here. Moreover, the Fli-1-FOG-GATA-1 combination resulted in the highest levels of transactivation of several Mk-reporter plasmids. These findings suggest that the Fli-1-GATA-1 interaction does not preclude FOG from binding GATA-1. As pointed out by Wang et al., the mechanism through which Fli-1 mediates a change in FOG function is unclear (35). Our findings that Fli-1 and GATA-1 interact and bind DNA in a cooperative fashion complement those of Wang et al. (35) and provide further clues as to the mechanism of Fli-1-GATA-1-FOG complex formation.
One of the hallmark features of Mk-specific promoters is the presence of GATA and Ets binding sites, often in close proximity to one another. Our findings that Fli-1 and GATA-1 synergize to activate the Mk-specific promoters may provide an explanation for the close association of GATA and Ets elements in Mk promoters. The observed synergy between Fli-1 and GATA-1 in our GPIX and GPIb reporter assays is likely to result, at least in part, from the direct protein-protein interaction between GATA-1 and Fli-1, as Western blot analysis ruled out potential effects of Fli-1 on GATA-1 expression levels and vice versa.
One mechanism through which synergistic gene expression can be achieved is via cooperative DNA binding. We have examined the consequences of the combined interactions of GATA-1 and Fli-1 with DNA on the distribution of GATA-1 in equilibrium mixtures of GATA-1, Fli-1, and DNA by performing quantitative gel shift assays. The findings are summarized in Fig. 10 and are concordant with the behavior for a binding model in which a ternary GATA-1-Fli-1-DNA complex is formed. The assembly of the ternary complex occurs randomly through the interactions of all three possible intermediate binary complexes. Upon comparison of the constants for formation of the three binary complexes, the higher affinity of the Fli-1-DNA interaction (0.94 nM) compared to the affinities of the other two binary complexes of GATA-1-Fli-1 and GATA-1-DNA (at 4.2 and 2.3 nM, respectively), indicates that the assembly of the ternary complex will occur predominantly through the interaction of an intermediate Fli-1-DNA binary complex with GATA-1 (Fig. 10). The analysis also indicates that GATA-1 binds the Fli-1-DNA binary complex with ∼26-fold higher affinity than the interaction of GATA-1 with DNA alone (0.16 compared with 4.2 nM). Thus, Fli-1 and GATA-1 exhibit substantial DNA-binding cooperativity. These data provide a mechanistic explanation for the observed transcriptional synergy between Fli-1 and GATA-1. Moreover, it is consistent with a model in which DNA-bound Fli-1 acts to facilitate the recruitment and binding of GATA-1 to the GPIX promoter.
Taking into account our observations of the protein-protein interaction between Fli-1 and GATA-1, the resulting synergistic effects upon the GPIX and GPIbα promoters, the induced expression of endogenous Mk-specific genes in cells engineered to express Fli-1 in the presence of endogenous GATA-1, and quantitative in vitro studies suggesting facilitated recruitment of GATA-1 to the GPIX promoter by Fli-1, the present study proposes the Fli-1-GATA-1 combination as an important determinant for the high-level expression of Mk-specific genes.
Acknowledgments
We thank Leonie Gaudry for assistance in conducting the flow cytometry experiments.
This work is supported by a program grant from the National Health and Medical Research Council of Australia and an infrastructure grant from the NSW Department of Health. M.E. is supported by an Australian Postgraduate Award. L.M.K. is a Principal Research Fellow of the NHMRC.
REFERENCES
- 1.Athanasiou, M., P. A. Clausen, G. J. Mavrothalassitis, X. K. Zhang, D. K. Watson, and D. G. Blair. 1996. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 7:1525-1534. [PubMed] [Google Scholar]
- 2.Athanasiou, M., G. Mavrothalassitis, L. Sun-Hoffman, and D. G. Blair. 2000. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14:439-445. [DOI] [PubMed] [Google Scholar]
- 3.Bastian, L. S., B. A. Kwiatkowski, J. Breininger, S. Danner, and G. Roth. 1999. Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood 93:2637-2644. [PubMed] [Google Scholar]
- 4.Behre, G., L. T. Smith, and D. G. Tenen. 1999. Use of a promoterless Renilla luciferase vector as an internal control plasmid for transient co-transfection assays of Ras-mediated transcription activation. BioTechniques 26:24-26, 28. [DOI] [PubMed] [Google Scholar]
- 5.Ben-David, Y., E. B. Giddens, and A. Bernstein. 1990. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc. Natl. Acad. Sci. USA 87:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, J. K., J. A. Lopez, M. C. Berndt, I. Dawes, C. N. Chesterman, and B. H. Chong. 1998. Quinine-dependent antibodies bind a restricted set of epitopes on the glycoprotein Ib-IX complex: characterization of the epitopes. Blood 92:2366-2373. [PubMed] [Google Scholar]
- 7.Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. [DOI] [PubMed] [Google Scholar]
- 8.Crossley, M., M. Merika, and S. H. Orkin. 1995. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol. Cell. Biol. 15:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisbacher, M., L. M. Khachigian, T. H. Khin, M. L. Holmes, and B. H. Chong. 2001. Inducible expression of the megakaryocyte-specific gene glycoprotein IX is mediated through an Ets binding site and involves upstream activation of extracellular signal-regulated kinase. Cell Growth Differ. 12:435-445. [PubMed] [Google Scholar]
- 10.Farr, A., and A. Roman. 1992. A pitfall of using a second plasmid to determine transfection efficiency. Nucleic Acids Res. 20:920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, A. H., K. Kowalski, G. F. King, J. P. Mackay, and M. Crossley. 1998. Key residues characteristic of GATA N-fingers are recognized by FOG. J. Biol. Chem. 273:33595-33603. [DOI] [PubMed] [Google Scholar]
- 12.Gaines, P., J. N. Geiger, G. Knudsen, D. Seshasayee, and D. M. Wojchowski. 2000. GATA-1- and FOG-dependent activation of megakaryocytic alpha IIB gene expression. J. Biol. Chem. 275:34114-34121. [DOI] [PubMed] [Google Scholar]
- 13.Goetz, T. L., T. L. Gu, N. A. Speck, and B. J. Graves. 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol. Cell. Biol. 20:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart, A., F. Melet, P. Grossfeld, K. Chien, C. Jones, A. Tunnacliffe, R. Favier, and A. Bernstein. 2000. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13:167-177. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, M., J. Turner, A. Fox, O. Chisholm, M. Crossley, and B. Chong. 1999. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem. 274:23491-23498. [DOI] [PubMed] [Google Scholar]
- 16.Kawada, H., T. Ito, P. N. Pharr, D. D. Spyropoulos, D. K. Watson, and M. Ogawa. 2001. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int. J. Hematol. 73:463-468. [DOI] [PubMed] [Google Scholar]
- 17.Lemarchandel, V., J. Ghysdael, V. Mignotte, C. Rahuel, and P. H. Romeo. 1993. GATA and Ets cis-acting sequences mediate megakaryocyte-specific expression. Mol. Cell. Biol. 13:668-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, H., X. Mao, E. T. Olejniczak, D. G. Nettesheim, L. Yu, R. P. Meadows, C. B. Thompson, and S. W. Fesik. 1994. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat. Struct. Biol. 1:871-875. [DOI] [PubMed] [Google Scholar]
- 19.Martin, D. I., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 20.Martin, D. I., L. I. Zon, G. Mutter, and S. H. Orkin. 1990. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature 344:444-447. [DOI] [PubMed] [Google Scholar]
- 21.Merika, M., and S. H. Orkin. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 23.Newton, A., J. Mackay, and M. Crossley. 2001. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem. 276:35794-35801. [DOI] [PubMed] [Google Scholar]
- 24.Orkin, S. H. 1995. Transcription factors and haematopoietic development. J. Biol. Chem. 270:4955-4958. [DOI] [PubMed] [Google Scholar]
- 25.Pevny, L., M. C. Simon, E. Robertson, W. H. Klein, S. F. Tsai, V. D'Agati, S. H. Orkin, and F. Costantini. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349:257-260. [DOI] [PubMed] [Google Scholar]
- 26.Rao, V. N., T. Ohno, D. D. Prasad, G. Bhattacharya, and E. S. Reddy. 1993. Analysis of the DNA-binding and transcriptional activation functions of human Fli-1 protein. Oncogene 8:2167-2173. [PubMed] [Google Scholar]
- 27.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shivdasani, R. A. 2001. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells 19:397-407. [DOI] [PubMed] [Google Scholar]
- 29.Shivdasani, R. A., Y. Fujiwara, M. A. McDevitt, and S. H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16:3965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shivdasani, R. A., and S. H. Orkin. 1996. The transcriptional control of haematopoiesis. Blood 87:4025-4039. [PubMed] [Google Scholar]
- 31.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang, A. P., Y. Fujiwara, D. B. Hom, and S. H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 34.Visvader, J., and J. M. Adams. 1993. Megakaryocytic differentiation induced in 416B myeloid cells by GATA-2 and GATA-3 transgenes or 5-azacytidine is tightly coupled to GATA-1 expression. Blood 82:1493-1501. [PubMed] [Google Scholar]
- 35.Wang, X., J. D. Crispino, D. L. Letting, M. Nakazawa, M. Poncz, and G. A. Blobel. 2002. Control of megakaryocyte-specific gene expression by GATA-1 and FOG-1: role of Ets transcription factors. EMBO J. 21:5225-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wasylyk, B., and A. Nordheim. 1997. Ets transcription factors: partners in the integration of signal responses, p. 253-275. In A. Papavassiliou (ed.), Transcription factors in eukaryotes, 1st ed. Landes Bioscience, Georgetown, Tex.
- 37.Weiss, M. J., and S. H. Orkin. 1995. GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23:99-107. [PubMed] [Google Scholar]
- 38.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]