Abstract
Thioglucoside glucohydrolase 1 (TGG1) is one of two known functional myrosinase enzymes in Arabidopsis. The enzyme catalyzes the hydrolysis of glucosinolates into compounds that are toxic to various microbes and herbivores. Transgenic Arabidopsis plants carrying β-glucuronidase and green fluorescent protein reporter genes fused to 0.5 or 2.5 kb of the TGG1 promoter region were used to study spatial promoter activity. Promoter activity was found to be highly specific and restricted to guard cells and distinct cells of the phloem. No promoter activity was detected in the root or seed. All guard cells show promoter activity. Positive phloem cells are distributed in a discontinuous pattern and occur more frequent in young tissues. Immunocytochemical localization of myrosinase in transverse and longitudinal sections of embedded material show that the TGG1 promoter activity reflects the position of the myrosinase enzyme. In the flower stalk, the myrosinase-containing phloem cells are located between phloem sieve elements and glucosinolate-rich S cells. Our results suggest a cellular separation of myrosinase enzyme and glucosinolate substrate, and that myrosinase is contained in distinct cells. We discuss the potential advantages of locating defense and communication systems to only a few specific cell types.
Arabidopsis ecotype Columbia has been shown to contain 23 different glucosinolates (Hogge et al., 1988). Myrosinase (EC 3.2.3.1), also known as β-thioglucoside glucohydrolase, catalyzes the hydrolysis of glucosinolates into Glc and an unstable intermediate that undergoes nonenzymatic rearrangement to form sulfate and isothiocyanates, thiocyanates, nitriles, epithioalkanes, or elementary sulfur dependent on the concentration of H+, metal ions, epithiospecifier protein, and/or other cofactors (Bones and Rossiter, 1996; Foo et al., 2000). The complexity of the myrosinase-glucosinolate system suggests a diverse and multifunctional role in the cruciferous plants. Glucosinolates are a diverse group of sulfur-containing glycosides that may serve as a sink for nitrogen and sulfur, and the hydrolysis products may have important roles in the defense of the plant against microorganisms and insects (Bones and Rossiter, 1996; Rask et al., 2000). Previous studies have shown that myrosinase is located in idioblasts named myrosin cells, which have been found in several species of Brassicacea, including oilseed rape (Brassica napus), white mustard (Sinapis alba), cauliflower (B. oleracea), and Chinese cabbage (B. campestris; Bones and Iversen, 1985; Thangstad et al., 1990, 1991; Bones et al., 1991; Höglund et al., 1991, 1992; Geshi et al., 1998). Myrosin cells occur as scattered cells in radicles, stems, leaves, petioles, seeds, and seedlings (Bones and Rossiter, 1996; Rask et al., 2000). The myrosinase enzyme has been localized to myrosin cells by immunocytochemical methods (Bones et al., 1991; Thangstad et al., 1990, 1991; Höglund et al., 1991; Geshi et al., 1998), and myrosinase transcripts have been detected by in situ hybridization (Lenmann et al., 1993; Falk et al., 1995).
The cellular localization of glucosinolates has been subjected to considerable debate, and the involvement of these compounds in growth, development, and defense could be more clearly defined following their cellular and subcellular localization (Kelly et al., 1998). Thangstad et al. (2001) used microautoradiography to study the in vivo localization of a titrated labeled desulfoglucosinolate precursor after pulse-chase feeding of oilseed rape plants. A cell-specific localization was found in radicles and cotyledons of the maturing embryo resembling the pattern of the myrosin cells known to contain myrosinase (Bones et al., 1991). Based on energy-dispersive x-rays and cell sap analysis, Koroleva et al. (2000) reported that specific cells next to the phloem bundles of the Arabidopsis flower stalk (S cells) were rich in glucosinolates—substrates of myrosinase.
The existence of multiple forms of myrosinase genes have been shown in various Brassicacea species (Xue et al., 1992; Thangstad et al., 1993; Falk et al., 1995). In Arabidopsis, two myrosinase functional myrosinase-encoding genes, thioglucoside glucohydrolase 1 (TGG1) and TGG2, have been reported (Chadchawan et al., 1993; Xue et al., 1995).
In this study, we used β-glucuronidase (GUS) and green fluorescent protein (GFP) reporter genes, and immunocytochemical localization to examine the expression pattern of the TGG1 myrosinase in Arabidopsis. Our results show that the TGG1 promoter directs a high level of reporter gene expression in guard cells and phloem cells, and that both cell types are myrosin cells. It is interesting that myrosin phloem cells in flower stalk are located as the nearest neighbors to S cells reported to be rich in glucosinolates (Koroleva et al., 2000). This supports a cellular separation of the enzyme and the substrate in Arabidopsis flower stalk.
RESULTS
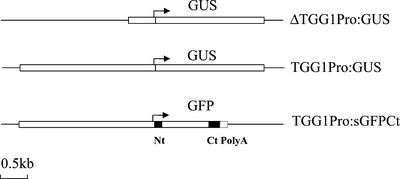
To investigate the spatial expression pattern of the TGG1 myrosinase gene, transgenic plants carrying constructs containing 0.5 or 2.5 kb of the TGG1 gene promoter region fused to the GUS gene were made (constructs were named ΔTGG1Pro:GUS and TGG1Pro:GUS, respectively). In addition, plants carrying a PBI construct containing the full TGG1 2.5-kb promoter fused to the GFP was made and named TGG1Pro:sGFPCt. The TGG1Pro:sGFPCt construct encodes a GFP:TGG1 fusion protein that contains amino acids from the N-terminal (38 amino acids) and C-terminal (58 amino acids) part of the TGG1 myrosinase. In addition, the construct contains 3′-untranslated region signals to allow correct cellular trafficking and three poly adenylation sites (Fig. 1). Promoter activity was detected as histochemical GUS staining in material pretreated in acetone for better GUS substrate penetration and as GFP fluorescence in living plants. Myrosinase-containing cells were immunocytochemical localized using the polyclonal K089 and monoclonal 3D7 myrosinase antibodies (Thangstad et al., 1990; Höglund et al., 1992).
Figure 1.
Schematic presentation of the TGG1 promoter GUS and GFP fusion gene constructs. TGG1 promoter sequence (0.5 or 2.5 kb). Black boxes, Partial TGG1 protein encoding sequences. Broken box, TGG1 poly(A) signal. Nt, N-terminal; Ct, C-terminal; PolyA, poly(A) signal. Scale bar = 0.5 kb.
Spatial Promoter Activity
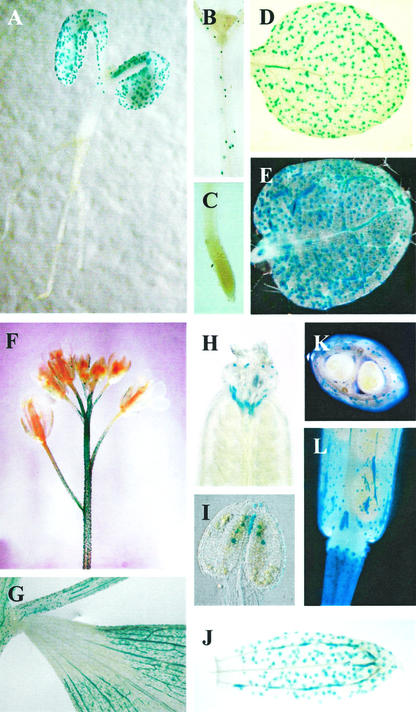
Six independent transgenic lines carrying TGG1Pro:GUS construct and 10 independent transgenic lines plants carrying the ΔTGG1Pro:GUS construct with the partial 0.5-kb TGG1 promoter were investigated. The plants containing the full promoter were studied for three subsequent generations. No GUS expression was observed in plants containing the partial promoter. The distribution of cells showing TGG1 promoter-directed GUS expression at various stages and tissues is presented in Figure 2. GUS-positive guard cells are present in the hypocotyl, cotyledon, and rosette, and positive phloem cells are present in the cotyledon and rosette leaves in seedlings (Fig. 2, A, B, D, and E). No GUS-expressing cells are detected in root tissue (Fig. 2C). An identical cellular expression pattern is observed in adult tissues examined. GUS-expressing guard cells and phloem cells are present in the stem, cauline leaf, pedicle, gynoecium, pollen sack, sepal, and silique, respectively (Fig. 2, F–K). The developing seeds are GUS negative, and GUS expression appears only in the phloem cells of the silique (Fig. 2, K and L). The lack of GUS expression in plants carrying the partial TGG1 promoter verifies that it is the TGG1 promoter that directs GUS expression in guard and phloem cells.
Figure 2.
Spatial distribution of cells showing TGG1 promoter-directed GUS expression in various stages and tissues of Arabidopsis. A through E, Seedling 5 to 12 d after germination. F through K, Plants 1 to 2 weeks after bolting. A, Five day seedling; B, hypocotyl; C, root tip; D, cotyledon; E, rosette leaf; F, inflorescence; G, cauline leaf; H, gynoecium; I, pollen sack; J, sepal; K, transverse section of silique with seeds; L, silique.
M Cells in Phloem
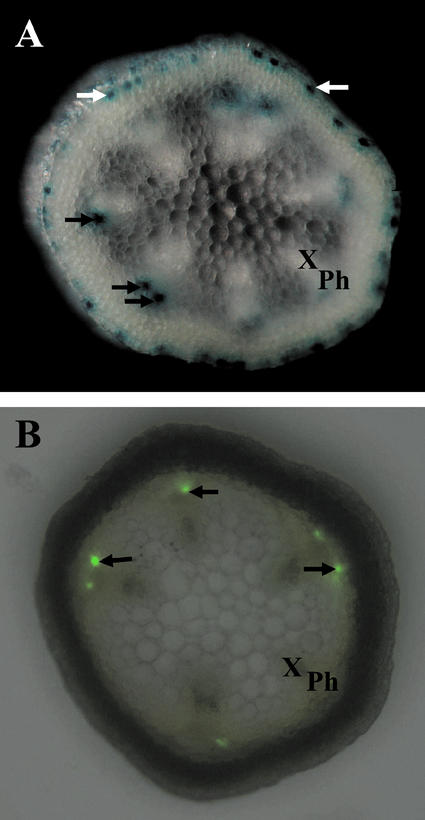
The recent report from Koroleva et al. (2000) that glucosinolate-rich S cells are located close to the phloem bundle in the upper flower stalk led us to focus on GUS-positive phloem cells in this region. As a first step, hand-cut transverse sections were made (approximately 0.5 mm thick) and the location of GUS- and GFP-expressing cells relative in phloem was examined. In the sampled region, the phloem bundles appear clearly visible and vary between five and eight in number (Fig. 3). At the site of the phloem bundle, GUS expression appears in phloem cells facing the endodermis, and the pattern in which they appear is discontinuous. No GUS expression is observed in the phloem sieve element/companion cell complexes. Figure 3A shows distinct GUS-expressing phloem cells in association with individual phloem bundles. An identical location of GFP-expressing cells in phloem is observed in plants containing the TGG1Pro:sGFPCt construct (Fig. 3B). The apparent difference in intensity of staining or level of fluorescence observed between the individual phloem cells is due to the use of intact plant tissue where some cells are out of the focal plane.
Figure 3.
Transverse section of the upper main flower stalk showing TGG1 promoter-directed GUS and GFP expression. A, Hand-cut section of a GUS plant. B, Hand-cut section of a GFP plant combined with the respective lightfield image. White arrows, Myrosin guard cells; black arrows, myrosin phloem cells. Ph, Phloem; X, xylem.
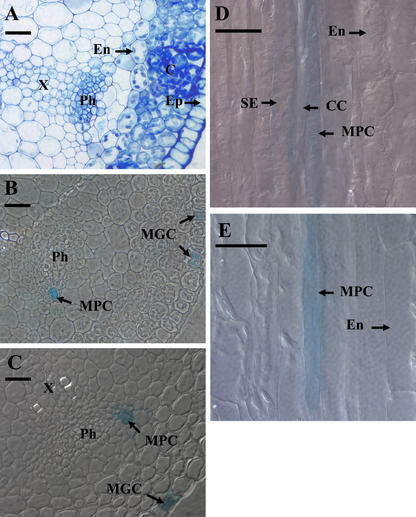
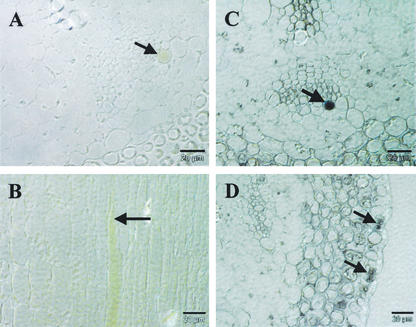
For a higher resolution view, nonstained and GUS-stained material from the upper flower stalk was embedded in LR white and semi-thin sections were made (2–3 μm), some being stained with toluidine blue for better identification of the different cell types (Fig. 4). The toluidine blue-stained section in Figure 4A shows the cell types at the site of the phloem bundle. On the inner side of the clearly visible plastid-rich endodermis layer, cells with a phenotype of the glucosinolate-rich S cells described by Koroleva et al. (2000) are located. These cells are large and irregularly shaped, and are only weakly stained with toluidine blue. No plastids are observed. Next to the potential S cells opposite to the endodermis layer are the phloem parenchyma cells and the phloem sieve element/companion cell complexes, respectively. The GUS-positive phloem cells are located at specific positions in the phloem bundle overlapping the phloem parenchyma cells (Fig. 4, B and C). Therefore, the GUS-positive phloem cells are believed to be a subpopulation of phloem parenchyma cells. Based on convention, positive guard cells were named myrosin guard cells (MGCs) and positive phloem cells were named myrosin phloem cells (MPCs). Most of the observed GUS-positive MPCs are larger in diameter and length than the phloem sieve element/phloem companion cell complexes (up to 90 μm in length and 15 μm wide), but are smaller than the observed potential S cells (up to 500 μm long and 20 μm wide).
Figure 4.
Semi-thin section of the Arabidopsis flower stalk. A, Transverse section of wild-type flower stalk stained with toluidine blue. B and C, Transverse sections from GUS-expressing plants. D and E, Longitual sections from GUS-expressing plants. C, Cortex; CC, phloem companion cell; En, endodermis; Ep, epidermis; Ph, phloem; S, sulfur-rich cell (Korolova et al., 2000); SE, phloem sieve element; X, xylem. Bars = 20 μm.
Immunocytochemical Localization of Myrosinase
To verify that MPCs showing TGG1 promoter activity are true myrosin cells (myrosinase-containing cells), material from the upper flower stalk was sectioned and immunocytochemical labeled using the anti-myrosinase antibodies K089 and 3D7. MPCs show immunolabeling with K089 and 3D7 antibodies (Fig. 5, A–C). MGCs only show immunolabeling with the K089 antibody (Fig. 5D). No stained cells were observed in sections incubated under identical conditions without the K089 antibody (result not shown). In addition to MPCs and MGCs, distinct cells of the cortex, endodermis, and xylem were also labeled with the K089 antibody.
Figure 5.
Immunocytochemical localization of myrosinase in transverse and longitual sections of Arabidopsis flower stalks using the anti-myrosinase antibodies K089 and 3D7. A, B, and D, Transverse sections. C, Longitudinal section. A and B, Sections immunolabeled with the 3D7 antibody. C and D, Sections immunolabeled with the K089 antibody. Arrows indicate myrosinase-containing MGCs and MPCs. Bars = 20 μm.
DISCUSSION
Our results show that the TGG1 myrosinase promoter is specifically expressed in guard cells and phloem cells at all the investigated developmental stages of the Arabidopsis plant. Immunocytochemical localization of myrosinase in cells of the flower stalk shows that the TGG1 promoter activity reflects the localization of the myrosinase enzyme in MGCs and MPCs. From Figure 4D, it appears that the sieve element and companion cells next to the MPCs also show GUS expression. From the transverse section (Fig. 4C), it is clear that these cells lack GUS staining in cytoplasm. Taken together with the lack of immunoreactivity of the sieve element and companion cells toward the myrosinase antibodies used, these cells are not myrosin cells, but they appear stained because of leakage of the stain from the neighboring MPCs.
The finding that the K089 antibody also show immunoreactivity toward other cell types than the GUS-expressing MGCs and MPCs suggests that additional myrosinases are expressed in the Arabidopsis flower stalk. The presence of myrosinase in other cell types than those expressing TGG1 is likely considering that a total of six myrosinase or myrosinase-like genes (of which two likely are pseudogenes) are present in the Arabidopsis genome.
Koroleva et al. (2000) reported that S cells of the Arabidopsis flower stalk are rich in glucosinolates. These cells are situated between endodermis and the cells belonging to the bundles of vascular phloem. Each of these bundles contains a phloem zone with phloem parenchyma and sieve element/companion cell complexes. The S cells were reported to be up to 1,000 μm long and 30 μm wide (Koroleva et al., 2000). Next to the MPCs in the Arabidopsis flower stalk, we found cells with an S cell appearance, which appears slightly smaller than described by Koroleva et al. (2000). We found these cells to be up to 20 μm wide and 500 μm long. Taken together, our observations indicate a location of myrosinase enzymes and glucosinolates in different but neighboring cells in the flower stalk of Arabidopsis.
Further support for glucosinolate-containing cells closely associated with phloem comes from the spatial expression pattern observed for the CYP79F1 and CYP79F2 genes involved in glucosinolate synthesis (Reintanz et al., 2001). These genes are exclusively expressed in or close to vascular phloem of the Arabidopsis, including the flower stalk. It is unfortunate that the identity of these cells was not described.
The role of glucosinolates and their hydrolysis products in plant defense is widely accepted (Bones and Rossiter, 1996; Rask et al., 2000). Fungistatic and bacteriostatic effects of glucosinolates have been reported, and the deterrence of nonspecialist pests has been well documented (Chew, 1998; Wallsgrove and Bennett, 1995; Tierens et al., 2001). Many specialized pests have overcome the toxicity of the myrosinase-glucosinolate system and even use glucosinolates and isothiocyanates to locate and identify their glucosinolate-containing host plants. The aphid specialist Brevicoryne brassicae can be induced to feed on nonhost plants when leaves are treated with 2-propenyl glucosinolate (Nault and Styler, 1972), and its intrinsic growth rate depends on the concentration of glucosinolates (Cole, 1997). The B. brassicae even expresses large amounts of its own endogenous myrosinase, which is believed to play a role in the insects defense and signaling against predators (Jones et al., 2001, 2002; Bridges et al., 2002).
The positioning of myrosinase-containing cells together with glucosinolate-containing S cells outside the phloem sieve elements is ideally suited for the protection of the phloem and its content against microbes and insects. During insect attachment, myrosinase and glucosinolate mix, resulting in the release of toxic hydrolysis products. The phloem parenchyma cells appear to be specialized in delivery of photosynthetic assimilate products such as Suc from the bundle sheath into the sieve tract (Haritatos et al., 2000). Similar mechanisms may enable efficient loading of products such as glucosinolates or hydrolyzed products into the phloem. The reactive, volatile, and/or cytotoxic effect of some of these products may serve different functions such as signaling and defense. Thangstad et al. (2001) showed that the glucosinolate precursor (desulphoglucosinolate) accumulates in specific cells in the embryo radicle and cotyledon, in a pattern that matches the distribution of myrosin cells in the embryo. This study did not address whether myrosinase enzyme and glucosinolate substrate were colocalized in the same cells or if they were located in different but adjacent cells, but it did provide evidence for a specific transport system for glucosinolates or desulfoglucosinolates.
There is now emerging evidence that the myro-sinase-glucosinolate system also is involved in processes other than plant defense. Knockout of the CYP79F1 gene results in a mutant where synthesis of short chain aliphatic glucosinolates is abolished, resulting in a bushy phenotype (Reintanz et al., 2001), indicating a role for glucosinolates in control of plant growth and development.
Our results show that the TGG1 myrosinase promoter is active in most organs of Arabidopsis except in the root and seeds. Promotor activity is especially high in young leaves, upper flower stalk, pedicle, and the transition zone between the pedicle and silique. In older tissue, TGG1 promoter activity is considerable lower. High myrosinase activity in young developing tissue may have dual functions. Myrosinase activity generates toxic components that may be beneficial to express in cells at the point of entry of microorganisms. The location of myrosinase in phloem and guard cells may indicate a role in communication, internally by hydrolysis of glucosinolates and transport in phloem, and externally by distribution of volatile degradation products from guard cells. The fact that myrosinase activity is especially high in developing tissue may also suggest that myrosinase is involved in the control of plant growth and development.
MATERIALS AND METHODS
Constructs
Isolation of the TGG1 Promoter DNA Sequence
A genomic library of Arabidopsis ecotype Columbia in λGEM11 (generously provided by Dr. Chris Somerville, Department of Plant Biology, Carnegie Institution, Stanford, CA) was screened using TGG1 cDNA, nucleotides 120 to 1,462 (Chadchawan et al., 1993), as a probe. Ten positive clones were isolated, and one clone showing the restriction sites matching the TGG1 cDNA (Chadchawan et al., 1993) was subcloned and verified by sequencing. The 5′-upstream region of TGG1 used for the TGG1 promoter fusion gene constructs was sequenced in both directions.
TGG1 promoter:GUS Fusion Construct, TGG1Pro:GUS
A λgt11 genomic subclone containing approximately 10 kb upstream of the TGG1 gene and 1 kb of the 5′ region of TGG1 gene was digested with BamHI (in the promoter region) and SstI (within the coding region), yielding a 2.5-kb fragment that was cloned into pBluescript II KS+. The TGG1 coding region was verified by sequencing. This clone was then digested with BamHI and HindIII (at the putative translation start site). The BamHI-HindIII fragment was filled with dATP and dGTP, and was then cloned into PBI101.2 (CLONETECH, Palo Alto, CA), which was digested with SalI and XbaI and filled with dTTP and dCTP (generating the compatible two-based overhang at both ends). Restriction analysis verified that the promoter was at the 5′ end of the GUS coding sequence in the proper orientation, and the ligated junction of the chimeric gene was sequenced to confirm the appropriate reading frame.
TGG1 Promoter:GFP Fusion Gene Construct, TGG1Pro:sGFPCt
The pBI-TGG1Pro:sGFPCt construct was made by substituting the GUS gene of the pBI-TGG1PRO:GUS construct with the GFP:TGG1 fusion gene sequence described below.
Cloning of GFP:TGG1 fusion gene
The bacterial artificial chromosome clone (T1N24) serving as a template for the TGG1 signal peptide and C-terminal sequences was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus). Forward primer 5′-TTGAAAAGCAAAGCAGTCCGAA-3′ and reverse primer 5′-GTTTGGATCCAAGTGAATGGCTCGT-3′ (BamHI-site is underlined) was used to amplify a 339-bp PCR fragment. The PCR fragment was digested with HindIII and BamHI, and the 98-bp DNA restriction fragment encoding 32 amino acids of the TGG1 N-terminal (including the 19-amino acid signal peptide) was cloned between the HindIII and BamHI site of a pBluescript II KS+ containing a GFP reporter gene, pTGG1sGFP. The GFP gene was derived from mGFP4 (Haseloff et al., 1997) and mGFP5-ER (from Jim Haseloff, Department of Plant Sciences, University of Cambridge, UK), removing the N-terminal signal peptide from the Arabidopsis basic chitinase gene and the C-terminal HDEL amino acid endoplasmic reticulum-retention signal. A new SphI and SacI site was added C-terminally to the GFP DNA sequence to allow insertion of other C-terminal sequences. Forward primer 5′-GTCAGCATGCGACTAAGTTACGTTGA-3′ (SphI site is underlined) and reverse primer 5′-TGGATGAGCTCTATTTGGTTCTTTC-3′ (SacI site is underlined) were used to amplify a 366-bp PCR product, which was digested with SphI and SacI and cloned in between the SphI and SacI sites of the pTGG1sGFP vector made above. In this cloning procedure, a GFP:TGG1 fusion gene encoding 32 N-terminal and 58 C-terminal TGG1 amino acids of the TGG1 protein and 183 bp of the TGG1 3′-untranslated region was made, pTGG1sGFPCt. The pTGG1sGFPCt vector was digested with HindIII and SacI and the TGG1:GFP fusion gene sequence cloned between the HindIII and SacI sites of the PBI vector containing the full 2.5-kb TGG1 promoter sequence (TGG1Pro:GUS) substituting the GUS gene. The TGG1Pro:sGFPCt and TGG1Pro:GUS fusion gene constructs are presented in Figure 1.
PCR amplification and sequencing
One hundred nanograms of plasmid DNA was used as a template for the PCR amplifications (four cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C, followed by 26 cycles of 1 min at 94°C, 1 min at 54°C, and 2 min at 72°C using a Pfu DNA polymerase [Stratagene, La Jolla, CA]) and a PE 480 thermal cycler (Applied Biosystems, Foster City, CA). The partial TGG1 encoding DNA sequences contained in the TGG1sGFPCt vector was checked by sequencing using T3 forward primer, GFP-specific reverse primer 5′-TGCC-CATTAACATCACCATCT-3′, and T7 reverse primer. Sequencing was performed using BigDye chemistry on the PE 480 thermal cycler and reactions were run on a ABI 377 sequencer (Applied Biosystems).
Plants
Plant Transformation
Agrobacterium tumefaciens pMP90 strain GW3101 (Koncz and Schell, 1986) was transformed with the GFP containing TGG1Pro:sGFPCt plasmid using electroporation. Arabidopsis plants were transformed and selected by kanamycin resistance according to Clough and Bent (1998).
Plant Growth and Sampling
Seeds from Arabidopsis ecotype Colombia homozygous for the pBI-TGG1Pro:GUS or TGG1Pro:sGFPCt constructs were grown on soil under a 16-h light (25°C) and 8-h dark (20°C) regime in controlled environment rooms until flowering. Seedlings were sampled 5 and 12 d after germination and flower stalks over a period of 2 weeks after bolting.
Histochemical GUS Detection
The histochemical detection of GUS expression was performed essentially as described by Jefferson et al. (1987) with one important adjustment. Prior to incubation in substrate solution, plant material was incubated for 5 min in acetone at −20°C, air dried, and incubated for an additional 30 min in 0.1 m phosphate buffer, pH 7.0. GUS staining was performed using 0.1 to 0.2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic-acid and sodium salt (DUCHEFA, Haarlem, The Netherlands) at 37°C for 3 to 5 h (with the 0.5- to 1.5-mm sections) or 24 h (with the 1.0- to 1.5-cm sections), with an initial 3-min vacuum infiltration. After staining, the samples were examined and photographed using a stereo microscope (SMZ1500; Nikon, Tokyo) equipped with a digital camera (Coolpix 990; Nikon) or a research microscope (Eclipse 800; Nikon) equipped with a cooled digital camera (SPOT RT; Diagnostic Instruments, Burroughs, MI).
GUS-stained flower stalk material was embedded in LR white as described by Peleman et al. (1989). Sections (2–3 μm) were made on a ultramicrotome (type 4801A; LKB, Uppsala) and were placed on a film of water. The sections were transferred to a glass slide on a drop of water, dried, and mounted using DPX mountant (British Drug House; Laboratory Supplies, Poole, UK). Differential interference contrast images were obtained using a research microscope (Eclipse 800; Nikon).
GFP Fluorescence Detection
GFP fluorescence in the Arabidopsis flower stalk was monitored in intact tissues or in hand-cut transverse sections (approximately 0.5 mm). Care was taken not to allow the transverse sections to dry due to development of autofluorescence in dried samples. GFP fluorescence was visualized in live plant tissue using a GFP(R)-BP (EX 460–500 nm/DM 505 nm/BA 510–560 nm) filter.
Immunocytochemistry
Flower stalks were sampled 1 week after start of bolting, were cut into 0.5- to 2.0-mm pieces with a razor blade, and were fixed overnight in 4% (w/v) paraformaldehyde in 50 mm phosphate buffer (pH 7.2) at 4°C. After washing in 50 mm phosphate buffer, the pieces were dehydrated through graded series of ethanol, embedded in paraffin wax, and sectioned (6 μm) as described by Thangstad et al. (1990). The sections were transferred to poly-l-Lys-coated (0.1%, w/v) slides and were incubated overnight at 37°C and at 55°C for 15 min before wax removal. The wax was removed by a 10-min incubation in xylene followed by two washes in absolute ethanol. The sections were rehydrated through graded series of ethanol followed by a 10-min incubation in phosphate-buffered saline (PBS) prior to immunocytochemical labeling. The marker used for localization was a rabbit polyclonal antibody K089 (Thangstad et al., 1991) or a mouse monoclonal antibody 3D7 (Lenmann, 1990). To prevent nonspecific binding, sections were incubated in 10% (w/v) normal serum and 5% (w/v) bovine serum albumin (BSA) in PBS-0.5% (v/v) Tween 20 (PBS-T) for 15 min. Immunohistochemistry was performed at 37°C including 5% (w/v) BSA in the PBS-T/antibody solutions. The sections were incubated for 45 min with primary myrosinase antibody K089 diluted 1/10,000 or 3D7 diluted 1/500, 30 min with biotinylated secondary antibody anti-rabbit Ig diluted 1/500 or anti-mouse Ig diluted 1/300, and 30 min in an Avidin:Biotynilated enzyme complex solution (VECTASTAIN ABC kit, Vector Laboratories, Inc., Burlingame, CA). Finally, immunoreactive cells were visualized by a 10-min incubation in DAB-solution (3,3′-diaminobenzidine, Vector Laboratories, SK-4100) at room temperature. Cover slips were mounted using VectaMount (Vector Laboratories). Differential interference contrast photography was performed as described above.
ACKNOWLEDGMENTS
We thank Dr. Jim Haseloff for the mGFP4 and mGFP5-ER, Dr. Chris Somerville for the Arabidopsis genomic DNA library, Arabidopsis Biological Resource Center for the TGG1 gene containing bacterial artificial chromosome clone (T1N24), Dr. Lars Rask for the 3D7 monoclonal antibody, Kjell Evjen for sectioning of LR white-embedded material, and Eli Marie Johannessen for sectioning the paraffin wax embedded material.
Footnotes
This work was supported by Nordisk Kontaktorgan for Jorbruksforkning (grant no. 104).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010925.
LITERATURE CITED
- Bones AM, Iversen T-H. Myrosin cells and myrosinase. Isr J Bot. 1985;34:351–375. [Google Scholar]
- Bones AM, Rossiter JT. The myrosinase-glucosinolate system, its organization and biochemistry. Physiol Plant. 1996;97:194–208. [Google Scholar]
- Bones AM, Thangstad OP, Haugen OA, Espevik T. Fate of myrosin cells: characterization of monoclonal antibodies against myrosinase. J Exp Bot. 1991;42:1541–1549. [Google Scholar]
- Bridges M, Jones AME, Bones AM, Hodgson C, Cole R, Bartlett E, Wallsgrove R, Karapapa VK, Rossiter JT. Spatial organisation of the glucosinolate-myrosinase system in Brassica specialist aphids is similar to that of the host plant. Proc Royal Soc Biol Sci. 2002;269:187–191. doi: 10.1098/rspb.2001.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadchawan S, Bishop J, Thangstad OP, Bones AM, Mitchell-Olds T, Bradley D. Arabidopsis cDNA sequence encoding myrosinase. Plant Physiol. 1993;103:671–672. doi: 10.1104/pp.103.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew FS. Searching for defensive chemistry in the Cruciferae, or, do glucosinolates always control interactions of Cruciferae with their potential herbivores and symbionts? No! In: Spencer KC, editor. Chemical Mediation of Coevolution. San Diego: Academic Press; 1998. pp. 45–88. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method of Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cole RA. The relative importance of glucosinolates and amino acids to the development of two aphid pests, Brevicoryne brassicae and Myzus perciae on wild and cultivated Brassica species. Entomol Exp App. 1997;85:121–133. [Google Scholar]
- Falk A, Ek B, Rask L. Characterization or a new myrosinase in Brassica napus. Plant Mol Biol. 1995;27:863–874. doi: 10.1007/BF00037015. [DOI] [PubMed] [Google Scholar]
- Foo HL, Grønning LM, Goodenough L, Bones AM, Danielsen BE, Whiting DA, Rossiter JT. Purification and characterization of epithiospecifier protein from Brassica napus enzymatic intermolecular sulfur addition with alkenyl thio-hydroximates derived from alkenyl glucosinolate hydrolysis. FEBS Lett. 2000;468:243–246. doi: 10.1016/s0014-5793(00)01176-5. [DOI] [PubMed] [Google Scholar]
- Geshi N, Andreasson E, Meijer J, Rask L, Brandt A. Colocalization of myrosinase- and myrosinase-binding proteins in grains of myrosin cells in cotyledon of Brassica napus seedlings. Plant Physiol Biochem. 1998;36:583–590. [Google Scholar]
- Haritatos E, Medville R, Turgeon R. Minor vein structure and sugar transport in Arabidopsis thaliana. Planta. 2000;211:105–111. doi: 10.1007/s004250000268. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogge LR, Reed DW, Underhill EW, Haughn GW. HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid-chromatography mass-spectrometry. J Chromatogr Sci. 1988;26:551–556. [Google Scholar]
- Höglund A-S, Lenman M, Falk A, Rask L. Distribution of myrosinase in rapeseed tissues. Plant Physiol. 1991;95:213–221. doi: 10.1104/pp.95.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglund A-S, Lenman M, Rask L. Myrosinase is localized to the interior of myrosin grains and is not associated to the surrounding tonoplast membrane. Plant Sci. 1992;85:165–170. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AME, Bridges M, Bones AM, Cole R. Purification and characterization of a non-plant myrosinase from the cabbage aphid Brevicoryne brassicae (L.) Insect Biochem Mol Biol. 2001;31:1–5. doi: 10.1016/s0965-1748(00)00157-0. [DOI] [PubMed] [Google Scholar]
- Jones AME, Winge P, Bones AM, Cole R, Rossiter JT. Characterization and evolution of a myrosinase from the cabbage aphid Brevicoryne brassicae. Insect Biochem Mol Biol. 2002;32:275–284. doi: 10.1016/s0965-1748(01)00088-1. [DOI] [PubMed] [Google Scholar]
- Kelly PJ, Bones A, Rossiter JT. Sub-cellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta. 1998;206:370–377. doi: 10.1007/s004250050412. [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of the TL-DNA 5 gene controls the tissue specific expression of chimeric gene carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Koroleva OA, Davies A, Deeken R, Thorpe MR, Tomos AD, Hedrich R. Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 2000;124:599–608. doi: 10.1104/pp.124.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenmann M, Falk A, Rödin J, Höglund A-S, Ek B, Rask L. Differential expression of myrosinase gene families. Plant Physiol. 1993;103:703–711. doi: 10.1104/pp.103.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenmann M, Rödin J, Josefsson LG, Rask L. Immunological characterization of rapeseed myrosinase. Eur J Biochem. 1990;194:747–753. doi: 10.1111/j.1432-1033.1990.tb19465.x. [DOI] [PubMed] [Google Scholar]
- Nault LR, Styler WE. Effects of sinigrin on host plant selection by aphids. Entomol Exp App. 1972;5:423–437. [Google Scholar]
- Peleman J, Boerjan W, Engler G, Seurinck J, Botterman J, Alliotte T, Van Montagu M, Inzé D. Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell. 1989;1:81–93. doi: 10.1105/tpc.1.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicacea. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangstad OP, Bones AM, Holtan S, Moen HL, Rossiter JT. Microautoradiographic localization of glucosinolate precursor to specific cells in Brassica napus L. embryos indicate a separate transport pathway in myrosin cells. Planta. 2001;213:207–213. doi: 10.1007/s004250000491. [DOI] [PubMed] [Google Scholar]
- Thangstad OP, Evjen K, Bones A. Immunogold-EM localization of myrosinase in Brassicacea. Protoplasma. 1991;161:85–93. [Google Scholar]
- Thangstad OP, Iversen T-H, Slupphaug G, Bones AM. Immunocytochemical localization of myrosinase in Brassica napus L. Planta. 1990;180:245–248. doi: 10.1007/BF00194003. [DOI] [PubMed] [Google Scholar]
- Thangstad OP, Winge P, Husebye H, Bones AM. The thioglucoside glucohydrolase (myrosinase) gene family in Brassicacea. Plant Mol Biol. 1993;23:511–524. doi: 10.1007/BF00019299. [DOI] [PubMed] [Google Scholar]
- Tierens KFM-J, Thomma BPHJ, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BPA, Broekaert WF. Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 2001;125:1688–1699. doi: 10.1104/pp.125.4.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallsgrove RM, Bennett RN. The biosynthesis of glucosinolates in Brassica. In: Wallsgrove RM, editor. Amino Acids and Their Deviates in Higher Plants. Cambridge, UK: Cambridge University Press; 1995. pp. 243–259. [Google Scholar]
- Xue J, Lenman M, Falk A, Rask L. The glucosinolate-degrading enzyme myrosinase in Brassicacea is encoded by a gene family. Plant Mol Biol. 1992;18:387–398. doi: 10.1007/BF00034965. [DOI] [PubMed] [Google Scholar]
- Xue J, Philgren U, Rask L. Temporal, cell-specific, and tissue-preferential expression of myrosinase genes during embryo and seedling development in Sinapis alba. Planta. 1995;191:95–101. [Google Scholar]