Abstract
Starch-branching enzymes (SBEs) catalyze the formation of α(1→6) glycoside bonds in glucan polymers, thus, affecting the structure of amylopectin and starch granules. Two distinct classes of SBE are generally conserved in higher plants, although the specific role(s) of each isoform in determination of starch structure is not clearly understood. This study used a heterologous in vivo system to isolate the function of each of the three known SBE isoforms of maize (Zea mays) away from the other plant enzymes involved in starch biosynthesis. The ascomycete Brewer's yeast (Saccharomyces cerevisiae) was employed as the host species. All possible combinations of maize SBEs were expressed in the absence of the endogenous glucan-branching enzyme. Each maize SBE was functional in yeast cells, although SBEI had a significant effect only if SBEIIa and SBEIIb also were present. SBEI by itself did not support glucan accumulation, whereas SBEIIa and SBEIIb both functioned along with the native glycogen synthases (GSs) to produce significant quantities of α-glucan polymers. SBEIIa was phenotypically dominant to SBEIIb in terms of glucan structure. The specific branching enzyme present had a significant effect on the molecular weight of the product. From these data we suggest that SBEs and GSs work in a cyclically interdependent fashion, such that SBE action is needed for optimal GS activity; and GS, in turn, influences the further effects of SBE. Also, SBEIIa and SBEIIb appear to act before SBEI during polymer assembly in this heterologous system.
The crystalline architecture of starch granules is generally similar in all plants (Jenkins et al., 1993). Starch structure presumably is an essential aspect of the physiological processes by which Glc units derived from photosynthesis are stored for hours or months in resistant form, and then later released and used when needed for nonphotosynthetic metabolism. Granule formation is largely dependent on the semicrystalline properties of amylopectin (Ap), the branched glucan polymer that provides about 75% of the granule mass (for reviews, see Myers et al., 2000, and refs. therein). Ap comprises α(1→4)-linked “linear” chains of Glc units, which are joined to each other by α(1→6) glycosidic bonds, i.e. “branch” linkages. The architectural arrangement of Ap, including the length of the linear chains, the frequency of α(1→6) linkages, and the placement of branches relative to each other and the ends of the chains, is likely to determine the ability of Ap to crystallize into granules. This presumption is derived, in part, from consideration of the structure of glycogen, which has the same chemical features as Ap but, because of more regularly spaced branch points and shorter linear chain lengths, is fully soluble and noncrystalline (Calder, 1991; Lomako et al., 1993; Alonso et al., 1995). Also, certain mutant plants accumulate soluble glucan polymers within the same cells and organelles that also contain starch granules comprising crystalline Ap (Zeeman et al., 1998). These soluble glucans exhibit linear chain lengths and branch frequencies that are distinct from those of the crystalline material.
The biosynthetic system that produces starch granules must be able to act with a certain degree of architectural specificity, although how the structural characteristics of Ap are precisely determined is not well understood. Just as starch structure is highly conserved in the plant kingdom, so are the sequences of the enzymes responsible for assembly of Glc units into Ap (Smith et al., 1997; Myers et al., 2000). Starch synthases (SS) catalyze the formation of linear chains, using ADP-Glc as the hexose donor and connecting Glc units through α(1→4) glycosidic bonds. Starch-branching enzymes (SBEs) catalyze the formation of α(1→6) bonds by means of cleavage within a linear chain and transfer of the free reducing end to a C6 hydroxyl. A priori, it seems possible that the enzymatic specificities of SSs and SBEs could explain how Ap is synthesized with its specific chemical architecture.
Glycogen synthesis in animals, fungi, and prokaryotes typically requires a single isoform of the glucan synthase and branching enzyme activities. Starch biosynthesis appears to be more complex, however, as indicated by the existence of multiple isoforms for each of these enzymes. In maize (Zea mays), there are at least five SS isoforms and three SBE isoforms called SBEI, SBEIIa, and SBEIIb (Smith et al., 1997; Myers et al., 2000). The three different SBEs have been defined clearly by biochemical fractionation (Boyer and Preiss, 1978a, 1978b) and by their primary sequences determined from cDNA clones (Fisher et al., 1993, 1995; Gao et al., 1997). SBEI is clearly distinguishable as a distinct isoform, whereas SBEIIa and SBEIIb are very closely related over most of their sequence but differ at their amino termini.
Isoform-specific functions are indicated by the facts that all four sequence classes of SS (I, II, III, and GBSS) and both SBE classes (I and II) are broadly conserved in plants (Smith et al., 1997; Cao et al., 1999; Li et al., 1999). Mutations in genes that code for specific SSs or SBEs often result in altered starch structure, which also indicates nonoverlapping functions of the multiple isoforms (Shannon and Garwood, 1984). Examples from maize are mutations of the dull1 (du1) gene that codes for the SSIII of this species (Gao et al., 1998) and mutations of the ae gene that codes for SBEIIb (Stinard et al., 1993). In addition to the multiple isoforms of SS and SBE, another level of complication that must be considered in addressing the mechanism of Ap biosynthesis is the involvement in this process of α(1→6) glucosidases, i.e. starch-debranching enzymes (Myers et al., 2000, and refs. therein).
Identifying the specific role(s) of each SS or SBE with regard to determination of Ap structure is complicated, in part, because of pleiotropic effects of mutations in genes coding for a specific enzyme. The classical genetic approach has been employed, with the strategy of eliminating one isoform by mutation and then characterizing Ap structure to identify resultant changes. In many instances, however, more than one starch biosynthetic enzyme is affected by a single mutation. For example, maize du1- mutations that directly affect the SSIII also have an indirect effect of significantly reducing the activity of SBEIIa present in total extracts of endosperm tissue (Boyer and Preiss, 1978b). Mutations in the du1 locus also cause an increase of another SS activity in total extracts, most likely SSI (Cao et al., 1999). Attributing a certain structural change to the loss of a particular enzyme, therefore, often is not possible using this approach.
A different approach is to isolate specific isoforms away from the full complement of plant starch biosynthetic enzymes, by means of expression in heterologous host organisms. If it were possible to convert a microbial cell that produces glycogen into an organism that produces Ap with crystalline properties, through the addition of specific plant enzymes, then the functions necessary for producing a crystallization-competent polymer would be identified definitively. Using this strategy, Guan et al. (1995) replaced the native glycogen-branching enzyme (GBE) of Escherichia coli with the maize isoforms SBEI and SBEIIb. The recombinant bacteria produced a soluble polymer resembling glycogen, as opposed to an Ap-like molecule with semicrystalline properties. By themselves, therefore, these two SBE isoforms by themselves do not account for Ap architecture.
In this study, the heterologous expression strategy was used with the ascomycete Brewer's yeast (Saccharomyces cerevisiae) as the host. The complete sequence of the yeast genome reveals all of the relevant glycogen biosynthetic genes. These are GSY1 and GSY2, coding for two very closely related forms of glycogen synthase (GS; Farkas et al., 1991), and GLC3, coding for GBE (Rowen et al., 1992; Thon et al., 1992). In contrast to plant SSs, the two yeast GS isoforms are functionally redundant (Farkas et al., 1991).
The three maize SBE isoforms were expressed in Brewer's yeast, singly and in all possible combinations, in a common genetic background. Chemical analysis of the product glucans revealed synergistic effects between the SBEs and the native GSs, such that each distinct combination of activities gave rise to different polymer structures. The ability of SBEI to engender modifications in the product glucan polymers depended on the simultaneous presence of both SBEIIa and SBEIIb. To explain these results, we suggest that either SBEII function is needed to generate a substrate that is suitable for SBEI in this in vivo system, or that the SBEII isoforms directly regulate the activity of SBEI.
RESULTS
Partial Complementation of glc3 by Maize SBEs
Transgenic yeast strains were constructed in a genetic background completely lacking GLC3, which is the only host gene that codes for a GBE. Maize cDNAs coding for various SBEs were expressed from the strong, constitutive promoter of the yeast TPI1 gene. Yeast colonies were grown on yeast peptone dextrose (YPD) plates for 2 d and then exposed to iodine vapor for 1 min to reveal stainable glucan-iodine complexes. In agreement with previous results (Thon et al., 1992), the glc3::KANR strain BSY-01, which does not contain any branching enzyme gene, did not stain appreciably and remained yellow-white (Fig. 1A). GS activity in total soluble extracts is known not to be affected by deletion of GLC3 (Thon et al., 1992), and the level of an epitope-tagged version of Gsy2p as detected by immunoblot analysis did not vary in the glc3::KANR mutant compared with the wild-type parental strain (data not shown). Glc3p, therefore, is required for production of iodine-stainable glucan polymer, even though GS accumulation and activity are unaltered.
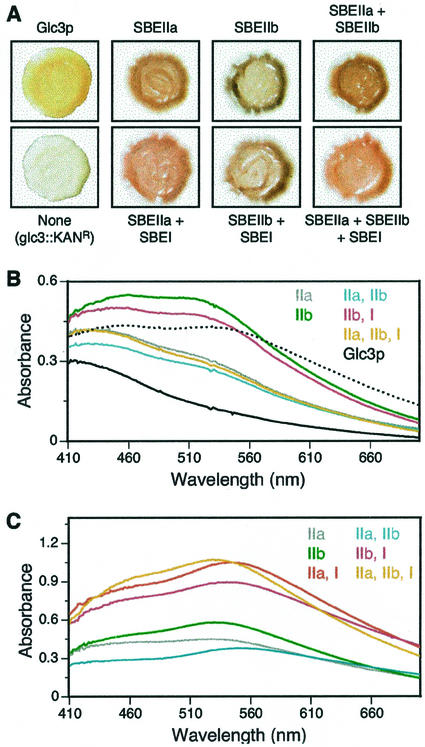
Figure 1.
Characterization of α-glucans by staining with iodine. A, α-Glucans in yeast colonies. Yeast colonies were grown on YPD medium and exposed to I2 vapor. All the strains share a common genetic background, differing only with respect to the genes that code for glucan-branching enzymes. The particular GBE, SBE, or combination of SBEs present is indicated for each photograph. Strains containing maize SBEs are all glc3::KANR mutants, thus, lacking any endogenous branching enzyme. The strains are as follows (refer to Table I): Glc3p, Y-2159; glc3::KANR, BSY-01, SBEIIa, BSY-03; SBEIIb, BSY-04, SBEIIa + SBEIIb, BSY-07; SBEIIa + SBEI, BSY-05; SBEIIb + SBEI, BSY-06; SBEIIa + SBEIIb + SBEI, BSY-08. B, Absorbance spectra of total soluble glucans (200 μg) extracted by the hot alkali method and stained with I2/KI. The dotted line shows the spectrum obtained for commercial maize Ap. The solid black line shows the spectrum obtained for glycogen from nonmutant yeast strain Y-2159, produced using Glc3p as the branching enzyme. The colored lines show spectra obtained for the α-glucans from glc3::KANR strains expressing the particular SBE or combination of SBEs indicated by the labels of corresponding color. The identity of each SBE-containing strain is indicated in the legend for A. C, Absorbance spectra of size-fractionated glucans (100 μg) extracted by the mechanical disruption method and stained with I2/KI. Strains are as in B, with the addition of BSY-05 (SBEIIa + SBEI).
Transformation of the glc3::KANR strain BSY-01 with integrative plasmids that express either SBEIIa or SBEIIb restored the ability to accumulate iodine-stainable glucan polymers (Fig. 1A). The SBEIIa strain BSY-03 produced a reddish-brown color, whereas the SBEIIb strain BSY-04 stained blue. In all instances, the particular iodine-staining phenotype cosegregated with the prototrophic marker linked to the SBE expression gene in progeny tetrads from crosses to tester strain Y-2158 (data not shown). Restoration of glucan polymer synthesis, therefore, can be attributed specifically to expression of the maize SBE. These results indicate that SBEIIa and SBEIIb can provide whatever function is lacking in the glc3::KANR mutant that allows GS to form a polymer or that allows glucan accumulation.
Expression of maize SBEI did not complement the defect in iodine-stainable glucan accumulation conditioned by the glc3::KANR mutation (data not shown). Co-expression of both SBEIIa and SBEIIb in strain BSY-07 resulted in the reddish-brown staining color similar to that resulting from SBEIIa alone (Fig. 1A), indicating phenotypic dominance of SBEIIa over SBEIIb. None of the other double-isoform combinations nor the triple-isoform combination yielded a difference from either the SBEIIa or SBEIIb result that could be distinguished by iodine vapor staining (Fig. 1A).
The quantity of total soluble glucan extracted from various strains by extraction with hot alkali was measured as a function of time in culture (data not shown). Expression of either SBEIIa or SBEIIb resulted in about 40% of the amount of glycogen seen in the nonmutant standard. The SBEI strain did not produce any detectable glucan polymer. The growth rate of the culture was not affected by expression of any of the maize proteins (data not shown).
Branching Enzyme Activity
From the restoration of glucan biosynthesis in the glc3::KANR host strain, it can be inferred that at least some of the maize SBEs are expressed in active form in this heterologous system. The level of debranching enzyme activity was tested directly in total soluble cell extracts using the phosphorylase a stimulation assay (Guan and Preiss, 1993). Table I indicates that branching enzyme activity present in extracts of the glc3::KANR strain is reduced to near background levels compared with the GLC3 control strain. Expression of SBEIIa in the glc3::KANR host restored approximately 40% of the branching activity seen in the nonmutant control strain, whereas expression of SBEIIb restored about 15% of the activity. Introduction of the SBEI plasmid did not result in an increased level of branching enzyme activity measured in total soluble cell extracts above that of the glc3::KANR host strain.
Table I.
Branching enzyme activity assays
| Strain | Genotype | Maize SBE | Branching Enzyme Activitya
|
|||
|---|---|---|---|---|---|---|
| Assay 1 | Assay 2 | Assay 3 | Mean | |||
| Y-2159 | GLC3 | – | 17,292 | 20,362 | 20,511 | 19,388 |
| BSY-01 | glc3∷KANR | – | 166 | 264 | 222 | 217 |
| BSY-02 | glc3∷KANR | I | 227 | 258 | 234 | 240 |
| BSY-03 | glc3∷KANR | IIa | 6,268 | 10,555 | 8,886 | 8,570 |
| BSY-04 | glc3∷KANR | IIb | N.D.b | 2,159 | 3,791 | 2,975 |
| BSY-05 | glc3∷KANR | I, IIa | 5,952 | 6,964 | 6,003 | 6,306 |
| BSY-06 | glc3∷KANR | I, IIb | 1,636 | N.D. | 1,311 | 1,474 |
| BSY-07 | glc3∷KANR | IIa, IIb | 8,786 | 7,055 | 7,970 | 7,937 |
| BSY-08 | glc3∷KANR | I, IIa, IIb | 7,303 | 9,225 | 8,580 | 8,396 |
Each assay was determined from a separate culture of yeast cells.
N.D., A value was not determined in that particular assay set.
Glucan Size Distribution
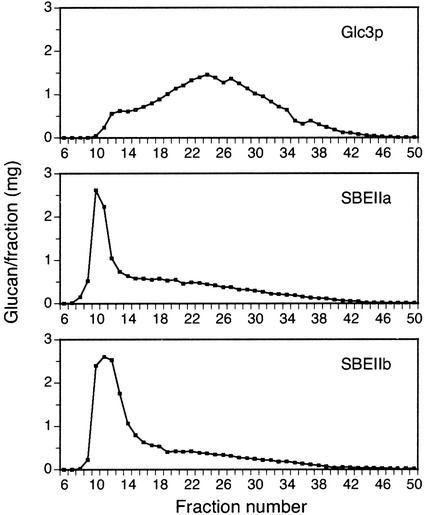
To compare the sizes of the glucan polymers produced by each combination of biosynthetic enzymes, the carbohydrates were isolated and fractionated by gel permeation chromatography. A standard method of boiling cells in hot alkali released a water-soluble glucan fraction that eluted from a TSK-HW50S column in a single narrow peak (data not shown). Further analysis of this fraction (see below) revealed the presence of a nonglucan contaminant in some of the samples. A different method was developed in which cells were mechanically disrupted and extracted in 1% (w/v) SDS. Total glucans from this extraction were chromatographed on Sepharose CL-2B. In this procedure various glucan size fractions were resolved, and the pattern differed significantly between the nonmutant and transgenic strains.
Glycogen from the nonmutant strain eluted from the CL-2B column in a very broad peak (Fig. 2). A small shoulder at the beginning of the peak indicated a distinct minority population of glucans that are larger in Mr than most of the molecules. In contrast, glucans from glc3::KANR strains expressing either SBEIIa or SBEIIb eluted predominantly in the peak of large material, and a minority of the polymers eluted in the broad tail of smaller Mr (Fig. 2). Thus, the particular enzyme responsible for introduction of branch linkages can be a specific determinant of the molecular size of the glucan polymer produced.
Figure 2.
Glucan size distributions. Glucans from the indicated yeast strains were prepared by the mechanical disruption method and fractionated by gel permeation chromatography on Sepharose CL-2B. The label for each panel indicates the glucan-branching enzyme that is present in otherwise congenic yeast strains. The strains are as follows (refer to Table I): Glc3p, Y-2159; SBEIIa, BSY-03; SBEIIb, BSY-04.
Visible Spectra of Glucan-Iodine Complexes
Soluble glucans extracted by the hot alkali method and purified by TSK-HW50S chromatography were complexed with iodine, and absorbance spectra were recorded (Fig. 1B). Using equal quantities of glucan, distinct spectra were determined for the maize Ap standard, glycogen from nonmutant yeast, and the glucans produced in yeast by SBEIIa or SBEIIb. These results are in agreement with the distinct colors seen in the colony stains of the nonmutant, SBEIIa, and SBEIIb strains (Fig. 1A). Glucan from the strain expressing both SBEIIa and SBEIIb yielded a spectrum similar to the SBEIIa strain, again revealing the dominance of SBEIIa. Co-expression of SBEI along with SBEIIa, SBEIIb, or SBEIIa + SBEIIb in combination did not significantly alter the absorption spectrum from that obtained with the SBEII isoform(s) alone.
Spectra also were recorded for the relatively large glucans extracted by the mechanical disruption method and purified in the peak fraction of the Sepharose CL-2B chromatographs (see Fig. 2). SBEIIa and SBEIIb again produced distinct spectra (Fig. 1C). In this instance, however, the combination of SBEIIa and SBEIIb in the same host strain yielded a spectrum distinct from either enzyme alone. Co-expression of SBEI resulted in a specific change in the absorbance spectrum. Because equal quantities of glucan were analyzed, the absorption coefficient can be standardized to moles of Glc equivalents. This value is significantly higher for all combinations containing SBEI compared with the corresponding strain in which SBEI is lacking (Fig. 1C). These data suggest that co-expression of SBEI does affect the structure of the product, even though SBEI by itself does not support glucan accumulation.
Chain-Length Distributions
Glucans prepared by the hot alkali method were debranched by treatment with Pseudomonas sp. isoamylase, an α(1→6)-specific glucosidase. The resultant populations of linear chains were then separated and quantified using high performance anion-exchange chromatography with post-column enzymatic digestion of glucans to Glc and pulsed-amperometric detection (HPAEC-ENZ-PAD; Wong and Jane, 1995, 1997). An unusual result was obtained specifically from samples expressing SBEIIb alone, in that an unidentified molecule(s) significantly increased the baseline PAD signal in the region where chains with degree of polymerization (DP) of 13 and larger normally elute (data not shown). The same PAD profile was obtained even after the glucan sample was digested completely to Glc by amyloglucosidase before HPAEC-ENZ-PAD analysis (data not shown). Thus, a nonglucan molecule copurifies with the glucan specifically from yeast strains expressing SBEIIb. This molecule was not observed in the SBEIIa + SBEIIb strain, which is another indication of the phenotypic dominance of SBEIIa.
The mechanical disruption method of glucan isolation, which includes boiling the solution in 1% (w/v) SDS and Sepharose CL-2B as the gel permeation matrix, yielded a glucan from all strains that gave the expected HPAEC-ENZ-PAD baseline and, thus, was not associated with the nonglucan contaminant. Further analysis of glucans from the yeast strains expressing maize SBEs was performed on the material in the peak fractions of the Sepharose CL-2B chromatographs (Fig. 2). An exception is nonmutant yeast glycogen, which was prepared by the hot alkali procedure.
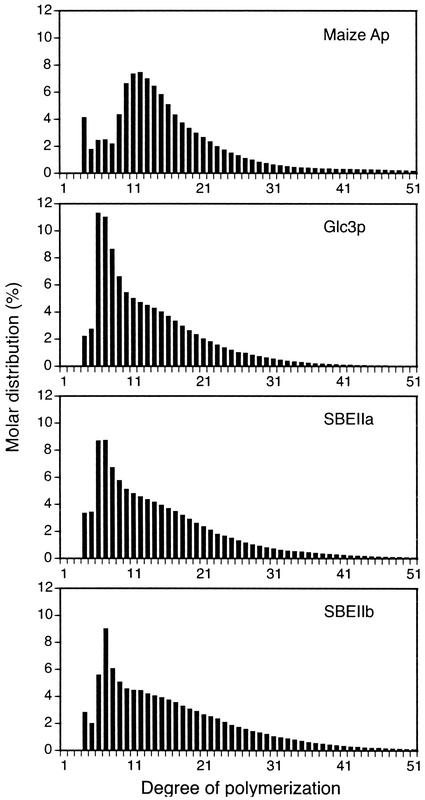
Sample data from the HPAEC-ENZ-PAD analysis, which affords quantitative determination of the molar frequency of linear chains of specific length, are shown in Figure 3. Data are shown only for chains of DP4 or greater, because the quantities of Glc, maltose, and maltotriose were not reproducible. For easier visualization of differences in these frequency distributions, samples were compared pair wise by subtracting the value for each chain length of one sample from the other, and the results were plotted as a function of DP. In this analysis, two nearly identical samples would yield a bar graph with only very slight variations from the baseline.
Figure 3.
Chain-length distributions. Size-fractionated glucans from the indicated yeast strains were completely debranched, and the resultant linear chains were separated and quantified by HPAED-ENZ-PAD. The quantitative distribution of each chain length up to DP50 is plotted. Representative data of the type used to generate the subsequent difference plots are shown. Data are shown for commercial maize Ap and for size-fractionated glucans isolated by the mechanical disruption procedure from yeast strains in which the indicated GBE or SBE is the only glucan-branching enzyme present in otherwise congenic strains. The yeast strains are as follows (refer to Table I): Glc3p, Y-2159; SBEIIa, BSY-03; SBEIIb, BSY-04.
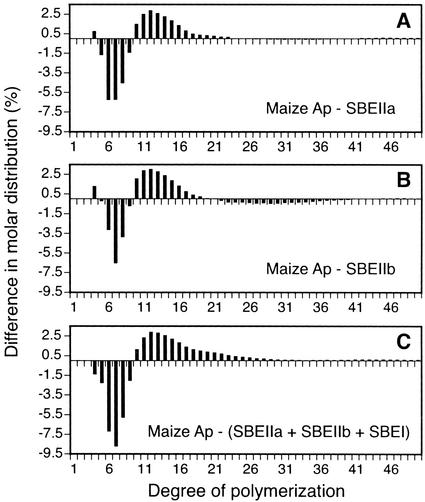
Figure 4 compares the structure of native maize Ap to those obtained from the maize SBEs working in combination with the yeast GSs. These data illustrate that maize SBEs are not entirely responsible for the Ap chain lengths, confirming the conclusion reached previously when SBEIIb and SBEI were co-expressed in E. coli (Guan et al., 1995). Here, the similar approach was extended to examine the effects of expressing all three known maize SBEs in a heterologous system. Thus, Figure 4C can be considered as a comparison between the three maize SBEs working in combination with yeast GSs and the condition in which the same three isozymes are working together with the full complement of maize SSs.
Figure 4.
Chain-length distribution comparisons using commercial maize Ap as a standard. For each chain length, the quantitative molar distribution in the polymer from the yeast strain expressing the indicated SBE or combination of SBEs was subtracted from the corresponding value obtained for commercial maize Ap. A, Chain-length frequencies in commercial maize Ap compared with those in the glucans from strain BSY-03 (SBEIIa). B, Chain-length frequencies in commercial maize Ap compared with those in the glucans from strain BSY-04 (SBEIIb). C, Chain-length frequencies in commercial maize Ap compared with those in the glucans from strain BSY-08 (SBEI + SBEIIa + SBEIIb).
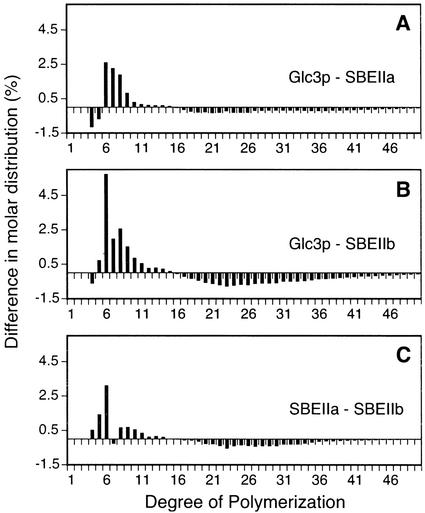
Figure 5 shows the differences in chain-length distribution that resulted when the specific branching enzymes present were varied in cells containing the native complement of GSs. Comparison of the product of the yeast GBE with that of maize SBEIIb reveals significantly more short chains (DP5–DP11) produced by Glc3p and fewer longer chains (DP18–DP36; Fig. 5B). A similar but less-pronounced effect is seen in comparison of the native GBE product with that of maize SBEIIa (Fig. 5A). Comparison of the SBEIIa product with that of SBEIIb reveals significantly more short chains, especially DP5 and DP6, produced by SBEIIb (Fig. 5C). These differences may account for the distinct colony stain colors and absorption spectra (Fig. 1).
Figure 5.
Chain-length distribution comparisons between glucans produced by various single branching enzymes. For each chain length, the molar distribution in the polymer produced by the second indicated branching enzyme in the pair was subtracted from the value obtained for the polymer produced by the first branching enzyme in the pair. A, Chain-length frequencies in the glucans from strain Y-2159 (Glc3p) compared with those from strain BSY-03 (SBEIIa). B, Chain-length frequencies in the glucans from strain Y-2159 (Glc3p) compared with those from strain BSY-04 (SBEIIb). C, Chain-length frequencies in the glucans from strain BSY-03 (SBEIIa) compared with those from strain BSY-04 (SBEIIb).
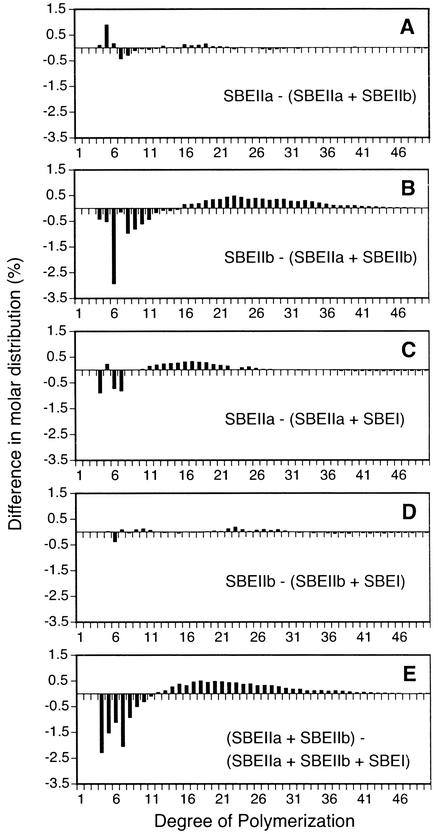
In Figure 6 the effects of varying the combination of maize SBEs are shown. The SBEIIa + SBEIIb combination produces a glucan virtually identical to that produced by SBEIIa alone (Fig. 6A). In contrast, the SBEIIa + SBEIIb product is quite distinct from that produced by SBEIIb alone (Fig. 6B). Thus, the dominance of SBEIIa over SBEIIb observed in the colony stains (Fig. 1A) and absorption spectra (Fig. 1B) is also reflected at the level of chain-length distribution.
Figure 6.
Chain-length distribution comparisons between glucans produced by various single SBE or combination of SBEs. For each chain length, the molar distribution in the polymer produced by the second indicated SBE or combination thereof was subtracted from the value obtained for the polymer produced by the first SBE or combination of SBEs. A, Chain-length frequencies in the glucans from strain BSY-03 (SBEIIa) compared with those from strain BSY-07 (SBEIIa + SBEIIb). B, Chain-length frequencies in the glucans from strain BSY-04 (SBEIIb) compared with those from strain BSY-07 (SBEIIa + SBEIIb). C, Chain-length frequencies in the glucans from strain BSY-03 (SBEIIa) compared with those from strain BSY-05 (SBEIIa + SBEI). D, Chain-length frequencies in the glucans from strain BSY-04 (SBEIIb) compared with those from strain BSY-06 (SBEIIb + SBEI). E, Chain-length frequencies in the glucans from strain BSY-07 (SBEIIa + SBEIIb) compared with those from strain BSY-08 (SBEIIa + SBEIIb + SBEI).
Addition of SBEI has virtually no effect on the structure of the glucans produced by SBEIIb (Fig. 6D) and only a slight effect on the SBEIIa product (Fig. 6C). A significant effect, however, was observed when all three maize SBEs were present compared with the product of the SBEIIa + SBEIIb combination (Fig. 6E). Thus, SBEI clearly is able to exert an influence on the structure of the glucan produced in yeast cells, but apparently it does so readily only when the other two SBEs are also present.
Branch Linkage Frequency
Branch linkage frequencies were determined for each glucan sample by comparing the quantity of free-reducing ends obtained after specific hydrolysis of α(1→6) bonds to that observed after complete hydrolysis of both α(1→6) and α(1→4) linkages. Table II shows the results obtained from an Ap standard and from yeast glucans isolated by the mechanical disruption procedure and size-fractionated by chromatography on Sepharose CL-2B. As expected, the native yeast glycogen branch frequency was significantly higher than that observed for maize Ap. In general the glucans produced by maize SBEs, when expressed either alone or in combination, exhibited values intermediate between those obtained for maize Ap and wild-type yeast glycogen. Thus, the SBEs by themselves cannot be the sole determinants of branch linkage frequency in Ap.
Table II.
Frequency of α(1→6) glycosidic bonds in α-glucans
| Source | SBE(s) or GBE Present | Branch Frequency |
|---|---|---|
| Maize Apa | Native maize | 4.77 |
| Y-2159 | Glc3p | 7.28 |
| BSY-03 | SBEIIa | 5.53 |
| BSY-04 | SBEIIb | 5.91 |
| BSY-05 | SBEIIa + SBEI | 6.18 |
| BSY-06 | SBEIIb + BEI | 5.31 |
| BSY-07 | SBEIIa + SBEIIb | 5.32 |
| BSY-08 | SBEIIa + SBEIIb + SBEI | 6.88 |
A-7780, lot no. 53H0260 (Sigma).
A comparison of the results obtained for strains BSY-07 and BSY-08 indicates that SBEI can have a significant effect on the branch linkage frequency if SBEIIa and SBEIIb are also present. In this instance, SBEI accounted for an increase of approximately 1.5% in the branch frequency. SBEI caused a slighter effect on this parameter if only SBEIIa is present in addition (compare strains BSY-03 and BSY-05). If only SBEIIb is present in addition, then SBEI expression resulted in a decrease in the observed branch frequency (compare strains BSY-04 and BSY-06).
DISCUSSION
Expression of maize SBEs in yeast confirmed the previous observation (Guan et al., 1995) that these enzymes are not entirely responsible for the chain-length distribution of Ap. This was observed even when all three known SBEs were present. Various synergistic effects involving yeast and maize SBEs were observed. (a) Yeast GBE is needed for accumulation of iodine-stainable glucans, even though GS is active. In the absence of Glc3p, maize SBEIIa and SBEIIb provide the function necessary for glucans to accumulate. SBEI, however, did not support glucan accumulation. (b) SBEIIa and SBEIIb produced glucans with distinct chain-length distributions and iodine complex absorption spectra. Expression of these two isoforms together resulted in SBEIIa-specific characteristics. (c) Glucans produced in yeast by maize SBEIIa or SBEIIb were larger than those of wild-type cells, indicating a role of SBEs in determining Mr. (d) SBEI can have a significant effect on chain-length distribution, branch linkage frequency, and the absorption spectrum, particularly when both SBEIIa and SBEIIb are present. These dominant effects prove that active SBEI is present and rule out lack of expression as an explanation for the fact that SBEI alone does not support glucan accumulation.
To explain these observations, we suggest that in this heterologous system, there is a sequential action of SBEs on glucan polymers under construction. After initiation, the GSs must elongate the polymer to an extent such that SBEs are able to catalyze chain cleavage and formation of branch linkages. This activity provides additional nonreducing ends for further action of the synthases. Some degree of GBE function may be needed in yeast to prepare an intermediate polymer that serves as an effective substrate for GS. We suggest that GS prepares the substrate for SBE and SBE prepares the substrate for GS in a cyclic relationship. SBEI does not support glucan accumulation, possibly because the initial precursor produced by the GSs is not a suitable substrate for this isoform, thus, breaking the cycle. In vitro, SBEI transfers longer chains than does SBEIIb (Takeda et al., 1993). It is possible that, in the absence of GBE, GS cannot produce polymers of sufficient length to serve as a SBEI substrate.
The fact that the SBE identity specifically effects the Mr of the product glucans is consistent with the hypothesis that SBEs influence activity of GSs. SBEs do not add any Glc units to the polymer; they only catalyze rearrangement of the existing polymerized chains. Continued elongation by GSs may be possible using the intermediate structures produced by the maize SBEs, whereas the products of the native Glc3p may cease to be suitable substrates of GS at a relatively earlier point in the construction process.
To explain the fact that SBEI effects were most noticeable when both SBEIIa and SBEIIb also were present, we suggest that the SBEII isoforms act on precursor polymers before SBEI. This idea is supported by the observation that the addition of SBEI to the SBEIIa + SBEIIb combination significantly increased the branch frequency. The action of the SBEII isoforms may result in a structure that is required of a substrate for SBEI. As an alternative, SBEII action may be needed to allow GS to extend the chains to reach the length required for SBEI. The concept that SBEIIa and/or SBEIIb activity is required for SBEI to have an effect on glucan structure, at least in this heterologous system, is a major finding from this study.
The results of this study with regard to SBEI activity are different from those obtained in previous studies using E. coli as the heterologous host system (Kossmann et al., 1991; Guan et al., 1995). In those instances, various plant SBEs, including SBEI from maize, were able to support the synthesis in of a glycogen-like polysaccharide in an E. coli host lacking a functional glgB gene, which codes for the GBE. A simple explanation for the difference between the two host species may be that the very low level of expression of SBEI in yeast (Table I) is insufficient to support glucan synthesis. The effect of expressing SBEI in the presence of SBEIIa and SBEIIb (Fig. 6E; Table II), however, argues that there is sufficient SBEI activity to have a significant impact on the branching pattern and branch linkage frequency of the glucan product. Another possibility, which is consistent with the synergistic effects observed in this study, is that SBEI interacts differently with the E. coli GS than it does with the yeast form of this enzyme. For example, the bacterial enzyme may be able to synthesize longer chains that are suitable as SBEI substrates, or SBEI activity may produce a precursor that is particularly suitable for E. coli GS but not the synthase from yeast.
SBEIIa dominating over SBEIIb with regard to the glucan structure when both enzymes are present may be explained by an ability of SBEIIa to act on outer chains to a greater extent than does SBEIIb. The two isoforms produce glucans that are similar to each other except for an excess of DP5 and DP6 in the SBEIIa product. We suggest that SBEIIa and SBEIIb have overlapping functions and properties during the earlier stages of glucan construction. If SBEIIa, however, can retain its activity toward a precursor glucan on which SBEIIb can no longer act, then mixing the two enzymes would result in the same final product as that formed by SBEIIa alone. The differences in DP distribution, especially if they are in the outer chains of the molecule, could result in different colors and spectra. This hypothesis appears to be inconsistent with the observation that loss the of SBEIIa in maize endosperm has no effect on Ap structure (Blauth et al., 2001). The native environment, however, differs from yeast in many regards including the complement of SSs and the fact that the polymers crystallize only in plants.
The preceding hypothesis is based on the presumption that SBEs and GSs act independently and that the observed synergistic effects result from substrate alterations in a sequential construction process. As an alternative, the activity of one enzyme may be directly dependent on the physical presence of another. The glucan structure produced by SBEIIb + SBEIIa, in terms of chain-length distribution and absorbance spectrum, is the same as that produced by SBEIIa alone. When SBEI is added, however, it affects the product structure much more significantly when both SBEIIa and SBEIIb are present than when only SBEIIa is operative. Thus, substrate structure may not be the only determinant of whether SBEI can act. The presence of the SBEIIa and SBEIIb proteins possibly has some direct effect on SBEI activity.
The requirement of a SBE for glucan accumulation could also result from direct interaction, in this instance between GS and GBE. This hypothesis has been suggested previously (Cannon et al., 1994), based on the fact that glc3 point mutations allow accumulation of an abnormal glucan, whereas deletions cause loss of detectable glycogen. In addition, certain point mutations of glc3 are partially dominant to wild type, again suggestive of multisubunit interactions within the biosynthetic system.
The results obtained from this heterologous system have advanced our understanding of the high level of complexity of what, at first consideration, could be viewed as a relatively straightforward biosynthetic system. Additional combinations of maize enzymes, in particular isolating specific SSs with particular SBEs, are likely to shed further light on the roles of each specific isoform.
MATERIALS AND METHODS
Recombinant Plasmids
cDNAs coding for SBEI and SBEIIb were obtained from plasmids pET23d-MBEI (Guan et al., 1994a) and pET23d-MBEII (Guan et al., 1994b). A SBEIIa cDNA clone was provided by Dr. M. Guiltinan (Pennsylvania State University, University Park). cDNAs were cloned in integrative vectors (pYX, Novagen, Madison, WI) containing the TPI1 promoter and specific selectable markers to form pBEI-022 (HIS3), pBEIIa-012 (URA3), and pBEIIb-042 (LEU2). Transcription initiates within the TPI1 promoter. The cDNAs were cloned into the expression vectors such that the amino terminus of each expressed SBE matches that of the mature protein within amyloplasts (Fisher et al., 1995; Gao et al., 1997). All expressed SBEs extend to the native C terminus. Plasmids were linearized by restriction enzyme cleavage within the marker gene before yeast transformation.
Strains and Growth Conditions
Standard methods were used for maintaining yeast cultures, constructing strains by transformation, and genetic analysis (Ausubel et al., 1989). Yeast strains in this study (Table III) are congenic in the D273–10B/A1 genetic background (Tzagoloff et al., 1975). The glc3::KANR strain BSY-01 was constructed from nonmutant Y-2159 by replacing the entire GLC3 open reading frame with the kanamycin resistance gene kanr from plasmid pFA6-kanMX4 (Wach et al., 1994). The gene replacement in the yeast chromosome was confirmed by PCR. SBE-expressing strains were obtained by transforming BSY-01. Yeast was grown at 30°C on YPD complete medium or synthetic dextrose selective medium supplemented as required. YPD plus 200 mg L−1 geneticin was used to select transformants. YPD liquid cultures for glucan isolation were inoculated with 0.1 volume of a saturated starter culture and grown in a 30°C shaker.
Table III.
Brewer's yeast strains used in this study
| Strain | Relevant Genotype | Derivation |
|---|---|---|
| Y-2159 | Matα his3 ura3 leu2 | Five backcrosses to D273-10B/A1 (Tzagoloff et al., 1975) |
| Y-2158 | Matα his3 ura3 leu2 | Same as Y-2159 |
| BSY-01 | MATα his3 ura3 leu2 glc3∷KANR | Transformation of Y-2159 |
| BSY-02 | MATα his3 ura3 leu2 glc3∷KANR his3∷SBEI∷HIS3 | Transformation of BSY-01 |
| BSY-03 | MATα his3 ura3 leu2 glc3∷KANR ura3∷SBEIIa∷URA3 | Transformation of BSY-01 |
| BSY-04 | MATα his3 ura3 leu2 glc3∷KANR leu2∷SBEIIb∷LEU2 | Transformation of BSY-01 |
| BSY-05 | MATα his3 ura3 leu2 glc3∷KANR his3∷SBEI∷HIS3 ura3∷SBEIIα∷URA3 | Transformation of BSY-02 |
| BSY-06 | MATα his3 ura3 leu2 glc3∷KANR his3∷SBEI∷HIS3 leu2∷SBEIIb∷LEU2 | Transformation of BSY-02 |
| BSY-07 | MATα his3 ura3 leu2 glc3∷KANR ura3∷SBEIIa∷URA3 leu2∷SBEIIb∷LEU2 | Transformation of BSY-03 |
| BSY-08 | MATα his3 ura3 leu2 glc3∷KANR his3∷SBEI∷HIS3 ura3∷SBEIIa∷URA3 leu2∷SBEIIb∷LEU2 | Transformation of BSY-06 |
Purification of α-Glucan Polymers
Cells were collected from culture by centrifugation (5,000g, 4°C, 5 min) and washed twice with water. The two different glucan purification schemes used are described as follows.
The hot alkali procedure described previously (Gunja-Smith et al., 1977) was used as follows. Cells were suspended in 20% (w/v) KOH at 0.05 to 0.40 g wet weight mL−1, boiled for 1 h, cooled to room temperature, and adjusted to pH 6 to 7 with concentrated HCl. Two volumes of 100% (v/v) ethanol was added. Precipitates were collected by centrifugation at 5,000g, 4°C, for 5 min and washed twice with 67% (v/v) ethanol. These were suspended in water and then heated and vortexed to obtain a fine suspension. The suspension was centrifuged at 8,000g, 4°C, for 50 min. The supernatant was mixed with 1.5 volumes of 100% (v/v) ethanol. After 20 min at 0°C, the mixture was centrifuged at 5,000g, 4°C, for 10 min. The pellet was suspended in 2 mL of water, heated briefly at 80°C, cooled to room temperature, and then adjusted to 10 mm NaOH. The solution was applied to a TSK-HW50S gel permeation column (1.7 × 80 cm, TosoHaas, Montgomeryville, PA), which was eluted with 10 mm NaOH (10 mL h−1; 2.5-mL fractions). Samples of each fraction (0.15 mL) were mixed with 0.3 mL of 0.01 m I2/0.5 m KI in 1 mL of total volume. Glucan-containing fractions indicated by dark color, typically four or five in each fractionation, were pooled, dialyzed into water, and lyophilized.
A mechanical disruption procedure also was employed. Cell pellets from 500-mL cultures were frozen in liquid N2, lyophilized, ground to powder in a mortar and pestle, suspended in 15 mL of 1% (w/v) SDS, and shaken for 15 min. The lysate was centrifuged at 4°C, 5,000g, for 5 min, and this pellet fraction “a” was saved. The supernatant was boiled for 15 min, cooled to room temperature, and centrifuged. This pellet fraction “b” also was saved. The supernatant was mixed with 3 to 4 volumes of cold ethanol, and the mixture was kept on ice for 10 min. The pellet obtained after centrifugation was suspended in 100% (v/v) dimethyl sulfoxide, boiled for 30 min, and cooled to room temperature. After centrifugation, the supernatant fraction “c” was saved. Pellet fractions a and b were re-extracted with 100% (v/v) dimethyl sulfoxide, pooled, and centrifuged to yield supernatant fraction “d.” Supernatants c and d were pooled and dialyzed into water. Three to 4 volumes of 100% (v/v) ethanol was added, and the mixture was kept overnight at 4°C. The precipitate was collected by centrifugation at 7,000g, 4°C, for 10 min. Glucans in the pellet were suspended in 10 mm NaOH and chromatographed as above, except the matrix was Sepharose CL-2B and the column size was 2.8 × 21 cm.
Analysis of α-Glucan Polymers
Absorbance spectra of glucan-iodine complexes were determined by mixing 100 to 200 μg of purified glucans with 150 μL of I2/KI solution in a total volume of 1 mL. A microtitre plate assay was used to quantify glucans. Samples (30 μL) were mixed with 30 μL of 100 mm sodium acetate, pH 5.0, containing 0.012 unit of Aspergillus niger amyloglucosidase (Megazyme International, Bray, County Wicklow, Ireland) and incubated at 37°C for 2 h. Free Glc was then quantified by the Glc oxidase method, using a commercial reagent kit (510A, Sigma, St. Louis).
Chain-length distributions were determined by HPAEC-ENZ-PAD (Wong and Jane, 1995, 1997). Glucans (1 mg) were debranched at 45°C for 90 min in a 1-mL volume of 10 mm sodium acetate, pH 4, containing 1 μL of Pseudomonas amyloderamosa isoamylase (0.2 unit, Megazyme). Test analyses showed no significant difference in the chain-length distributions obtained from the 90-min digestion compared with the data obtained after 24 h of isoamylase treatment, indicating that the debranching reactions had proceeded to completion (data not shown). The debranching reactions were neutralized by the addition of 1.4 μL of 6.5 m NaOH, boiled for 10 min, and centrifuged in a microfuge for 10 min. A sample of 0.7 mL of the neutralized digest was applied for HPAEC-ENZ-PAD analysis. The mass values indicated by PAD peak areas were divided by the Mr of each chain, yielding relative molar values. Data are presented for each chain length as the percentage of the total molar value summed from DP1 to DP50. The native maize Ap analyzed was a commercial preparation (A7780, lot no. 53H0260, Sigma). Ap and glycogen from wild-type yeast were analyzed independently four times, and the experimental samples were all analyzed at least twice. In no instance was a significant difference observed in chain-length distribution between the independent analyses of each sample.
Branch frequency was determined from the reducing end concentration after hydrolysis of α(1→6) bonds by isoamylase and compared with that after hydrolysis of α(1→6) and α(1→4) bonds by amyloglucosidase (Fox and Robyt, 1991). Maltose was used as the reducing sugar standard. Debranching reactions were the same as those used for HPAEC-ENZ-PAD, and amyloglucosidase digestions were the same as those used to quantify total glucan.
Branching Enzyme Assays
Yeast cultures were grown in 10 mL of YPD medium for 17 h at 30°C, reaching early stationary phase (A600 > 2.5). Cells were collected by centrifugation, washed in water, suspended in 1 mL of breaking buffer (20 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 5% [v/v] glycerol, 0.3 m ammonium sulfate, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [P2714, Sigma]), and broken by vortexing in the presence of glass beads. The supernatant was collected after centrifugation for 10 min in a microfuge, and the protein concentration was determined. Branching enzyme assays were performed as follows, according to the method of Guan and Preiss (1993). The 100-μL reaction volume contained 2 mg mL−1 phosphorylase a, 50 mm [14C]Glc-1-P (9 cpm nmol−1), 1 mm ATP, and 40 μg of total soluble extract. Reactions were incubated for 5 h at 30°C, after which 100 μL of a 10 mg mL−1 solution of glycogen was added as carrier and glucan polymers were precipitated with methanol. Incorporation of radioactivity into the glucans was quantified by liquid-scintillation counting. Each yeast strain was analyzed two or three times starting with separate cultures.
ACKNOWLEDGMENTS
We thank Jay-lin Jane for providing the HPAEC-ENZ-PAD apparatus and Tracie Bierwagen for performing the tetrad analysis. We also thank Dr. Hanping Guan and Dr. Mark Guiltinan for providing cDNA clones.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–9982555 to A.M.M.). This article is journal paper no. J–14,349 of project no. 3,593 of the Iowa Agriculture and Home Economics Experiment Station (Ames).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010756.
LITERATURE CITED
- Alonso MD, Lomako J, Lomako WM, Whelan WJ. A new look at the biogenesis of glycogen. FASEB J. 1995;9:1126–1137. doi: 10.1096/fasebj.9.12.7672505. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1989. [Google Scholar]
- Blauth SL, Yao Y, Klucinec JD, Shannon JC, Thompson DB, Guiltinan MJ. Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol. 2001;125:1396–1405. doi: 10.1104/pp.125.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer CD, Preiss J. Multiple forms of (1,4)-α-d-glucan-6-glucosyl transferase from developing Zea mays L. kernels. Carbohydr Res. 1978a;61:321–334. [Google Scholar]
- Boyer CD, Preiss J. Multiple forms of starch branching enzyme of maize: evidence for independent genetic control. Biochem Biophys Res Commun. 1978b;80:169–175. doi: 10.1016/0006-291x(78)91119-1. [DOI] [PubMed] [Google Scholar]
- Calder PC. Glycogen structure and biogenesis. Int J Biochem. 1991;23:1335–1352. doi: 10.1016/0020-711x(91)90274-q. [DOI] [PubMed] [Google Scholar]
- Cannon JF, Pringle JR, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM. Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol. 1999;120:205–216. doi: 10.1104/pp.120.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I, Hardy TA, Goebl MG, Roach PJ. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991;266:15602–15607. [PubMed] [Google Scholar]
- Fisher DK, Boyer CD, Hannah LC. Starch branching enzyme II from maize endosperm. Plant Physiol. 1993;102:1045–1046. doi: 10.1104/pp.102.3.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DK, Kim K-N, Gao M, Boyer CD, Guiltinan MJ. A cDNA encoding starch branching enzyme I from maize endosperm. Plant Physiol. 1995;108:1314. doi: 10.1104/pp.108.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JD, Robyt JF. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal Biochem. 1991;195:93–96. doi: 10.1016/0003-2697(91)90300-i. [DOI] [PubMed] [Google Scholar]
- Gao M, Fisher DK, Kim K-N, Shannon JC, Guiltinan MJ. Independent genetic control of maize starch-branching enzymes IIa and IIb. Plant Physiol. 1997;114:69–78. doi: 10.1104/pp.114.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wanat J, Stinard PS, James MG, Myers AM. Characterization of dull1, a maize gene coding for a novel starch synthase. Plant Cell. 1998;10:339–412. doi: 10.1105/tpc.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Baba T, Preiss J. Expression of branching enzyme I of maize endosperm in Escherichia coli. Plant Physiol. 1994a;104:1449–1453. doi: 10.1104/pp.104.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Baba T, Preiss J. Expression of branching enzyme II of maize endosperm in Escherichia coli. Cell Mol Biol. 1994b;40:981–985. [PubMed] [Google Scholar]
- Guan H, Kuriki T, Sivak M, Preiss J. Maize branching enzyme catalyzes synthesis of glycogen-like polysaccharide in glgB-deficient E. coli. Proc Natl Acad Sci USA. 1995;92:964–967. doi: 10.1073/pnas.92.4.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H, Preiss J. Differentiation of the properties of the branching isozymes from maize (Zea mays) Plant Physiol. 1993;102:1269–1273. doi: 10.1104/pp.102.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunja-Smith Z, Patil NB, Smith EE. Two pools of glycogen in Saccharomyces. J Bacteriol. 1977;130:818–825. doi: 10.1128/jb.130.2.818-825.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PJ, Cameron RE, Donald AM. A universal feature in the starch granules from different botanical sources. Starke. 1993;45:417–420. [Google Scholar]
- Kossmann J, Visser RGF, Muller-Rober B, Willmitzer L, Sonnewald U. Cloning and expression analysis of a potato cDNA that encodes branching enzyme: evidence for co-expression of starch biosynthetic genes. Mol Gen Genet. 1991;230:39–44. doi: 10.1007/BF00290648. [DOI] [PubMed] [Google Scholar]
- Li Z, Chu X, Mouille G, Yan L, Kosar-Hashemi B, Hey S, Napier J, Shewry P, Clarke B, Appels R et al. The localization and expression of the class II starch synthases of wheat. Plant Physiol. 1999;120:1147–1156. doi: 10.1104/pp.120.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomako J, Lomako WM, Whelan WJ, Dombro RS, Neary JT, Norenberg MD. Glycogen synthesis in the astrocyte: from glycogenin to proglycogen to glycogen. FASEB J. 1993;7:1386–1393. doi: 10.1096/fasebj.7.14.8224611. [DOI] [PubMed] [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress towards understanding biosynthesis of the amylopectin crystal. Plant Physiol. 2000;122:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen DW, Meinke M, LaPorte DC. GLC3 and GHA1 of Saccharomyces cerevisiae are allelic and encode the glycogen branching enzyme. Mol Cell Biol. 1992;12:22–29. doi: 10.1128/mcb.12.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JC, Garwood DL. Genetics and physiology of starch development. In: Whistler RL, BeMiller JN, Paschall EF, editors. Starch: Chemistry and Technology. San Diego: Academic Press; 1984. pp. 25–86. [Google Scholar]
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:67–87. doi: 10.1146/annurev.arplant.48.1.67. [DOI] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS. Genetic isolation, cloning, and analysis of a Mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell. 1993;5:1555–1566. doi: 10.1105/tpc.5.11.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Guan H, Preiss J. Branching of amylose by the branching isozymes of maize endosperm. Carbohydr Res. 1993;240:253–263. [Google Scholar]
- Thon VJ, Vigneron-Lesens C, Marianne-Pepin T, Montreuil J, Decq A, Rachez C, Ball SG, Cannon JF. Coordinate regulation of glycogen metabolism in the yeast Saccharomyces cerevisiae: induction of glycogen branching enzyme. J Biol Chem. 1992;267:15224–15228. [PubMed] [Google Scholar]
- Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system: characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wong KS, Jane J. Effects of pushing agents on the separation and detection of branched amylopectin by high performance anion exchange chromatography with pulsed amperometric detection. J Liq Chromatogr. 1995;18:63–80. [Google Scholar]
- Wong K-S, Jane J-L. Quantitative analysis of debranched amylopectin by HPAEC-PAD with postcolumn enzyme reactor. J Liq Chromatogr. 1997;20:297–310. [Google Scholar]
- Zeeman SC, Umemoto T, Lue W-L, Au-Yeung P, Martin C, Smith AM, Chen J. A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell. 1998;10:1699–1712. doi: 10.1105/tpc.10.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]