Abstract
Rice (Oryza sativa) accumulates prolamins and glutelins as storage proteins. The latter storage protein is synthesized on the endoplasmic reticulum (ER) as a 57-kD proglutelin precursor, which is then processed into acidic and basic subunits in the protein storage vacuole. Three esp2 mutants, CM1787, EM44, and EM747, contain larger amounts of the 57-kD polypeptide and corresponding lower levels of acidic and basic glutelin subunits than normal. Electron microscopic observation revealed that esp2 contained normal-appearing glutelin-containing protein bodies (PB-II), but lacked the normal prolamin-containing PB (PB-I). Instead, numerous small ER-derived PBs of uniform size (0.5 μm in diameter) and low electron density were readily observed. Immunoblot analysis of purified subcellular fractions and immunocytochemistry at the electron microscopy level showed that these new PBs contained the 57-kD proglutelin precursor and prolamin polypeptides. The 57-kD proglutelin was extracted with 1% (v/v) lactic acid solution only after removal of cysteine-rich prolamin polypeptides, suggesting that these proteins form glutelin-prolamin aggregates via interchain disulfide bonds within the ER lumen. The endosperm of esp2 mutants contains the lumenal chaperones, binding protein and calnexin, but lacks protein disulfide isomerase (PDI) at the protein and RNA levels. The transcript of PDI was expressed in the seed only during the early stage of seed development in the wild type. These results suggest that PDI plays an essential role in the segregation of proglutelin and prolamin polypeptides within the ER lumen.
All plants utilize storage proteins as a reserve of nitrogen, sulfur, and carbon in the form of salt-soluble globulins or alcohol-soluble prolamins (Shewry and Casey, 1999). In addition to the alcohol-soluble prolamins typically found in cereals (Shewry and Tatham, 1999), rice (Oryza sativa) also accumulates glutelins, which are proteins homologous to the 11S globulin of soybean (Glycine max) and pea (Pisum sativum; Zhao et al., 1983; Takaiwa et al., 1987; Shotwell and Larkins, 1989). Although both storage proteins are initially synthesized on the endoplasmic reticulum (ER) membrane (Yamagata et al., 1982) and are translocated into the ER lumen, they are stored in morphologically distinct protein bodies (PB; Tanaka et al., 1980; Krishnan et al., 1986; Yamagata and Tanaka, 1986). Prolamins are stored as intracisternal inclusion granules within the ER lumen (PB-I), whereas glutelins are packaged in a protein storage vacuolar compartment (PB-II; Tanaka et al., 1980; Krishnan et al., 1986). These PBs are readily distinguishable at the light and electron microscopy levels. The prolamin-containing PB-I is spherical, with a diameter of about 1 to 2 μm, and exhibits concentric rings of varying electron density (Bechtel and Juliano, 1980; Tanaka et al., 1980; Krishnan et al., 1986; Yamagata and Tanaka, 1986). In contrast, the glutelin-containing PB-II is larger (3–4 μm), irregularly shaped, and of highly uniform staining density.
The cellular processes responsible for PB-I and PB-II formation are poorly understood. Although prolamin polypeptides are retained and assembled as intracisternal inclusion granules within the ER lumen, these proteins lack the usual ER retrieval/retention peptide signal at their C terminus. The rice prolamins may contain an unidentified ER retention signal like those proposed in a previous report to exist for the ER retention of other cereal prolamins (Altschuler et al., 1993). In an alternate manner, PB-I formation is the result of protein-protein interactions when the lumenal concentration of prolamins attains a critical level. Cellular processes have been identified that would elevate the local concentration of prolamin polypeptide within specific ER regions (subdomains). First, prolamin RNAs are not randomly distributed on the ER; rather, they are localized to the ER membranes that delimit PB-I (Li et al., 1993a; Choi et al., 2000). Second, although the lumenal chaperone binding protein (BiP) is an excellent marker for ER, it is not randomly distributed within this membrane complex in rice endosperm cells (Li et al., 1993b; Muench et al., 1997). Instead, BiP is highly enriched on the periphery of PB-I (Li et al., 1993b; Muench et al., 1997). BiP is associated with the nascent prolamin polypeptide chain associated with polysomes, with free prolamin polypeptides, and with the inclusion body itself, suggesting that this lumenal chaperone not only facilitates transport of the nascent polypeptide across the ER membrane and polypeptide folding, but also helps retain and assemble prolamins into an intracisternal inclusion granule (Li et al., 1993b; Okita and Rogers, 1996; Muench and Okita, 1997; Muench et al., 1997; Okita et al., 1998). On the other hand, glutelins are initially synthesized as a 57-kD precursor and are sorted into the vacuole, where they are proteolytically processed into acidic and basic subunits (Yamagata et al., 1982). Very little is known about the gene that regulates the biochemical and cellular processes responsible for the formation of PBs.
Rice mutants that show variations in the profile patterns of storage proteins have been identified and assigned into four classes (Kumamaru et al., 1988; Ogawa et al., 1989). Two classes, esp1 and Esp4 had modified prolamin protein profiles with apparent changes in glutelin content. A third class, esp3, contains reduced amounts of the Cys-rich prolamins without any effect on glutelin content. A fourth class, esp2, accumulates high amounts of the 57-kD polypeptide and corresponding low amounts of the glutelin acidic and basic subunits (Kumamaru et al., 1988). Because the phenotype of F1 and F2 seeds derived from a cross between esp2 and the wild type of rice do not show a gene dosage effect (Kumamaru et al., 1987), esp2 is likely not a structural gene of the 57-kD polypeptide, rather it is a gene involved in the folding and transport of 57-kD proglutelin from the ER to the vacuole or in the proteolytic processing of the 57-kD glutelin precursor into acidic and basic subunits.
In the present study, to clarify the function of the gene related to the pathway of glutelin biosynthesis, we analyzed the esp2 mutation characterized by a high elevation of the 57-kD polypeptide and a decrease of glutelin acidic and basic subunits.
RESULTS
The esp2 Mutant Accumulates Large Quantities of the 57-kD Glutelin Precursor
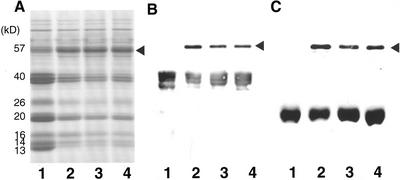
The major storage proteins resolved by SDS-PAGE of the rice variety Kinmaze consist of the 40-kD acidic and 20-kD basic glutelin subunits, 26-kD globulin polypeptide, and 16-, 14-, 13-, and 10-kD prolamin polypeptides (Tanaka et al., 1980; Fig. 1A). Small amounts of the 57-kD glutelin precursor are also present. Seed protein extracts from three esp2 mutant lines, CM1787, EM44, and EM747, which were generated independently, contain increased quantities of a 57-kD polypeptide (Fig. 1A, arrowhead) and corresponding reduced levels of the 40-kD acidic and 20-kD basic glutelin subunits in comparison with the parental line var. Kinmaze.
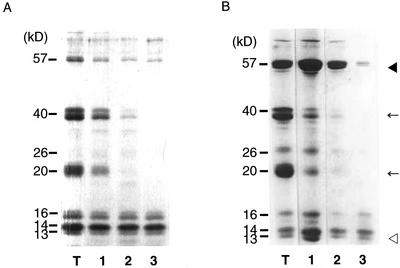
Figure 1.
Immunoblot analysis of the 57-kD polypeptide in the endosperm of wild-type var. Kinmaze and three esp2 mutants. Protein extracts from wild-type var. Kinmaze and from the esp2 mutants CM1787, EM44, and EM747 were separated by an SDS 15% to 25% (w/v) gradient gel and were stained with Coomassie Blue (A) or transferred to nitrocellulose membrane and incubated with glutelin acidic subunit antibody (B) and glutelin basic subunit antibody (C). The black arrowhead indicates the 57-kD polypeptide. Lane 1, Wild- type var. Kinmaze; lane 2, CM1787; lane 3, EM44; lane 4, EM747.
Glutelins are initially synthesized as a 57-kD precursor and are transported into the vacuole where they are proteolytically processed into acidic and basic subunits (Yamagata et al., 1982). To determine whether the 57-kD polypeptide in esp2 mutant is the glutelin precursor, we performed immunoblot analysis using monospecific antibodies against the 40-kD acidic or 20-kD basic glutelin subunits. As shown in Figure 1, B and C, the 57-kD polypeptide reacted strongly with both glutelin antibodies. There was almost no difference in the accumulation level of proglutelin among the three lines. These results indicate that the 57-kD polypeptide accumulated in esp2 mutants is the glutelin precursor.
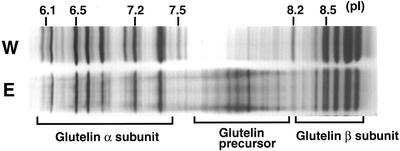
The glutelins are encoded by a multigene family consisting of at least six distinct classes (Takaiwa et al., 1999). To determine whether the esp2 mutation affected all of the glutelin polypeptides or only a few select species, we analyzed seed protein extracts from var. Kinmaze and esp2 by isoelectric focusing (IEF). Glutelin acidic and basic subunits from the parental line var. Kinmaze were separated into 11 and nine bands, respectively, by IEF analysis (Fig. 2). The esp2 mutant contained elevated amounts of six proglutelin polypeptide bands in the pI range of 7.8 to 8.2 in comparison with the wild type of rice (Fig. 2). The polypeptide bands of glutelin acidic and basic subunits were reduced almost uniformly in the esp2 mutant, indicating that the accumulation of 57-kD polypeptides is at the expense of glutelin subunits. These observations indicate that the esp2 mutation is not a glutelin structural gene (Kumamaru et al., 1987), but is likely a gene that affects the efficient processing of glutelin precursor to mature glutelin subunits.
Figure 2.
Analysis of the glutelin composition from Kinmaze (W) or esp2 (E) by IEF. The pI of the 57-kD polypeptide and glutelin acidic and basic subunits are depicted.
The Loss of PB-I and the Appearance of a New Type of PB in esp2
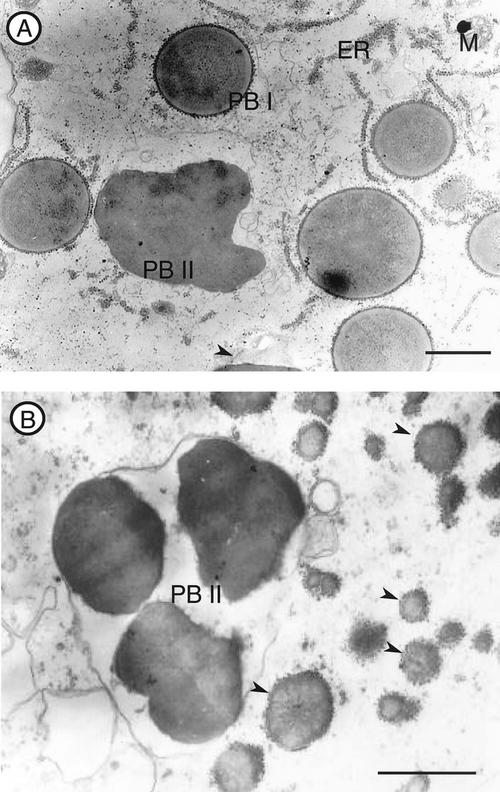
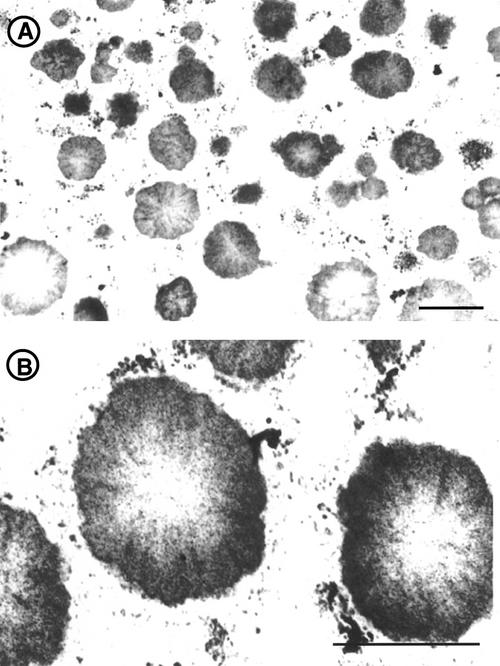
Developing endosperms were observed by an electron microscope to determine the deposition site of proglutelin in esp2. In the parent line var. Kinmaze, two types of PBs, PB-I and PB-II, are readily discernible (Fig. 3A). The prolamin containing PB-I is spherical, 1 to 2 μm in diameter, with a lamellar structure surrounded by rough ER. The glutelin-containing PB-II is a larger (3–4 μm in diameter), irregularly shaped structure that exhibits uniform staining (Tanaka et al., 1980; Krishnan et al., 1986).
Figure 3.
Electron microscopic observation of developing endosperm of Kinmaze (A) and esp2 (B). In esp2 endosperm, only PB-II is seen, along with replacement of PB-I by a new PB type (arrowhead). M, Mitochondria; bars = 1 μm.
Figure 3B depicts the types of PB observed in esp2. Normal-appearing PB-II, but not PB-I, can be seen. Instead of PB-I, a new type of PB is readily evident. Similar to PB-I, this new PB is spherical and has polysomes attached to the surface of it, but it is much smaller (0.5 μm in diameter) and lacks the lamellar structure in the typical PB-I. The attachment of the polysome on the surface of the PB suggests that the new type of PB is derived from the same ER that gave rise to PB-I.
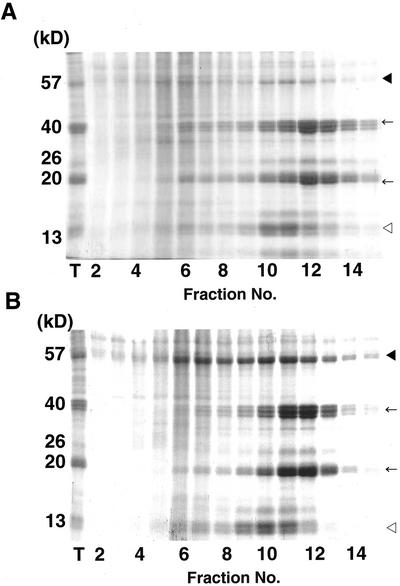
To obtain direct evidence for the deposition site of proglutelin, we resolved the PBs of an esp2 extract by Suc density gradient (SDG) centrifugation and we analyzed the fractions for their protein composition by SDS-PAGE (Fig. 4). The protein in wild-type rice was resolved into the prolamins peaking in fractions 10 through 12 and the glutelin subunits peaking in fractions 11 through 13, which corresponded to PB-I and PB-II, respectively, as reported by Tanaka et al. (1980). In esp2, the distribution of glutelin subunits and prolamin polypeptides was similar to the distribution in wild-type rice. The glutelin precursors showed a much wider distribution across the SDG, indicating that this PB containing the proglutelin possessed various densities.
Figure 4.
Resolution of PBs and membranes by SDG centrifugation. SDS-PAGE analysis of the fractions was obtained by SDG centrifugation. The black arrowhead, arrow, and white arrowhead indicate the 57-kD glutelin precursor, mature glutelin subunits, and prolamin polypeptides, respectively. A, Wild-type var. Kinmaze; B, esp2. T, Total protein.
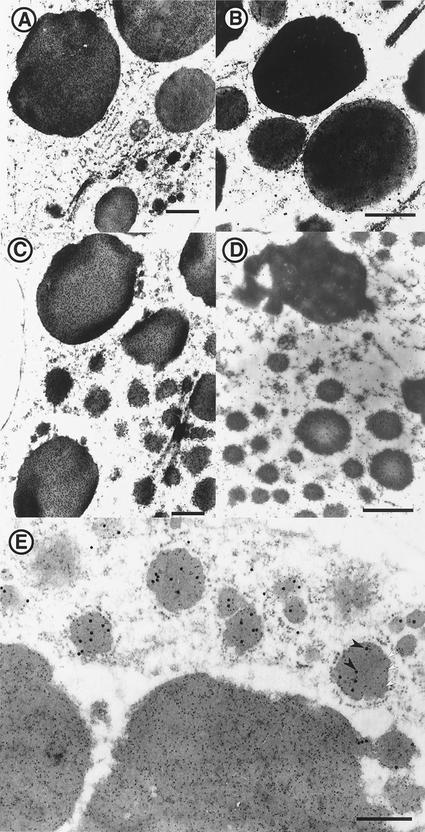
In addition to their physical features and protein composition, PB-I and PB-II can be readily distinguished by their sensitivity to pepsin digestion (Ogawa et al., 1987). Prolamin in PB-I is resistant to pepsin digestion, whereas glutelin in PB-II is readily digested by this protease in wild-type rice (Fig. 5A). To determine the sensitivity of the new PB observed in esp2, we treated PB fractions isolated from developing endosperm in esp2 with different concentrations of pepsin solution. As shown in Figure 5B, the glutelin precursor was much more resistant to pepsin digestion than were the glutelin subunits (lane 2), but it was less resistant than prolamin (lane 3). To determine the deposition site of the glutelin precursor, we analyzed the pepsin-treated PBs (Fig. 5B, lane 2) by transmission electron microscopy (Fig. 6). Only a single type of PB was evident, with morphology corresponding to the new type PB in Figure 3B. These results suggest that the glutelin precursor is accumulated in the mutant type PB with prolamin polypeptide.
Figure 5.
SDS-PAGE analysis of PB fractions treated with pepsin solution. A, Wild-type var. Kinmaze; B, esp2. T, Total seed proteins. Lane 1, Total PB fraction; lane 2, PB fraction treated with 10−4 mg mL−1 pepsin solution; lane 3, PB fraction treated by 1 mg mL−1 pepsin solution. The black arrowhead, arrow, and white arrowhead indicate the 57-kD glutelin precursor, mature glutelin subunits, and prolamin polypeptides, respectively.
Figure 6.
Electron microscopic observations of PB treated by pepsin solution of esp2 mutant. A, Bar = 1 μm; B, bar = 500 nm.
To obtain direct evidence for the colocalization of glutelin precursor and prolamin polypeptides in the mutant type PB, immunocytochemical studies were conducted using the antibodies for glutelin and prolamin, respectively. In the wild type of rice, PB-I and PB-II reacted with anti-glutelin antibody and anti-prolamin antibody, respectively (Fig. 7, A and B). The new type PB contained antigens that reacted with both antibodies (Fig. 7, C and D), and double immunolabeling showed that the new type PB reacted with both antibodies (Fig. 7E). These results clarified that glutelin precursors are deposited with prolamin polypeptides in one protein body, resulting in the conversion of PB-I into a new type PB.
Figure 7.
Immunogold labeling of developing endosperm by anti-glutelin and antiprolamin. A, Wild-type var. Kinmaze reacted with antiglutelin basic subunit antibody; B, Wild-type var. Kinmaze reacted with anti-13a prolamin polypeptide antibody; C, esp2 mutant reacted with antiglutelin basic subunit antibody; D, esp2 mutant reacted with anti-13a prolamin polypeptide antibody; E, esp2 mutant reacted with anti-13a prolamin polypeptide (white arrowhead) and antiglutelin basic subunit antibody (black arrowhead). Bars = 500 nm.
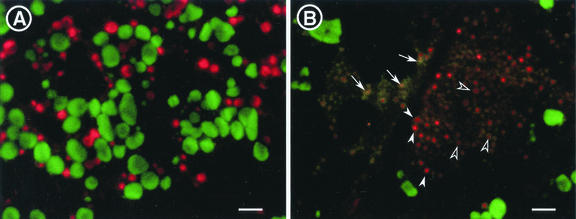
Double immunofluorescence studies were also conducted using secondary antibodies conjugated with FITC (glutelin) and rhodamine (prolamin) to analyze the deposition of the rice storage proteins (Fig. 8A). In the wild-type endosperm, the storage proteins are packaged into separate protein bodies, that is, in red spherical prolamin PBs and larger, irregularly shaped green glutelin PBs. However, in esp2, small orange signals were observed. The color of the small orange signals varied slightly from one instance to another, and we also observed grenadine particles (black arrowhead), dark-red particles (white arrowhead), and yellow particles (arrow). This result indicates that proglutelin and prolamin accumulates in the same PB, and that the ratio of proglutelin and prolamin in the PB varies among the PBs. This result agreed with the result of SDG centrifugation in which PBs containing proglutelin possessed various densities.
Figure 8.
Fluorescence microscopic observation of developing endosperm by antiglutelin and antiprolamin. A, Wild-type var. Kinmaze; B, esp2 mutant. In esp2 endosperm, grenadine signals (black arrowhead), dark-red signals (white arrowhead), and yellow signals (arrow) were observed. Bars = 3 μm.
The Glutelin Precursor Aggregated with Cys-Rich Prolamin
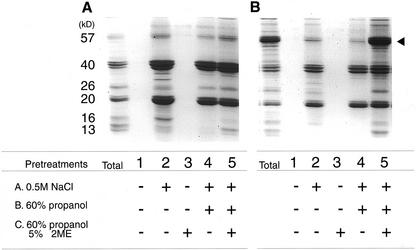
To determine the deposition state of proglutelin within the PBs in esp2, we examined the conditions for proglutelin extraction. The rice seed storage proteins, globulins, prolamins, and glutelins are readily resolved by selective extraction solution (Cagampang et al., 1966; Juliano, 1972). Glutelins were extracted only after the removal of globulins from endosperm tissue (Fig. 9A, lane 2). Glutelins were not extracted into 1% (v/v) lactic acid unless the seed was first treated with NaCl to extract the globulins (Fig. 9A, lane 1). In contrast, prior removal of the Cys-poor and Cys-rich prolamins had no effect on subsequent glutelin extraction (Fig. 9A, lanes 3 and 4). Given that glutelin and globulin are deposited in PB-II (Tanaka et al., 1980; Krishnan et al., 1992), these results suggest that the globulins may encase the glutelins in PB-II, thereby preventing the direct extraction of the glutelin by acidic solutions.
Figure 9.
SDS-PAGE analysis of proteins extracted by 1% (v/v) lactic acid from after pre-extraction with different solvents. A, Wild-type var. Kinmaze; B, esp2. T, Total protein; +, pretreatment, −, no pretreatment. Black arrowhead indicates the 57-kD polypeptide.
We next investigated the state of deposition of glutelin precursors in esp2 using the same experimental approach outlined above. Even though glutelin subunits in esp2 were readily extracted only after the removal of globulin polypeptides (Fig. 9B, lanes 1 and 2), as was the case for the wild type, the glutelin precursors were not extensively extracted under these conditions. Prior removal of the Cys-poor and Cys-rich prolamins or of the globulins and Cys-poor prolamins also had no effect on proglutelin extraction (Fig. 9B, lane 4). Only after the Cys-rich prolamin polypeptides were removed completely by 60% (v/v) n-propanol with 5% (v/v) 2-mercaptoethanol (2ME) following globulin extraction, the 57-kD glutelin precursor efficiently was extracted by 1% (v/v) lactic acid (Fig. 9B, lane 5). The necessity of the preremoval of Cys-rich prolamins for glutelin extraction suggests the possibility that proglutelin is aggregated with Cys-rich prolamin by a disulfide bond.
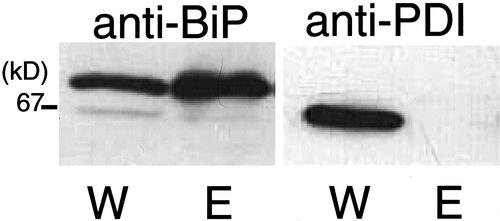
The esp2 Mutant Lacks Protein Disulfide Isomerase (PDI)
The anomalous interaction of proglutelin and prolamin polypeptides suggests a deficiency in protein folding. Therefore, we investigated the levels of the lumenal chaperones BiP, calnexin, and PDI by immunoblot analysis (Fig. 10). All three chaperones (results with calnexin are not shown) were readily detected in the wild type, var. Kinmaze, but only BiP and calnexin (results not shown) were evident in esp2. BiP was present at relatively higher levels in esp2 than in var. Kinmaze, whereas PDI was completely absent in esp2. Forty F2 seeds obtained from a cross between esp2 and wild-type rice were analyzed to investigate whether the accumulation of the glutelin precursor in esp2 occurred because of the lack of PDI protein. SDS-PAGE analysis shows that the normal-to-elevated 57-kD phenotypes segregated 3:1 in these F2 seeds. Immunoblot analysis showed that all seeds exhibiting the elevated 57-kD phenotype lacked PDI, whereas all seeds containing normal levels of the 57-kD precursor contained PDI (data not shown). These results suggest that the high level accumulation of the glutelin precursor in esp2 is related to the lack of PDI.
Figure 10.
Western-blot analysis of the protein from the immature esp2 mutant reacted with anti-PDI and anti-BiP antibodies. W, Wild type var. Kinmaze; E, esp2 mutant.
To obtain the homologous sequences expressed in rice endosperm, a λZAP cDNA library constructed from developing rice seed mRNA was probed with the castor bean (Ricinus communis) anti-PDI antibody. We identified one clone, 1,201 bp in length and containing a poly A tail, that contained a single open reading frame but lacked an ATG start codon (National Center for Biotechnology Information accession no. AB039278). The deduced amino acid sequence of the partially isolated PDI cDNA shows that rice, maize (Zea mays; Li and Larkins, 1996), barley (Hordeum vulgare; Chen and Hayes, 1994), wheat (Triticum aestivum; Shimoni et al., 1995a, 1995b), and castor bean (Coughlan et al., 1996) clones display 84.8%, 84.2%, 83.9%, and 62.5% sequence identity, respectively. Rice PDI contained the C-terminal tetrapeptide KDEL, the ER retention/retrieval signal characteristic of many ER resident proteins (Denecke et al., 1992).
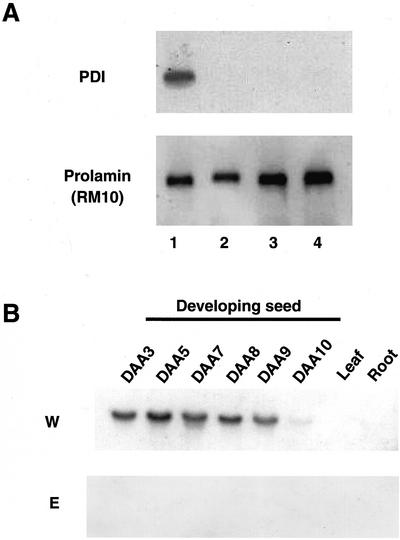
To confirm whether the expression of PDI is suppressed at the transcript level in esp2 mutant, we further investigated the RNA isolated from developing seeds of wild-type rice and three esp2 mutant lines, CM1787, EM44, and EM747, by northern-blot analysis using the rice PDI cDNA as a probe. The probe reacts with a 2.2-kb signal in the wild-type sample, but not in the esp2 samples (Fig. 11A). Prolamin mRNA was detected in the wild-type and the mutant lines at the same density. This result showed that the gene of PDI is not expressed in all three esp2 mutant lines in which it was induced independently, indicating that the lack of PDI protein is essential for the high accumulation of the glutelin precursor in esp2 mutants.
Figure 11.
Northern-blot analysis of PDI mRNA in developing seeds and some tissues. A, Three individual esp2 mutants. Lane 1, Wild type var. Kinmaze; lane 2, CM1787; lane 3, EM44; lane 4, EM747. B, Rice tissues and developing seeds collected from 3 to 10 d after anthesis (DAA). W, Wild type var. Kinmaze; E, esp2 mutant.
To characterize the PDI, we analyzed the tissue-specific expression of PDI gene and the level of transcription during seed development (Fig. 11B). The PDI mRNA was present only in var. Kinmaze developing endosperm, showing the seed-specific expression of PDI gene. PDI was not detected in all tissues in esp2. The PDI mRNA content was detected 3 DAA, and it increased from 3 to 7 DAA and then declined. These results suggest that the PDI plays a specific role in the accumulation of seed storage proteins.
DISCUSSION
In normal endosperm, prolamins and glutelins are synthesized on the ER, translocated into the ER lumen, and then packaged into separate compartments of PB-I and PB-II, respectively. Prolamins accumulate in the ER lumen to form PB-I, whereas the glutelin precursor is transported first to the Golgi body and then to a protein storage vacuole, which eventually forms PB-II. At the destination site, proglutelin is proteolytically cleaved to form acidic and basic subunits, resulting in low amounts of the precursor form.
The esp2 mutants accumulate substantial quantities of a 57-kD polypeptide. Based on results of the studies described herein, we demonstrate that the 57-kD polypeptide is the glutelin precursor. This abnormal accumulation of the glutelin precursor is also associated with a change in the intracellular location of this protein from the normal protein storage vacuolar site, PB-II, to an ER lumenal site of PB-I, where it interacts with the Cys-rich prolamins. Further, we show that overaccumulation of the glutelin precursor in esp2 is due to the lack of PDI, as viewed by immunoblot and northern-blot analysis. Current efforts are directed at determining whether esp2 is the structural gene for PDI or if it is a modifying gene.
The present study shows that the separation of prolamin and proglutelin polypeptides within the ER lumen does not operate normally in the esp2 endosperm. Two possible mechanisms can account for this mistargeting of glutelin precursors. As shown earlier by Li et al. (1993a), one process that may facilitate the routing of these storage proteins into different intracellular compartments is that their mRNAs are not randomly distributed on the ER. Instead, prolamin RNAs are localized on the ER that binds PB-I, and glutelin RNAs are enriched on the cisternal ER. The enrichment of prolamin RNAs on the PB-I surface has been suggested to help concentrate the newly synthesized prolamin polypeptides in the lumen and thereby facilitate assembly of these proteins into an intracisternal inclusion granule. Moreover, by segregating these mRNAs on separate ER domains, potential obstacles in protein trafficking to the Golgi body mediated by the prolamin intracisternal granule may be avoided. In esp2 endosperm, the RNA sorting processes may be disrupted, resulting in substantial localization of glutelin precursors to the PB-I compartment.
A second and more likely mechanism is that the deficiency in PDI forges conditions conducive for interaction of the glutelin precursor with prolamins, which would, in turn, prevent the sorting of the former to the Golgi body and later to PB-II. Although PDI is an essential protein that facilitates the isomerization of disulfide bonds (Chivers and Raines, 1997), especially in assisting the folding of synthesized proteins, not all proteins may require this chaperone. Given that normal-appearing PB-II and substantial amounts of acidic and basic subunits are evident in esp2 endosperm, PDI is not essential for the folding, transport to PB-II, or processing of proglutelin. Hence, it is likely that a deficiency of this lumenal chaperone has an adverse effect on the folding and maturation of prolamin polypeptides. Under normal conditions, prolamins fold properly and are self-assembled to form an intracisternal inclusion granule, processes likely assisted by BiP and PDI. Glutelin precursors, which transiently reside in PB-I due to their synthesis by mRNAs on the ER that binds PB-I, although at 10-fold lower levels than prolamin RNAs, and on adjacent cisternal-ER, are eventually transported to the Golgi body. In the absence of PDI, the prolamins may not fold properly, and they may not have the capacity to interact with transient glutelin precursors to form heterogeneous protein complexes. Evidence for elevated levels of unfolded prolamins in esp2 endosperm is suggested by the increased amounts of BiP (Fig. 10) and reduced levels of prolamin (Kumamaru et al., 1988), symptoms of the unfolding protein response in esp2 endosperm. Such a condition has been observed in the maize floury-2 mutant in which the accumulation of zeins with uncleaved signal peptides mediates severalfold elevated levels of BiP (Boston et al., 1991; Fontes et al., 1991; Marocco et al., 1991; Coleman et al., 1995).
Previous studies (Li et al., 1993b; Muench et al., 1997) have shown that folding and assembly of prolamin polypeptides is dependent on BiP. This lumenal chaperone binds to the nascent prolamin chain, to free prolamin polypeptides, and to the assembled intracisternal inclusion granule (Li et al., 1993b). The existence of these BiP-prolamin complexes suggests that the folding and assembly of prolamin on the protein body aggregate is not a cotranslational process; rather, it suggests that translation and protein assembly are sequentially independent events. The interaction of BiP with the prolamin polypeptide from the time it is synthesized until the time it is deposited suggests that BiP functions to retain prolamin in the ER in a competent state until it is deposited onto the protein body aggregate, at which time BiP is released (Li et al., 1993b). The correct formation of intra- and interchain disulfide bonds catalyzed by PDI may be required before assembly of prolamins can occur.
The esp2 mutation has a pronounced effect on PB-I formation and no apparent effect on PB-II. One explanation for this biased esp2-mediated effect on only PB-I is that the glutelins and prolamins have overlapping but distinct patterns of gene expression. Glutelins are expressed at an earlier stage of seed development, in contrast to the prolamins, which are preferentially expressed during the latter stages (Yamagata et al., 1986). Hence, substantial amounts of glutelin have already been synthesized and packaged into PB-II before the onset of prolamin accumulation.
F2 analysis of progeny obtained by a genetic cross between esp2 and var. Kinmaze shows that the 57-kD phenotype is always accompanied by a reduction in prolamin content (Kumamaru et al., 1987), indicating that the esp2 mutation also affects prolamin biosynthesis. Rice prolamin consist of two major polypeptides that migrate as a 13-kD protein containing two bands, 13a and 13b, and lower abundant classes at 10 and 16 kD (Ogawa et al., 1987). The 13b prolamin polypeptide is Cys-poor, whereas the 10- and 16-kD, and the bulk of the 13-kD prolamin polypeptides are Cys-rich (Kim and Okita, 1988a, 1988b; Hibino et al., 1989; Masumura et al., 1989, 1990; Shyur and Chen, 1990). In esp2, the content of the 13b Cys-poor prolamin is severely reduced, whereas the levels of the others remain unchanged (Kumamaru et al., 1987). Therefore, esp2 mutation affects the synthesis of only one of two prolamin classes.
The basis for this selective effect on the 13b class prolamins may be due to differences in their temporal expression during endosperm development. Prolamin synthesis is first detected as early as 8 to 10 d after flowering, and its synthesis continues throughout endosperm development (Yamagata et al., 1982, 1986). Analysis of the steady-state mRNA indicates that the Cys-rich prolamins are synthesized at the early and late stages, whereas the Cys-poor prolamins are preferentially synthesized only during the later stages of endosperm development (Kim et al., 1988b; Shyur et al., 1992; Kim et al., 1993). Hence, during the early stages of esp2 endosperm development, protein aggregation between the proglutelin and Cys-rich prolamin polypeptides is likely to occur, and it would disrupt the normal self-assembly process of prolamins and instead form a heterogeneous inclusion granule, which phenomenon is supported by the immunocytochemical data. If the assembly of Cys-rich prolamins is necessary for assembly of the Cys-poor prolamins, as suggested by the temporal accumulation patterns of their RNAs, then this interaction with proglutelins may inhibit this process and thereby result in the reduction of Cys-poor prolamins.
MATERIALS AND METHODS
Plant Material
In these experiments, we used esp2 mutant lines CM1787, EM44, and EM747, induced independently by N-methyle-N-nitrosourea treatment in which the high accumulation of substantial amounts of the 57-kD polypeptide have been described (Kumamaru et al., 1987, 1988). CM1787 was used as the representative line of esp2 unless otherwise mentioned. The mutants and the parental line, var. Kinmaze, were grown in the field, and the developing seeds at 10 to 20 d after flowering were used in biochemical and electron microscopic studies.
SDS-PAGE and Western-Blot Analysis
Storage proteins were extracted in 4% (w/v) SDS, 4 m urea, 5% (v/v) 2ME, and 0.125 m Tris-HCl, pH 6.8, from mature seed. SDS-PAGE analysis was conducted on 15% to 25% (w/v) polyacrylamide concentration gradient gels as described by Laemmli (1970). After electrophoresis, gels were stained with Coomassie Brilliant Blue.
For western-blot analysis, proteins resolved by SDS-PAGE were transferred to nitrocellulose membranes, which were then incubated in various antibodies. Antibody-antigen reactions were visualized using a commercial purple detection kit (Roche Molecular Biochemicals, Mannheim, Germany).
IEF Electrophoresis
Horizontal slab IEF gels were prepared and electrophoresed according to Brinegar and Peterson (1982). The polyacrylamide gel containing 6 m urea, 4% (w/v) acrylamide, 2% (w/v) Nonidet P-40, and 2% (v/v) ampholine (pH 3.5–10.0:pH 6.0–8.0:pH 8.0–10.5, at a 1:1:1 ratio) were used for IEF analysis. Glutelins, extracted from rice (Oryza sativa) endosperm as described below, were precipitated by the neutralization of the solution, and the pellet was dissolved in 8.5 m urea, 2% (w/v) Nonidet P-40, and 5% (v/v) 2ME (O'Farrell, 1975), then subjected to IEF electrophoresis according to the procedure of Wall et al. (1984). After IEF, the gel was incubated in 15% (w/v) trichloroacetic acid for 20 min, stained with 0.15% (w/v) Coomassie Brilliant Blue R-250 in 50% (v/v) ethanol and 10% (v/v) acetic acid, then destained with 25% (v/v) ethanol and 10% (v/v) acetic acid.
Purification of PBs by SDG Centrifugation
We homogenized 0.5 g of the dehulled developing rice seed using a mortar and pestle with 2 mL of 10 mm Tris-HCl, pH 7.5, containing 0.4 m Suc on ice. The homogenate was filtered through cheesecloth and was then centrifuged at 70g for 10 min. One milliliter of the supernatant was layered onto a 50%, 52%, 54%, 56%, 58%, 60%, 62%, and 64% (w/w) stepwise SDG and was centrifuged with an SW40-TI rotor at 21,000 rpm for 4 h in an ultracentrifuge (L-70; Beckman Coulter, Fullerton, CA). We collected 0.5-mL fractions from the top of the gradient.
PB Isolation by Pepsin Treatment
SDG purified PBs were incubated with different concentrations of pepsin (3,200–,3800 units mg−1; Sigma, St. Louis) at 37°C in 0.2 m sodium acetate-HCl, pH 1.7, for 1 h. After pepsin treatment, PB was collected by centrifugation for 10 min, and it was then washed with 10 mm Tris-HCl, pH 7.5, containing 0.4 m Suc. The precipitated PB was analyzed by SDS-PAGE and was observed by an electron microscope.
Transmission Electron Microscopic Observation
Transverse sections (1–2 mm thick) of developing seeds were fixed for 1 h in 1.5% (v/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde buffered at pH 7.2 with 20 mm PIPES [piperazine-N,N′-bis (2-ethane sulfuric acid]. The samples were thoroughly washed with the PIPES buffer, post-fixed for 2 h in 1% (v/v) osmium tetroxide, and dehydrated by a series of ethanol concentrations. Thin sections were embedded in Spurr's low viscosity resin as described by Baba et al. (l991), sectioned with an ultramicrotome (Reichert-Jung, NuBlock, Germany), then stained sequentially with uranyl acetate and lead citrate solution. Microscopical observations were carried out with a transmission electron microscope (JEM 200C; JEOL Ltd., Tokyo).
Immunocytochemical Observation
Transverse sections of developing grains of esp2 were fixed for 1 h in 4% (v/v) paraformaldehyde and 0.2% (v/v) glutaraldehyde buffered at pH 7.0 with 20 mm PIPES containing 0.5 mm CaCl2. The fixed samples were dehydrated, embedded in LR White resin (medium grade acryl resin; London Resin, Berkshire, UK), and then sectioned as described above.
Immunolocalization was accomplished by methods described previously (Baba et al., 1991). Grids were floated on a drop of phosphate-buffered saline (PBS; 0.15 m NaCl in 10 mm potassium phosphate, pH 7.2) containing 1% (w/v) bovine serum albumin (BSA; Sigma) for 15 min. The grids were then incubated for 30 min with the appropriate antibodies diluted with PBS and 1% (w/v) BSA. Nonspecifically bound antibodies were removed by washing the section several times in a drop of PBS. The grids were then floated for 15 min on a drop (5 μL of 5-nm-diameter protein A-gold or 15-nm-diameter protein A-gold; Funakoshi Chemical, Tokyo), diluted to one-twelfth strength in PBS-BSA and distilled water as described above, then sequentially stained with uranyl acetate and lead citrate.
Fluorescence Microscopic Observation
Sections were treated with blocking buffer containing 0.8% (w/v) BSA, 0.1% (w/v) gelatin, and 2 mm NaN3 in PBS, and were then reacted with the appropriate antibodies diluted with blocking buffer. Nonspecifically bound antibodies were removed by washing the section several times in PBS. The sections were reacted with secondary antibody (anti-rabbit FITC [green] or anti-rabbit rhodamine [red]; Funakoshi Chemical, Tokyo), and were then observed by microscope (AX 80; Olympus, Tokyo).
Protein Extraction
Rice glutelin and proglutelin were extracted by 1% (v/v) lactic acid after pretreatment. Pretreatments were 10 mm Tris-HCl, pH 7.5, containing 0.5 m NaCl for globulin removal; 60% (v/v) n-propanol for Cys-poor prolamin removal; and 60% (v/v) n-propanol containing 5% (v/v) 2ME for Cys-rich removal. Extraction in each solvent was accomplished by resuspending milled rice in the solvent solution and sonicating it for 1 min. After centrifugation, the residue was extracted twice more with the same solvent.
cDNA Cloning
A cDNA library was constructed from mRNA isolated from mid-developing rice endosperm, and the cDNA library was probed with radiolabeled castor bean (Ricinus communis) PDI antiserum as described by Kim and Okita (1988a).
RNA Extraction and Northern-Blotting Analysis
RNA was extracted from developing seeds, leaves, and roots and was resolved by formaldehyde-containing agarose gels according to the process described by Kim and Okita (1988a). RNA was blotted onto positively charged nylon membranes and was then probed with cDNA sequences for PDI or rice prolamin (RM10; Masumura et al., 1990) labeled using the enhanced chemiluminescence labeling system (Amersham Biosciences, Piscataway, NJ).
Antibodies
Antisera to rice BiP from castor bean were a gift from R.S. Boston (North Carolina State University, Raleigh). Antibody to PDI from castor bean was raised in rabbit with antigen by expressed cDNA encoding PDI in Escherichia coli, as described in Coughlan et al. (1996). Antibodies to glutelin acidic subunit and glutelin basic subunit were raised in rabbit with antigens. Antibodies to prolamin polypeptide were raised in mouse. Seed storage proteins were separated by SDS-PAGE. Each band was picked up and solubilized by preparative electrophoresis.
Footnotes
This work was supported by the Ministry of Education, Science and Culture of Japan (grant nos. 10660009 and 1213826) and by the National Science Foundation (grant no. IBN–9982483 to T.W.O.). This research study is part of Project 0590 of the Agricultural Research Center, Washington State University.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010624.
LITERATURE CITED
- Altschuler Y, Rosenberg N, Harel R, Galili G. The N- and C-terminal regions regulate the transport of wheat-gliadin through the endoplasmic reticulum in Xenopus oocytes. Plant Cell. 1993;5:443–450. doi: 10.1105/tpc.5.4.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K, Ogawa M, Nagano A, Kuroda H, Sumiya K. Developmental changes in the bark lectin of Sophora japonica L. Planta. 1991;183:462–470. doi: 10.1007/BF00197746. [DOI] [PubMed] [Google Scholar]
- Bechtel DB, Juliano BO. Formation of protein bodies in the starchy endosperm of rice (Oryza sativa L.): a re-investigation. Ann Bot. 1980;45:503–509. [Google Scholar]
- Boston RS, Fontes EBP, Shank BB, Worbel RL. Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell. 1991;3:497–505. doi: 10.1105/tpc.3.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinegar AC, Peterson DM. Separation and characterization of oat globulin polypeptides. Arch Biochem Biophys. 1982;219:71–79. doi: 10.1016/0003-9861(82)90135-7. [DOI] [PubMed] [Google Scholar]
- Cagampang GB, Lourders JC, Espiritus G, Remedios GS, Juliano BO. Studies on the extraction and composition of rice protein. Cereal Chem. 1966;43:145–155. [Google Scholar]
- Chen F, Hayes PM. Nucleotide sequence and developmental expression of duplicated genes encoding protein disulfide isomerase in barley (Hordeum vulgare L.) Plant Physiol. 1994;106:1705–1706. doi: 10.1104/pp.106.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers PT, Raines RT. General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry. 1997;36:15810–15816. doi: 10.1021/bi971504l. [DOI] [PubMed] [Google Scholar]
- Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Okita TW. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature. 2000;407:765–767. doi: 10.1038/35037633. [DOI] [PubMed] [Google Scholar]
- Coleman CE, Lopes MA, Gillikin JW, Boston RS, Larkins BA. A defective signal peptide in the maize high-lysine mutant floury 2. Proc Natl Acad Sci USA. 1995;92:6828–6831. doi: 10.1073/pnas.92.15.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan SJ, Hastings C, Winfrey RJ., Jr Molecular characterization of plant endoplasmic reticulum: identification of protein disulfide-isomerase as the major reticuloplasmin. Eur J Biochem. 1996;235:215–224. doi: 10.1111/j.1432-1033.1996.00215.x. [DOI] [PubMed] [Google Scholar]
- Denecke J, De Rycke R, Botterman J. Plant and mammalian sorting signals for protein retention in the endoplasmic reticulum contain a conserved epitope. EMBO J. 1992;11:2345–2355. doi: 10.1002/j.1460-2075.1992.tb05294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes EBP, Shank BB, Worbel RL, Moose SP, O'Brian GR, Wurtzel ET, Boston RS. Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell. 1991;3:483–496. doi: 10.1105/tpc.3.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Kidzu K, Masumura T, Ohtsuki K, Tanaka K, Kawabata M, Fujii S. Amino acid composition of rice prolamin polypeptide. Agric Biol Chem. 1989;53:513–518. [Google Scholar]
- Juliano BO. The rice caryopsis and its composition. In: Houston DF, editor. Rice Chemistry and Technology. American Association of Cereal Chemists, St. Paul. 1972. pp. 16–74. [Google Scholar]
- Kim WT, Li X, Okita TW. Expression of storage protein multigene families in developing rice endosperm. Plant Cell Physiol. 1993;34:595–603. [Google Scholar]
- Kim WT, Okita TW. Nucleotide and primary sequence of a major rice prolamin. FEBS Lett. 1988a;231:308–310. doi: 10.1016/0014-5793(88)80839-1. [DOI] [PubMed] [Google Scholar]
- Kim WT, Okita TW. Structure, expression, and heterogeneity of the rice seed prolamines. Plant Physiol. 1988b;88:649–655. doi: 10.1104/pp.88.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HB, Franceschi VR, Okita TW. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta. 1986;169:471–480. doi: 10.1007/BF00392095. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, White JA, Pueppke SG. Characterization and localization of rice (Oryza sativa L.) seed globulins. Plant Sci. 1992;81:1–11. [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M. Mutants for rice storage proteins 3: genetic analysis of mutants for storage proteins of protein bodies in the starchy endosperm. Jpn J Genet. 1987;62:333–339. doi: 10.1007/BF00288825. [DOI] [PubMed] [Google Scholar]
- Kumamaru T, Satoh H, Iwata N, Omura T, Ogawa M, Tanaka K. Mutants for rice storage proteins 1: screening of mutants for rice storage proteins of protein bodies in the starchy endosperm. Theor Appl Genet. 1988;76:11–16. doi: 10.1007/BF00288825. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the heads of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li CP, Larkins BA. Expression of protein disulfide isomerase is elevated in the endosperm of the maize floury-2 mutant. Plant Mol Biol. 1996;30:873–882. doi: 10.1007/BF00020800. [DOI] [PubMed] [Google Scholar]
- Li X, Franceschi VR, Okita TW. Segregation of storage protein m-RNAs on the rough endoplasmic reticulum membranes of rice endosperm cells. Cell. 1993a;72:869–879. doi: 10.1016/0092-8674(93)90576-c. [DOI] [PubMed] [Google Scholar]
- Li X, Wu Y, Zhang DZ, Gillikin JW, Boston RS, Franceschi VR, Okita TW. Rice prolamin protein body biogenesis: a BiP-mediated process. Science. 1993b;262:1054–1056. doi: 10.1126/science.8235623. [DOI] [PubMed] [Google Scholar]
- Marocco A, Santucci A, Cerioli S, Motto M, Fonzo ND, Thompson R, Salamini F. Three high-lysine mutations control the level of ATP-binding hsp70-like protein in the maize endosperm. Plant Cell. 1991;3:507–515. doi: 10.1105/tpc.3.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumura T, Shibata D, Hibino T, Kato T, Kawabe K, Takeba G, Tanaka K, Fujii S. cDNA cloning of an mRNA encoding a sulfur-rich 10 kDa prolamin polypeptide in rice seeds. Plant Mol Biol. 1989;12:123–130. doi: 10.1007/BF00020497. [DOI] [PubMed] [Google Scholar]
- Masumura T, Hibino T, Kidzu K, Mitsukawa N, Tanaka K, Fujii S. Cloning and characterization of a cDNA encoding a rice 13kDa prolamin. Mol Gen Genet. 1990;221:1–7. doi: 10.1007/BF00280360. [DOI] [PubMed] [Google Scholar]
- Muench DG, Okita TW. The storage proteins of rice and oat. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 404–412. [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Omura T, Iwata N, Omura T, Kasai Z, Tanaka K. Purification of protein body-I of rice seed and its polypeptide composition. Plant Cell Physiol. 1987;28:1517–1527. [Google Scholar]
- Ogawa M, Kumamaru T, Satoh H, Omura T, Park T, Shintaku K, Baba K. Mutants for rice storage proteins: isolation and characterization of protein bodies from rice mutants. Theor Appl Genet. 1989;78:305–310. doi: 10.1007/BF00265288. [DOI] [PubMed] [Google Scholar]
- Okita TW, Choi S-B, Ito H, Muench DE, Wu Y, Zhang F. Entry into the secretory system: the role of mRNA localization. J Exp Bot. 1998;49:1081–1090. [Google Scholar]
- Okita TW, Rogers JC. Compartmentation of proteins in the endomembrane system of plant cells. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:327–350. doi: 10.1146/annurev.arplant.47.1.327. [DOI] [PubMed] [Google Scholar]
- Shewry PR, Casey R. Seed proteins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 1–10. [Google Scholar]
- Shewry PR, Tatham AS. The characteristic, structures and evolutionary relationship of proteins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 11–33. [Google Scholar]
- Shimoni Y, Segal G, Zhu XZ, Galili G. Nucleotide sequence of a wheat cDNA encoding protein disulfide isomerase. Plant Physiol. 1995a;107:281. doi: 10.1104/pp.107.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Zhu XZ, Levanony H, Segal G, Galili G. Purification, characterization, and intracellular localization of glycosylated protein disulfide isomerase from wheat grains. Plant Physiol. 1995b;108:327–335. doi: 10.1104/pp.108.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotwell M, Larkins BA. The biochemistry and molecular biology of seed storage proteins. In: Marcus E, editor. The Biochemistry of Plants: A Comprehensive Treatise. Orlando, FL: Academic Press; 1989. pp. 296–345. [Google Scholar]
- Shyur LF, Chen CS. Nucleotide sequence of two rice prolamin cDNA. Nucleic Acids Res. 1990;18:6683. doi: 10.1093/nar/18.22.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyur LF, Wen TN, Chen CS. cDNA cloning and gene expression of the major prolamins of rice. Plant Mol Biol. 1992;20:323–326. doi: 10.1007/BF00014501. [DOI] [PubMed] [Google Scholar]
- Takaiwa F, Kikuchi S, Oono K. A rice glutelin gene family: a major type of glutelin mRNAs can be divided into two classes. Mol Gen Genet. 1987;208:15–22. [Google Scholar]
- Takaiwa F, Ogawa M, Okita TW. Rice glutelins. In: Shewry PR, Casey R, editors. Seed Proteins. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 401–425. [Google Scholar]
- Tanaka K, Sugimoto T, Ogawa M, Kasai Z. Isolation and characterization of two types of protein bodies in rice endosperm. Agric Biol Chem. 1980;44:1633–1639. [Google Scholar]
- Wall JS, Fey DA, Paulis JW. Improved two-dimensional electrophoretic separation of zein proteins: application to study of zein inheritance in corn genotypes. Cereal Chem. 1984;61:141–146. [Google Scholar]
- Yamagata H, Sugimoto T, Tanaka K, Kasai Z. Biosynthesis of storage proteins in developing rice seeds. Plant Physiol. 1982;70:1094–1100. doi: 10.1104/pp.70.4.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Tanaka K. The site of synthesis and accumulation of storage proteins. Plant Cell Physiol. 1986;27:135–145. [Google Scholar]
- Zhao WM, Gatehouse JA, Boulter D. The purification and partial amino acid sequence of a polypeptide from the glutelin fraction of rice grains: homology to pea legumin. FEBS Lett. 1983;162:96–102. [Google Scholar]