Abstract
Light-inducible promoters are able to respond to a wide spectrum of light through multiple photoreceptor systems. Several cis-acting elements have been identified as components of light-responsive promoter elements; however, none of these regulatory elements by itself appears to be sufficient to confer light responsiveness; rather, the combination of at least two elements seems to be required. Using phylogenetic structural analysis, we have identified conserved DNA modular arrays (CMAs) associated with light-responsive promoter regions that have been conserved throughout the evolutionary radiation of angiosperms. Here, we report the functional characterization of CMA5, a native 52-bp fragment of the Nicotiana plumbaginifolia rbcS 8B promoter, which contains an I- and a G-box cis-element. CMA5 behaves as a light-responsive minimal unit capable of activating a heterologous minimal promoter in a phytochrome-, cryptochrome-, and plastid-dependent manner. We also show that CMA5 light induction requires HY5 and that downstream negative regulators COP (constitutive photomorphogenic)/DET (de-etiolated) regulate its activity. Our results show that the simplest light-responsive promoter element from photosynthesis-associated genes described to date is the common target for different signals involved in light regulation. The possible mechanism involved in light-transcriptional regulation and tissue specificity of combinatorial elements units is discussed.
Photosynthetic organisms have evolved complex biochemical systems to perceive and respond to light of different wavelengths. Three classes of photoreceptors have been identified in higher plants: red light (RL)- and far-red light (FR)-absorbing phytochromes (PHYs), blue-light (BL) receptors, and UV light receptors (Kendrick and Kronenberg, 1994). In Arabidopsis, five members compose the PHY family of photoreceptors (PHY A–E) and at least three different BL photoreceptors have been identified (cryptochromes [CRYs], NPH1, and NPL1; Briggs and Huala, 1999; Smith, 2000; Jarillo et al., 2001). Light signals absorbed by these photoreceptors and transduced by associated molecular systems regulate the expression of many genes at the transcriptional and posttranscriptional level (Silverthorne and Tobin, 1984; Gallie, 1993).
Biochemical, physiological, and genetic approaches have revealed the high complexity of the light transduction signal network. These studies have shown that several phototransduction pathways coupled to different photoreceptors converge in their early steps, but also that independent pathways probably exist (Fankhauser and Chory, 1997; Deng and Quail, 1999). This complexity includes negative regulators, which repress photomorphogenesis and light-inducible gene expression in dark-grown seedlings (Hardtke and Deng, 2000).
Many genes have been reported to be activated through more than one photoreceptor (Kuno and Furuya, 2000). How the different signal transduction pathways elicited by light signals are orchestrated to regulate transcriptional gene activation is still poorly understood. It is conceivable that different light-activated transduction pathways target different transcription factors and/or cis-acting light responsive elements within the promoter of a given gene, but it is also possible that they target common light-responsive elements. To date, there are only two transcription factors for which there is enough evidence to suggest their importance in mediating light-regulation: the extensively studied basic Leu zipper factor HY5 (Oyama et al., 1997) and the more recently described bHLH factor PIF3 (Martínez-García et al., 2000). HY5 seems to be implicated in responses to PHY and CRY, whereas PIF3 seem to be mainly involved in PHYB signaling (Koornneef et al., 1980; Zhu et al., 2000).
Deletion and mutagenesis analysis of the promoter region of photosynthesis-associated nuclear genes (PhANGs), particularly those encoding the chlorophyll a/b-binding proteins (CAB) and the small subunit of the Rubisco (RBCS), led to the identification of a number of cis-acting elements involved in the control of transcription by light. Several of these motifs, such as the G-, I-, and GT1-boxes, are found in the promoter region of many light-regulated genes and have been experimentally shown to be important components in the light response (Giuliano et al., 1988; Green et al., 1988; Menkens et al., 1995).
Unequivocal experimental evidence, indicating an essential role in PHY responsiveness, exists only for the LS5-LS7 region from the Lemna gibba CAB19 gene (Kehoe et al., 1994) and for the CGF-1 factor-binding site from the Arabidopsis CAB2 gene (Anderson and Kay, 1995), which include the GATA and GT-1 sequences, respectively. However, these two regions are unable to activate transcriptional activity of homologous or heterologous minimal promoters (Anderson et al., 1994; Kehoe et al., 1994), suggesting that additional cis-regulatory elements are involved in mediating photoresponses in their respective promoters. These and many more experimental data (for review, see Terzaghi and Cashmore, 1995; Argüello-Astorga and Herrera-Estrella, 1998) have led to the general hypothesis that plant light-responsive elements (LREs) are actually complex elements formed by aggregates of cognate sequences for different transcription factors (Schulze-Lefert et al., 1989; Terzaghi and Cashmore, 1995). Furthermore, it has been shown that artificial sequences composed of paired combinations of tetrameric repeats of G- and GATA-boxes or GT1- and GATA-boxes, but not multimers of a single motif, can function as LREs, confirming the complex nature of these regulatory elements (Puente et al., 1996; Chattopadhyay et al., 1998b).
Genes that are activated by the same photoreceptor system display marked differences in their response to light, in terms of the intensity and spectral quality required for their activation (i.e. White et al., 1995). The different protein-protein or protein-DNA interactions that potentially can take place on composite LREs may explain this diversity of responses (Miner and Yamamoto, 1991). Knowledge of the minimal structure, functionality, and regulation of native light-responsive units will be an important basis for understanding the mechanisms by which light activates transcription initiation. However, the components and structural complexity of natural light-responsive units from PhANGs still remain to be determined.
To address the question of what exactly constitutes a native light-responsive sequence, we have analyzed the upstream sequences from more than 110 light-regulated plant PhANGs by means of a phylogenetic structural method (Argüello-Astorga and Herrera-Estrella, 1996). As a result of this analysis, 30 distinct conserved DNA modular arrays (CMAs) associated with light-responsive promoter regions were identified. These CMAs are composed of combinations of either a sequence related to the I-box core motif (GATAAGR) or its inverted version (YCTTATC), with a G- or GT1-box related element. Two main observations support the functional significance of CMAs: (a) the specific combination, spacing, and relative orientation of individual cis-acting elements constituting a CMA are conserved in evolution as a unit, and (b) all the promoter regions that have been experimentally shown to confer light responsiveness contain at least one CMA (Argüello-Astorga and Herrera-Estrella, 1996).
Here, we report the functional characterization of CMA5, the shortest native LRE of a PhANG capable of activating a minimal heterologous promoter in a PHY-, CRY-, repressor of photomorphogenesis-, and plastid signal-dependent manner.
RESULTS
CMA5 Is an Enhancer Capable of Activating a Heterologous Minimal Promoter
We have postulated that CMAs represent minimal light-regulated elements (LREs). One of these CMAs (CMA5) is present in the promoter of at least 13 rbcS genes from 10 plant species (Argüello-Astorga and Herrera-Estrella, 1996). CMA5 contains two of the DNA motifs that have been implicated in the regulation of PhANGs by light, namely the I-box (GATAAGA) and the G-box (CACGTGGC).
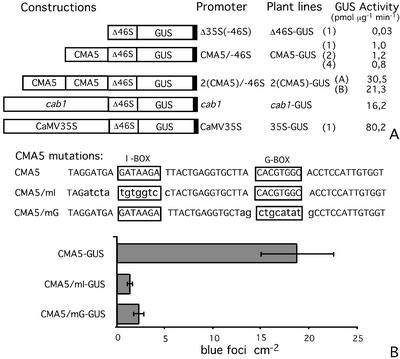
To assess whether CMA5 represents a minimal light-regulated element, one or two copies of CMA5, a 52-bp fragment of the Nicotiana plumbaginifolia rbcS 8B promoter (−260/−208) encompassing CMA5, were fused to the −46-truncated version of the 35S promoter of the cauliflower mosaic virus (CaMV) and the β-glucuronidase (GUS) reporter gene (Fig. 1A). These chimeric promoters were used to produce transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants (CMA5-GUS lines).
Figure 1.
CMA5 activates the minimal Δ35S(−46S) promoter expression. A, CMA5 has an enhancer activity on the minimal Δ35S(−46S) promoter. Diagram of the different promoter-GUS constructs used in this work and name of the corresponding transgenic line. GUS activity in 6-d-old transgenic plants grown in 16-/8-h light/dark cycle was measured. GUS activity in one to three independent transgenic lines (name in parentheses) is shown. The averages of three independent experiments, using three groups of 25 seedlings, are shown. B, Effect of mutations on the CMA5 enhancer activity. Top, Two mutated versions of CMA5 were synthesized, one lacking the I-box (CMA5/mI) and a second lacking the G-box (CMA5/mG). The original sequences were replaced by computer-generated random sequences lacking palindromes (highlighted in bold lowercase). Bottom, The mutated CMA5 derivatives were inserted upstream of the minimal Δ35S(−46S) promoter and used in transient expression experiments. The level of expression directed by CMA5/mI and CMA5/mG in transient expression assays is shown as the number of blue foci per square centimeter (left) recorded in histochemically stained leaves after bombardment. The averages and se of three independent experiments, using three groups of 12 leaf discs, are shown.
With the objective of testing the functionality of CMA5 as an enhancer able to activate the expression of the heterologous minimal Δ35S(−46S) promoter, we measured the level of GUS activity of at least 10 independent transgenic tobacco (N. tabacum) and Arabidopsis lines for each construct. Because the expression pattern of the gene constructs tested was very similar for all tested transgenic lines, we report results for one to three representative lines in each case. The GUS activity of 6-d-old transgenic Arabidopsis lines grown under a 16/8 h light-dark cycle was analyzed. As can be seen in Figure 1A, in lines containing the chimeric promoter with one copy of CMA5 (CMA5/−46S), this construct directs a 30-fold higher level of expression than the minimal Δ35S(−46S) promoter. The chimeric promoter containing two copies of CMA5 (2[CMA5]/−46S) directs 20 to 30 times higher activity than the construct containing one copy of CMA5, its expression level was similar or higher than that directed by the CAB1 promoter. Similar results for CMA5 were obtained in transgenic tobacco plants (data not shown).
The I- and G-Boxes Are Necessary for the Functionality of CMA5
CMA5 contains two well-characterized sequence motifs, the I- and the G-box. To determine whether these motifs are required for the functionality of CMA5, two mutated versions of CMA5 were produced, one lacking the I-box (CMA5/mI) and the other lacking the G-box (CMA5/mG; Fig. 1B). In both cases the length and sequence of CMA5 were preserved with the exception of the consensus sequence of either the I- or G-boxes, which were replaced by a computer-generated random sequence. The enhancer activity of these mutated CMA5 elements was evaluated in mature Arabidopsis leaf tissue, using a particle bombardment, transient expression assay. Both mutations abolish the capacity of CMA5 to activate the expression of the truncated Δ35S(−46S) promoter (Figs. 1B and 4, A–C). Similar results were obtained in transient expression assays using pea (Pisum sativum) and tobacco (N. tabacum) leaves and in transgenic tobacco (N. tabacum) plants (not shown). Therefore, the integrity of both the I- and G-box are an absolute requirement for the enhancer activity of CMA5.
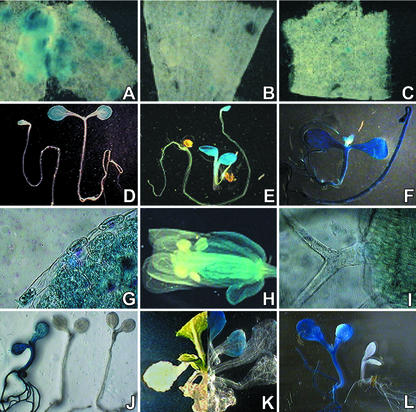
Figure 4.
Histochemical analysis of CMA5/−46S. Tissue-specific and plastid-dependent expression. A through C, Effect of I- or G-box mutations on the CMA5 enhancer activity. Mutated CMA5 derivatives affecting the I-(CMA5/mI) or G-box (CMA5/mG) were used in transient expression experiments. The level of expression directed by: A, CMA5; B, CMA5/mI; and C, CMA5/mG in histochemically stained mature Arabidopsis leaves after bombardment is shown. D through F, Expression pattern of representative 6-d-old etiolated and light-grown seedlings: D, CMA5-GUS-1; E, cab1-GUS; and F, 35S-GUS-1 lines. G through I, GUS histochemical analysis of CMA5-GUS-1 Arabidopsis transgenic plants at the flowering stage: G, leaf; H, reproductive organs; and I, CMA5-GUS-1 trichomes. J through L, Effect of norfluorazon (Nf) treatment. J, GUS staining of representative 4-d-old seedlings grown in media containing 1 μm Nf: 35S-GUS (left), CMA5-GUS-1 (right), and cab1-GUS (center) seedlings. K, CMA5-GUS-1 plant grown in medium supplemented with 1 μm Nf for 10 d and transferred to medium without Nf, before (left) and after (right) histochemical treatment. L, GUS expression directed by the 35S-GUS (left) or CMA5-GUS (right) gene constructs in the cla1 mutant background.
CMA5 Is a Light-Regulated Element (LRE)
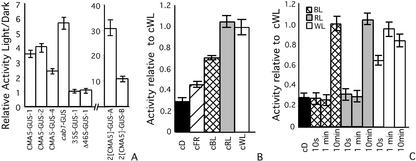
With the objective of testing the functionality of CMA5 as a sequence able to confer light responsiveness to a heterologous minimal promoter, the GUS activity present in CMA5-GUS Arabidopsis seedlings grown under a 16-/8-h light-dark cycle or in complete darkness was determined. The chimeric promoter containing one copy of CMA5 (CMA5/−46S) directs a 2- to 4-fold higher level of expression in light-grown plants than in plants grown in darkness (Fig. 2A). The CMA5/−46S light/dark relative GUS activity was similar to that obtained for the CAB1 promoter. Lines containing the chimeric promoter with two copies of CMA5 (2[CMA5]/−46S) directed the highest light/dark relative GUS activity. In contrast, no differences between light- and dark-grown plants were observed for the gene construct containing the complete CaMV35S promoter or the truncated version Δ35S(−46S) (Fig. 2A). Similar results for CMA5 were obtained in transgenic tobacco plants (not shown).
Figure 2.
CMA5 is a light-regulated element. A, Relative GUS activity in light- versus dark-grown 6-d-old transgenic seedlings. B, GUS activity of 6-d-old CMA5-GUS-1 grown in dark or continuous light of different spectral quality relative to GUS activity in continuous (c) white light (WL). D, dark. C, GUS activity of 4-d-old etiolated seedlings of CMA5-GUS-1 subjected to treatments with single pulses of WL, RL, or BL of different length, relative to GUS activity of plants grown in continuous WL. cD, control in continuous dark. The averages and ses over at least three independent experiments using three groups of 25 seedlings are shown. Statistical ANOVA and Tukey analyses indicate significant differences between the GUS activity of light-grown or light-treated versus dark-grown seedlings at 95% level confidence, for all transgenic lines except the Δ46S-, 35S-GUS line, and CMA5-GUS-1 treated with 10 s or 1 min of BL and RL.
In addition, we determined the level of GUS activity present in CMA5-GUS-1 seedlings germinated in continuous light of different spectral quality. It was observed that CMA5/−46S is activated by continuous WL, RL, FR, and BL treatments (Fig. 2B).
Considering the plethora of physiological and biochemical differences occurring between etiolated and nonetiolated leaves, we tested whether the same response could be observed on leaves having similar developmental status. With this aim, etiolated seedlings were subjected to single pulses of WL, RL, and BL at 200, 140, and 200 μmol m−2 s−1, respectively. It was observed that CMA5 is activated with pulses of WL, RL, and BL (Fig. 2C). However, shorter length pulses of WL than RL and BL are required to activate CMA5/−46S. Short pulses of RL or BL (5 s) activate CMA5/−46S when administrated one per day during 4 d (data not shown).
These results show that CMA5 is an LRE that can be activated by light of different spectral qualities with high sensitivity. To the best of our knowledge, CMA5 is the shortest native sequence from a PhANG that has been shown to act as an LRE enhancer element, capable of activating a heterologous minimal promoter.
CMA5 Is Activated by PHYs and CRYs
In higher plants, the PHY and CRY families of photoreceptors have been shown to mediate transcriptional activation by light (Thompson and White, 1991; Terzaghi and Cashmore, 1995). However, it is still unknown whether these two families of photoreceptors or even different members of the same family target common or different cis-acting elements. Because CMA5 functions as a minimal LRE, this native sequence can be used to determine whether different photoreceptor systems target a common regulatory element.
To address this question, the CMA5/−46S -, cab1-, and 35S-GUS constructs were introduced, by genetic crosses, into the hy1-1 (affected in the biosynthesis of the PHY chromophore), phyA and phyB (null mutants affected in the PHYA and PHYB apoprotein, respectively), and cry1 (a missense mutation affected in a key aminoacid for the CRY1 function) photoreceptor mutants (Somers et al., 1991; Ahmad and Cashmore, 1993; Parks and Quail, 1993; Muramoto et al., 1999). After crossing, homozygous lines for a combination of transgenes and mutations were identified by phenotypic analysis, reporter gene expression, and antibiotic resistance.
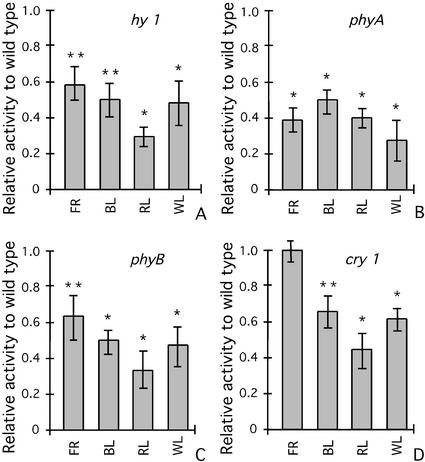
To determine the effect of photoreceptor mutants on the expression of CMA5/−46S, the CMA5/hy mutant seedlings were grown under continuous RL, FR, BL, or WL treatments and compared with GUS-positive, wild-type siblings obtained from the same crosses. It was observed that in the mutant background phyA plants, the expression of CMA5/−46S was significantly lower than in wild-type seedlings under all light treatments (Fig. 3B). In CMA5/phyB and CMA5/ hy1-1 lines, GUS activity was significantly lower in RL, BL, and WL than in wild-type seedlings (Fig. 3, A and C). No significant effect on expression of the CaMV35S promoter was detected in the hy1-1 or phyB backgrounds (data not shown).
Figure 3.
Effect of mutations in photoreceptor functionality on the light-induction of CMA5/−46S. The CMA5/−46S-GUS construct was introduced into the hy1, phyB, phyA, and cry1 mutants by crosses between CMA5-GUS-1 and the corresponding mutants. Mutants and wild-type siblings derived from each cross were grown simultaneously under continuous RL, FR, BL, or WL. Relative GUS activity in mutant versus wild-type 6-d-old seedlings was determined for each treatment. A, hy1; B, phyB; C, phyA; and D, cry1 mutant backgrounds. At least three independent lines for each cross were analyzed with similar results. Averages and ses over three experiments carried out with representative lines using three groups of 25 seedlings are shown. Statistical ANOVA and Tukey analysis indicate significant differences at 95% (*) or at 90% confidence level (**) in GUS activity for all light treatments and all mutants compared with wild-type siblings subjected to identical treatments.
In the cry1 mutant background, the response of CMA5 to BL, RL, and WL, but not that to FR, was significantly affected, although the effect was less notorious than in phy mutants (Fig. 3D). The cry1 mutation had no effect on CaMV35S promoter expression (data not shown).
The diminished responsiveness to light induction of CMA5/−46S in the phyA, phyB, and cry1 mutants suggests that signal transduction pathways activated by these three photoreceptors directly or indirectly target this minimal light-responsive unit. These results also suggest a functional interaction between the PHY and CRY photoreceptor systems.
A small but reproducible effect of the phyB mutation on the response of CMA5 to FR was found. Because in our FR filter system a small percentage (2%–5%) of RL is present, we were unable to determine whether this effect represents a physiological meaningful response of phyB to FR light.
CMA5 Enhancer Activity Is Tissue Specific and Dependent on Chloroplast-Derived Signals
To determine whether CMA5 confers a tissue-specific pattern of expression on the −46 truncated 35S promoter, Arabidopsis CMA5-GUS lines were subjected to GUS histochemical analysis. It was found that in light-grown seedlings, the CMA5/−46S promoter directs expression only in cotyledons, and that this expression is light dependent (Fig. 4D). The same is true for the CAB1 promoter (Fig. 4E), whereas the complete CaMV35S promoter triggers constitutive expression in both light- and dark-grown seedlings (Fig. 4F). In adult CMA5-GUS plants, GUS activity was detected in photosynthetically active tissues such as stomata and mesophyll cells (Fig. 4G) of leaves, stem, petioles, and sepals (Fig. 4H), but not in non-photosynthetic tissues such as epidermis, trichomes (Fig. 4, G and I), or vascular bundles. In mature leaves of flowering plants, GUS activity was more prominent in the region surrounding the hydathodes and secondary veins. CMA5-GUS transgenic tobacco plants showed a very similar pattern of GUS expression to that observed in Arabidopsis (data not shown).
It is well known that the expression of PhANGs is not only light regulated but also dependent on chloroplast development (Oelmüller et al., 1986). In albino mutants or plants treated with Nf (Norfluorazon, a herbicide that inhibits carotenoid biosynthesis and arrests chloroplast development), it has been shown that PhANGs are not expressed because a plastid-derived signal is missing (Oelmüller et al., 1986). Because CMA5 is derived from a PhANG, we tested the influence of the developmental stage of plastids on the expression mediated by this LRE. For this purpose, we germinated CMA5-GUS transgenic Arabidopsis lines in media containing Nf. GUS histochemical analysis revealed a clear effect of Nf on the level of GUS expression in CMA5-GUS-1 lines (Fig. 4J, right). Similar results were obtained for cab1-GUS (Fig. 4J, center). When Nf-treated CMA5-GUS-1 plants were transferred to Murashige and Skoog without Nf, GUS activity was restored in the new green leaves (Fig. 4K). In contrast, the CaMV35S promoter is expressed in seedlings germinated in Nf-containing media (Fig. 4J, left).
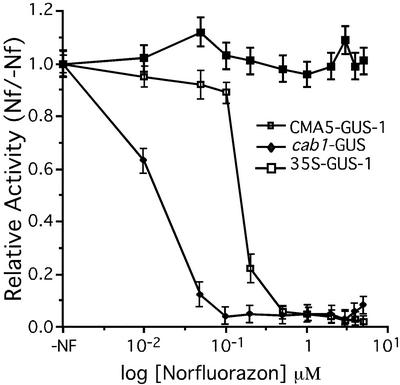
GUS fluorometric assays of seedlings growing in different Nf concentrations showed that the levels of GUS expression for both CMA5-GUS-1 and cab1-GUS plants were dependent on the concentration of Nf (Fig. 5). However, the expression of the CAB1 promoter was completely inhibited at significantly lower concentrations of Nf. In our hands, 50 nm of Nf is sufficient to induce complete bleaching in Arabidopsis, correlating with CAB1 promoter inhibition.
Figure 5.
Effect of Nf on the expression of CMA5-GUS. Relative GUS activity of plants germinated in media with different concentrations of Nf versus non-treated seedlings. Three-day-old seedlings were completely bleached at Nf concentrations of 50 nm or higher. Six-day-old seedlings were collected to determine GUS activity. Averages and ses over three independent experiments using three groups of 25 seedlings are shown. Statistical ANOVA and Tukey analysis indicate significant differences between GUS activity averages of Nf-treated and of non-treated seedlings, at 95% level confidence for all transgenic lines except the 35S-GUS line.
To corroborate that the reduction of GUS expression in CMA5-GUS and cab1-GUS plants was because of an effect of Nf on plastid development and not to an indirect toxic effect of this herbicide, we analyzed the expression of the CMA5/−46S chimeric promoter in the albino cla1 mutant background. In cla1 plants, chloroplast development is arrested at an early stage and expression of PhANGs is severely affected (Mandel et al., 1996). No detectable expression directed by CMA5/−46S was observed in the cla1 mutant background (Fig. 4L, right). In contrast, the 35S-GUS seedlings were completely stained in the same genetic background (Fig. 4L, left). Based on these results, we conclude that CMA5 enhancer activity is tissue specific and dependent on chloroplast development in a very similar manner to that previously reported for PhANG promoters.
The G-Box Binding Factor HY5 Is Required for the Light Induction of the CMA5-Mediated Expression
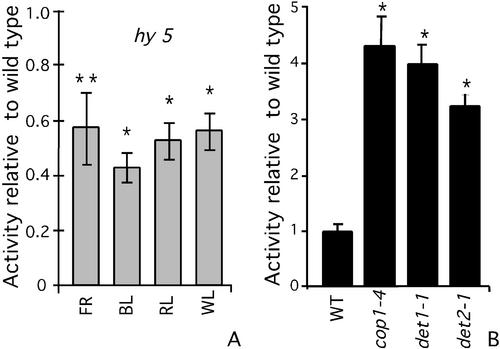
HY5 is a light-regulated nuclear basic Leu zipper transcription factor that has been implicated in mediating the BL and RL responses of photoregulated promoters by directly binding the G-box present in their promoter region (Koornneef et al., 1980; Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998a). Because CMA5 expression appears to be dependent on PHY and CRY signal transduction pathways and contains a G-box, we decided to evaluate the effect of the null hy5-1 mutation on the activity of CMA5/−46S. To this end, the CMA5/−46S construct was transferred to the hy5-1 mutant background by crosses. In this mutant background, the light responsiveness of CMA5 was significantly diminished in all light spectra: WL, FR, RL, and BL (Fig. 6A). The expression of the CaMV35S promoter was not affected by this mutation (data not shown).
Figure 6.
Effect of hy5 and cop/det mutations on the expression of CMA5-GUS. The CMA5/−46S construct was introduced into the hy5, cop1-4, det1-1, and det2-1 mutants by crosses between CMA5-GUS-1 and the corresponding mutants. Mutants and wild-type siblings derived from each cross were grown simultaneously under each treatment. GUS activity was measured in 6-d-old seedlings. A, Relative GUS activity of hy5*CMA5 grown under continuous RL, FR, BL, or WL versus activity in wild-type siblings grown simultaneously. B, Relative GUS activity of cop1-4*CMA5, det1-1*CMA5, and det2-1*CMA5 grown in darkness versus activity in wild-type siblings grown simultaneously. At least three independent lines for each cross were analyzed, with similar results. Averages and ses over three experiments carried out with representative lines using three groups of 25 seedlings are shown. Statistical ANOVA and Tukey analysis indicate significant differences at 95% (*) or at 90% confidence levels (**) in GUS activity for all light treatments and all mutants, compared with wild-type siblings subjected to the identical treatment.
HY5 directly or indirectly modulates CMA5 activity; however, because the hy5-1 allele is a null mutation and CMA5 in this genetic background retained 50% of its activity and is still light responsive, other transcription factors must exist in Arabidopsis that can, at least partially, replace the function of HY5.
CMA5 Activity Is Repressed in the Dark by Photomorphogenesis Repressors
The isolation of recessive constitutive photomorphogenic (cop) and de-etiolated (det) mutants revealed the existence of several loci involved in the repression of photomorphogenesis in darkness (Chory, 1993; Osterlund et al., 1999). These mutants show morphological characteristics of light-grown seedlings in darkness, including the expression of several light-regulated genes (e.g. Chory, et al., 1989; Deng et al., 1991). It is as yet unknown whether the negative regulatory mechanisms of all the photomorphogenesis repressors eventually converge to regulate transcriptional gene activity through the same cis-element. Therefore, we analyzed whether photomorphogenesis repressors affect CMA5-mediated expression. To this end, the CMA5/−46S construct was transferred to the cop1-4, det1-1, or det2-1 mutant backgrounds by crosses, and GUS activity measured in 6-d-old dark-grown mutants or wild-type siblings. It was found that in etiolated cop and det seedlings, CMA5 directs a 3- to 4-fold higher activity than in the wild-type controls (Fig. 6B). These results show that CMA5 enhancer activity is also regulated in dark-grown seedlings by COP/DET negative regulators. As expected, the expression directed by the CAB1 promoter in the dark is also higher for the mutants than for the wild-type siblings (Deng et al., 1991). The effect of the det mutations on the activity of CAB1 promoter in dark is at comparable levels with the effect on CMA5 unit activity, but the effect of cop1-4 mutant is 4 times higher on CAB1 promoter than on CMA5 unit activity (data not shown). The expression of the CaMV35S promoter was not affected in the cop and det mutant backgrounds (data not shown).
DISCUSSION
Although several cis-acting elements involved in light regulation have been characterized, all efforts to identify common DNA motifs that could act as a universal light switch have been unsuccessful. The general conclusion from these studies is that a combination of at least two different DNA motifs is required to confer light responsiveness (Schulze-Lefert et al., 1989; Terzaghi and Cashmore, 1995; Argüello-Astorga and Herrera-Estrella, 1998). The so-called Unit 1 present in the promoter of several CHS genes, composed of an H- and a G-box, has been shown to confer light regulation to a minimal promoter (Loake et al., 1992; Kaiser et al., 1995). However, minimal native light-responsive units from PhANGs, which differ in terms of tissue specificity and light spectrum regulatory properties from those involved in the phenylpropanoid biosynthetic pathway, remain to be identified.
CMA5 Is a Composite LRE Exhibiting PhANGs Regulatory Properties
Our results show that CMA5, a native regulatory unit containing only one copy of the I- and G-boxes, confers light-responsiveness in a PHY- and CRY-dependent manner. In addition to light responsiveness, CMA5 was found to share other functional characteristics with PhANG promoters, including tissue specificity, dependence on the developmental stage of the plastids, requirement of HY5 activity for its light responsiveness, and repression by cop/det genes. In addition, mutational analysis showed that both the I- and the G-boxes present in CMA5 are required for its enhancer activity, demonstrating the composite nature of minimal PhANGs LREs. Therefore, to the best of our knowledge, CMA5 is the most simplified native light-responsive enhancer element reported for a PhANG to date.
Different Photoreceptor Signals Converge on CMA5 by Interrelated Pathways
Considering the complexity of the signal transduction pathways that couple photoreception with transcriptional gene activation and the numerous potentially active DNA motifs present in light-regulated promoters, the identification of native minimal LREs represents a unique system to address important questions in plant photobiology. The findings that CMA5 can be activated by pulses of RL, FR, and BL and that the activity of this regulatory element is affected by mutations in PHYA, PHYB, and CRY1 show that CMA5 is targeted by the signal transduction pathways activated by at least three different photoreceptor systems. These results suggest that the PHYA, PHYB, and CRY1 signal transduction pathways converge at some point to act on a common light-responsive unit.
In low-fluence experiments (using light pulses), it was observed that the threshold for activation of CMA5 with WL is lower than that for RL or BL; in addition, the response to WL is diminished in phyA, phyB, and cry1 mutants. These results suggest that PHY and CRY1 photoreceptor systems need to be functional and probably activated simultaneously, to achieve a maximal CMA5 response to light, indicating a synergistic interaction between these photoreceptor systems.
Because the cry1 mutation affects the response of CMA5 to RL, we conclude that CRY1 is required for the PHYB-mediated activation of CMA5. The effect of phyA and phyB mutations on the responses of CMA5 to BL could be explained by the amplification of BL responses by PHYs (Gil et al., 2000). However, a mutual functional interdependence in which PHYs are also required for the activation of CMA5 by CRY1 cannot be excluded. The functional interactions between PHY and CRY systems have largely been documented through physiological responses (for review, see Casal, 2000). Our results show that functional interdependence between PHYA or PHYB and CRY1 photoreception systems also takes place at the level of transcriptional activation of LREs. The interaction between PHY and CRY systems to activate transcription could take place by direct interactions between the photoreceptors (Ahmad et al., 1998), at any step in the signal transduction pathways, or by acting on common transcription factor(s).
The Light and Plastid Signals Converge on the Minimal Photoresponsive CMA5 Unit
All data reported to date have shown that there is a strong correlation between light- and chloroplast-dependent expression of PhANG promoters. To date, all evidence suggests that these properties cannot be separated in the smallest light-responsive promoter fragments, nor in synthetic pair-wise combinations of cis-acting elements (Simpson et al., 1986; Kusnetsov et al., 1996). Our finding that expression of CMA5 is both light regulated and dependent on the developmental stage of plastids shows that even in a minimal light-responsive unit, these properties cannot be separated and that signals derived from different photoreceptor systems and plastids converge on a single regulatory unit.
Arabidopsis mutants, uncoupling light-regulated RBCS and CAB expression from chloroplast development, have been isolated (Susek et al., 1993). The altered response of these mutants suggests that plastid- and light-activated signal transduction pathways are at least partially independent. Our results show that although plastid and light signals might be separated, they converge to act on a single light-responsive unit. This convergence of environmental and tissue signals could occur at the signal transduction level and/or at the level of transcription factor(s) activation.
HY5 Heterodimerization Could Be Involved in the Regulation of Light-Responsive Units
The two native minimal light-responsive units described to date (unit 1 from CHS and CMA5 from RBCS) are composite units similar in terms of length, structural arrangement, and the presence of a G-box. However, they direct expression that differs in tissue specificity and responses to light of different wavelength (Kaiser and Batschauer, 1995; Fuglevand et al., 1996; this work). It is interesting that HY5 appears to be involved in the regulation of these two regulatory units (Ang et al., 1998; this work). Because HY5 is a GBF (Chattopadhyay et al., 1998a), it could modulate the activity of these two enhancer elements by binding to their G-box. If the same GBF binds to both units, an additional cell-specific factor is probably required to interact and/or activate HY5 to direct transcription in specific tissues. The most obvious possibility is that tissue-specific expression is related to transcription factors, as yet unknown, that bind the companion element of the G-box (the I-box in CMA5 and the H-box in the CHS-unit1). The finding that HY5 has no activation domain makes more appealing the possibility that this GBF could be acting on transcriptional activity through heterodimerization. This type of combinatorial control in transcriptional regulation has been previously postulated as a mechanism to confer plasticity to gene expression through composite cis-elements in combination with heterodimerization of transcriptional factors (Lamb and McKnight, 1991; Miner and Yamamoto, 1991).
COP/DET Participate in the CMA5 Transcriptional Activity
The loci participating in the repression of photomorphogenesis encode proteins involved in the regulation of light-responses (Chory 1993; Osterlund et al., 1999). Our results show that cop/det genes also regulate CMA5 activity, repressing its expression in etiolated seedlings. Because COP1 interacts directly with HY5 (Ang et al., 1998) through the WD40 repeats in the C-terminal domain (McNellis et al., 1994) and the cop1-4 mutant expresses a truncated COP1 lacking this domain, this photomorphogenesis repressor could regulate CMA5 through HY5.
It is interesting that DET2 and DET1 also regulate CMA5 activity in the dark. DET1 is a nuclear protein (Pepper et al., 1994), but the mechanism by which it participates in light regulation is still unclear. However, because HY5 acts as an extragenic suppressor of det1 (Pepper and Chory, 1997), DET1 could also regulate CMA5 enhancer activity through HY5. DET2 encodes an enzyme that participates in brassinosteroid synthesis; therefore, it is unlikely to participate in the same pathway of photomorphogenesis repression than COP1 or DET1 (Li et al., 1996). Because COP1, DET1, and DET2 affect the enhancer activity of a common regulatory unit, it is possible that different negative regulatory mechanisms converge, targeting different intermediates within a common network that regulates CMA5 activity.
MATERIALS AND METHODS
Arabidopsis Strains and Growth Conditions
Seeds of hy, cop, and det mutants (Koornneef et al., 1980; Chory et al., 1989; Deng et al., 1991; Parks and Quail, 1993) were obtained from the Arabidopsis Resource Center (Ohio State University, Columbus). The cla1 (Mandel et al., 1996) mutant was kindly provided by Dr. Patricia León (Universidad Nacional Autónoma de México, Cuernavaca, Morelos, Mexico). The cab1-GUS line containing the uidA (GUS) coding sequence fused to the Arabidopsis CAB1 gene promoter was kindly provided by Dr. Xing-Wang Deng (Yale University, New Haven, CT; Deng et al., 1991).
Arabidopsis seeds were surface sterilized and imbibed with sterile distilled water for 4 d at 4°C before being plated on Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc and 6.5 g L−1 agar (Phytagar, Gibco-BRL, Rockville, MD), and transferred to the appropriate experimental light conditions (details in figure legends).
For Nf treatments, seeds were germinated and grown in light at 100 μmol m2 s−1 in Murashige and Skoog medium supplemented with 1 μm Nf unless otherwise stated. Nf was kindly provided by the Sandoz Chemical Company (Des Plaines, IL).
Gene Constructs and Plant Transformation
Oligonucleotides encompassing the −260/-210 (CMA5) region of the Nicotiana plumbaginifolia rbcS8B promoter (Mazur and Chui, 1985) were synthetized and cloned in 1 and 2 copies in pBI146S. pBI146S was created by inserting the CaMV35S minimal promoter (-46/+8) upstream of the coding region of the uidA (GUS) reporter gene in pBI101 (Jefferson, 1989). For transient expression experiments, the chimeric promoter-GUS cassettes from the pBI vectors were transferred to pBlueScript SK.
Arabidopsis plants were transformed by the Agrobacterium tumefaciens-mediated vacuum infiltration method (ecotype Columbia; Clough and Bent, 1998). At least 10 independent transgenic lines homozygous for each construct were analyzed. The CMA5-1 line was selected for detailed characterization and crosses with mutants.
Genetic Crosses
Transgenes were introduced into mutant backgrounds by crossing wild-type transgenic lines with each of the mutants used in this study. After crossing, lines homozygous for a combination of transgenes and mutations were identified by phenotypic analysis, reporter gene expression, and antibiotic resistance. In all cases, the relative hypocotyl elongation behavior under WL, BL, FR, and RL of the selected GUS-positive lines was similar to that previously reported for hy1-1, phyA, phyB, or cry1.
At least three independent homozygous lines from each cross were analyzed and compared with siblings with wild-type phenotype. The significance of differences in GUS activity between mutants and wild-type siblings was determined using ANOVA. When ANOVA was significant, averages were also compared by the Tukey method.
Light Conditions
Continuous Light Treatments
WL treatments were under a 100 μmol m2 s−1 fluence at 24°C in a growth chamber (MOD AR-32L, Percival Scientific Inc., Boone, IA). RL was supplied by two 21-W fluorescent bulbs (daylight, SOLAR, Mexico) and one 100-W opaque incandescent bulb (SOLAR) filtered through two layers of red acetate (Lee 182; Hampshire, UK; red enriched between 650–800 nm). BL was supplied by two 21-W fluorescent bulbs filtered through two layers of blue acrylic (Lee 183; maximum transmittance at 450 nm and 800 nm). FR was supplied by two 21-W fluorescent bulbs and two 100-W incandescent bulbs filtered through a layer of red acrylic (Lee 182) and an additional layer of blue acrylic (Lee 172; maximum transmittance at 700 nm). Continuous RL, FR, and BL treatments were at 3.5, 2.7, and 2.3 μmol m−2 s−1, respectively. To ensure that seedlings in each experiment were exposed to identical growth conditions and light treatments, wild-type and mutant seedlings for each treatment were grown on the same plates. Dark-grown seedlings were harvested using green safe-light conditions at 0.01 μmol m−2 s−1.
Light Pulses
For light-pulses, 4-d-old seedlings were treated with a single pulse of 10 s, 1 min, or 10 min. WL, RL, and BL pulses were at 200, 140, and 200 μmol m−2 s−1, respectively. Seedlings were harvested 24 h after the onset of the last irradiation.
Transient Expression in Arabidopsis Leaves
Mature plant leaves were bombarded essentially as described (Cabrera-Ponce et al., 1997). After bombardment, leaves were incubated for 48 h under a 16-/8-h light/dark cycle and then assayed for GUS activity by histochemical staining and the number of blue foci recorded.
GUS Activity Measurement
Procedures previously described were used for GUS histochemical staining and quantitative GUS activity assays (Jefferson, 1989; Gallagher, 1992). Fluorescence was measured using a TKO 100 fluorometer (Hoefer Scientific Instruments, San Francisco). GUS activity is expressed as picomoles of methyl-umbelliferone per microgram of protein per minute.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
ACKNOWLEDGMENTS
We are grateful to Nacyra Assad for Arabidopsis transformation, Dr. Octavio Martínez for statistical analyses, and Drs. June Simpson, Alfredo Herrera, Luis Gonzalez, Jean-Phillipe Vielle, and Reynaldo Pless for critical reading of this manuscript.
Footnotes
This work was supported in part by the Howard Hughes Medical Institute (grant no. 75191–526901 to L.H.-E.) and by Consejo Nacional de Ciencia y Tecnología-México (doctoral fellowship to A.M.-H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010678.
LITERATURE CITED
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Anderson SL, Kay SA. Functional dissection of circadian clock- and phytochrome-regulated transcription of the Arabidopsis CAB2 gene. Proc Natl Acad Sci USA. 1995;92:1500–1504. doi: 10.1073/pnas.92.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SL, Teakle GR, Martino-Catt SJ, Kay SA. Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 1994;6:457–470. doi: 10.1046/j.1365-313x.1994.6040457.x. [DOI] [PubMed] [Google Scholar]
- Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- Argüello-Astorga G, Herrera-Estrella L. Evolution of light-regulated plant promoters. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:525–555. doi: 10.1146/annurev.arplant.49.1.525. [DOI] [PubMed] [Google Scholar]
- Argüello-Astorga GR, Herrera-Estrella LR. Ancestral multipartite units in light-responsive plant promoters have structural features correlating with specific phototransduction pathways. Plant Physiol. 1996;112:1151–1166. doi: 10.1104/pp.112.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Huala E. Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol. 1999;15:33–62. doi: 10.1146/annurev.cellbio.15.1.33. [DOI] [PubMed] [Google Scholar]
- Cabrera-Ponce JL, Lopez L, Assad-Garcia N, Medina-Arevalo C, Bailey-Moreno AM, Herrera-Estrella L. An efficient particle bombardment system for the genetic transformation of asparagus (Asparagus officinalis L.) Plant Cell Rep. 1997;16:255–260. doi: 10.1007/BF01088276. [DOI] [PubMed] [Google Scholar]
- Casal JJ. Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell. 1998a;10:673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Puente P, Deng XW, Wei N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998b;15:69–77. doi: 10.1046/j.1365-313x.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Chory J. Out of darkness: Mutants reveal pathways controlling light-regulated development in plants. Trends Genet. 1993;9:167–172. doi: 10.1016/0168-9525(93)90163-c. [DOI] [PubMed] [Google Scholar]
- Chory J, Peto Ch, Feinbaum R, Ausbel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;2:121–129. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- Deng XW, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fuglevand G, Jackson JA, Jenkins GI. UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell. 1996;8:2347–2357. doi: 10.1105/tpc.8.12.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. GUS Protocols: using the GUS gene as a reporter of gene expression. San Diego: Academic Press; 1992. [Google Scholar]
- Gallie DR. Posttranscriptional regulation of gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:77–105. [Google Scholar]
- Gil P, Kircher S, Adam E, Bury E, Kozma-Bognar L, Schafer E, Nagy F. Photocontrol of subcellular partitioning of phytochrome-B:GFP fusion protein in tobacco seedlings. Plant J. 2000;22:135–145. doi: 10.1046/j.1365-313x.2000.00730.x. [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA. 1988;85:7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Yong MH, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua NH. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988;7:4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Deng X-W. The cell biology of the COP/DET/FUS proteins: regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 2000;124:1548–1557. doi: 10.1104/pp.124.4.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Gabrys H, Capel J, Alonso JM, Ecker JR, Cashmore AR. Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature. 2001;410:952–954. doi: 10.1038/35073622. [DOI] [PubMed] [Google Scholar]
- Jefferson RA. The GUS reporter gene system. Nature. 1989;342:837–838. doi: 10.1038/342837a0. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Batschauer A. Cis-acting elements of the CHS1 gene from white mustard controlling promoter activity and spatial patterns of expression. Plant Mol Biol. 1995;28:231–243. doi: 10.1007/BF00020243. [DOI] [PubMed] [Google Scholar]
- Kaiser T, Emmler K, Kretsch T, Weisshaar B, Schafer E, Batschauer A. Promoter elements of the mustard CHS1 gene are sufficient for light regulation in transgenic plants. Plant Mol Biol. 1995;28:219–229. doi: 10.1007/BF00020242. [DOI] [PubMed] [Google Scholar]
- Kehoe DM, Degenhardt J, Winicov I, Tobin EM. Two 10-bp regions are critical for phytochrome regulation of a Lemna gibba Lhcb gene promoter. Plant Cell. 1994;6:1123–1134. doi: 10.1105/tpc.6.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants. Ed 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol Bod. 1980;100:147–160. [Google Scholar]
- Kuno N, Furuya M. Phytochrome regulation of nuclear gene expression in plants. Semin Cell Dev Biol. 2000;6:485–493. doi: 10.1006/scdb.2000.0205. [DOI] [PubMed] [Google Scholar]
- Kusnetsov V, Bolle C, Lubberstedt T, Sopory S, Herrmann RG, Oelmuller R. Evidence that the plastid signal and light operate via the same cis-acting elements in the promoters of nuclear genes for plastid proteins. Mol Gen Genet. 1996;252:631–639. doi: 10.1007/BF02173968. [DOI] [PubMed] [Google Scholar]
- Lamb P, McKnight SL. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development in Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Loake GJ, Faktor O, Lamb CJ, Dixon RA. Combination of H-box [CCTACC(N)7CT] and G-box (CACGTG) cis elements is necessary for feed-forward stimulation of a chalcone synthase promoter by the phenylpropanoid-pathway intermediate p-coumaric acid. Proc Natl Acad Sci USA. 1992;89:9230–9234. doi: 10.1073/pnas.89.19.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, Leon P. CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 1996;9:649–658. doi: 10.1046/j.1365-313x.1996.9050649.x. [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mazur BJ, Chui C-F. Sequence of a genomic DNA clone for the small subunit of ribulose bis-phosphate carboxylase-oxygenase from tobacco. Nucleic Acids Res. 1985;13:2373–2386. doi: 10.1093/nar/13.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng X-W. Genetic and moleculas analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens AE, Schindler U, Cashmore AR. The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20:506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- Miner JN, Yamamoto KR. Regulatory cross-talk at composite response elements. Trends Biochem Sci. 1991;16:423–426. doi: 10.1016/0968-0004(91)90168-u. [DOI] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM. The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell. 1999;11:335–348. doi: 10.1105/tpc.11.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for a rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. Expression of nuclear genes as affected by treatments acting on the plastids. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Ang L-H, Deng X-W. The role of COP1 in repression of Arabidopsis photomorphogenic development. Trends Cell Biol. 1999;9:113–118. doi: 10.1016/s0962-8924(99)01499-3. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus- induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. Plant Cell. 1993;5:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AE, Chory J. Extragenic supressors of the Arabidopsis det1 mutant identify elements of flowering-time and light-response regulatory phatways. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper A, Delaney T, Washburn T, Poole D, Chory J. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 1996;15:3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert P, Becker-Andre M, Schulz W, Hahlbrock K, Dangl JL. Functional architecture of the light-responsive chalcone synthase promoter from parsley. Plant Cell. 1989;1:707–714. doi: 10.1105/tpc.1.7.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverthorne J, Tobin E. Demonstration of transcription regulation of specific genes by phytochrome action. Proc Natl Acad Sci USA. 1984;81:1112–1116. doi: 10.1073/pnas.81.4.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J, VanMontagu M, Herrera-Estrella L. Photosynthesis-associated gene families: differences in response to tissue-specific and environmental factors. Science. 1986;233:34–38. doi: 10.1126/science.233.4759.34. [DOI] [PubMed] [Google Scholar]
- Smith H. Phytochromes and light signal perception by plants: an emerging synthesis. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. [Google Scholar]
- Thompson WF, White MJ. Physiological and molecular studies of light-regulated nuclear genes in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:423–466. [Google Scholar]
- White MJ, Kaufman LS, Horwitz BA, Briggs WR, Thompson WF. Individual members of the Cab gene family differ widely in fluence response. Plant Physiol. 1995;107:161–165. doi: 10.1104/pp.107.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tepperman JM, Fairchild CD, Quail PH. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci USA. 2000;97:13419–13424. doi: 10.1073/pnas.230433797. [DOI] [PMC free article] [PubMed] [Google Scholar]