Abstract
The Arabidopsis gene ATHCOR1, which encodes the CORI1 (coronatine-induced) protein, was expressed in bacterial cells. Soluble recombinant CORI1 was purified and shown to possess chlorophyllase (Chlase) activity in vitro. To determine its activity in vivo, wild-type Arabidopsis and coi1 mutant, which lacks ATHCOR1 transcripts, were transformed with sense and antisense forms of the gene. Wild-type and coi1 plants overexpressing ATHCOR1 showed increased contents of chlorophyllide (Chlide) without a substantial change in the total amount of the extractable chlorophyll (Chl). These plants presented high Chlide to Chl ratios in leaves, whereas antisense plants and nontransformed coi1 mutant showed undetectable ATHCOR1 mRNA and significantly lower Chlide to Chl ratios, relative to wild-type control. Overexpression of ATHCOR1 caused an increased breakdown of Chl a, as revealed by the Chlide a to b ratio, which was significantly higher in sense than wild-type, coi1 mutant, and antisense plants. This preferential activity of CORI1 toward Chl a was further supported by in vitro analyses using the purified protein. Increased Chlase activity was detected in developing flowers, which correlated to the constitutive expression of ATHCOR1 in this organ. Flowers of the antisense plant showed reduced Chlide to Chl ratio, suggesting a role of CORI1 in Chl breakdown during flower senescence. The results show that ATHCOR1 has Chlase activity in vivo, however, because coi1 flowers have no detectable ATHCOR1 mRNA and present Chlide to Chl ratios comparable with the wild type, an additional Chlase is likely to be active in Arabidopsis. In accordance, transcripts of a second Arabidopsis Chlase gene, AtCLH2, were detected in both normal and mutant flowers.
The chlorophyllase (Chlase) enzyme (chlorophyll-chlorophyllido hydrolase, EC 3.1.1.14) catalyzes the hydrolysis of the ester bond of the chlorophyll (Chl) molecule producing chlorophyllide (Chlide) and phytol (for review, see Gossauer and Engel, 1996; Matile et al., 1996, 1999). This reaction is considered to be the first step during Chl catabolism, because the products of the Chl breakdown in different plant species present mainly nonesterified structures (Amir-Shapira et al., 1987; Engel et al., 1991; Shioi et al., 1991).
Chlase has been purified from a variety of plants and shown to be a glycosylated protein associated to plastid membranes (McFeeters, 1975; Terpstra, 1981; Schellenberg and Matile, 1995; Brandis et al., 1996; Matile et al., 1997; Tsuchiya et al., 1997). However, despite the great availability of purified Chlases obtained in the past decades for N-terminal sequencing and antibody production, the identification of Chlase genes was reported only recently (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999; Takamiya et al., 2000).
Chlase activity has been correlated to reduced Chl contents in senescing leaves (Jenkins et al., 1981; Kura-Hotta et al., 1987; Rodríguez et al., 1987) and to respond to ethylene during fruit ripening (Trebitsh et al., 1993). However, Chlase activity has also been found in presenescent leaves, in greened tissues, and during periods of increased Chl synthesis, suggesting a role in Chl turnover (Tanaka et al., 1982; Matile et al., 1996; Minguez-Mosquera and Gallardo-Guerrero, 1996). It has been proposed that Chlase activity is latent because hydrolysis of endogenous Chl does not take place unless chloroplast membranes are disrupted or solubilized with detergents (Terpstra, 1980; Schoch and Brown, 1987). This latency has been attributed to a spatial separation between Chls bound to proteins in the thylakoid membrane and Chlase, which appears to be located in the plastid envelope (Matile et al., 1997). Therefore, Chlase activity in vivo and its regulation and physiological role during Chl catabolism and senescence are still not fully understood.
The Arabidopsis ATHCOR1 was first identified as a gene induced by coronatine (Benedetti et al., 1998), a chlorosis-inducing phytotoxin produced by various plant pathogenic bacteria (for review, see Bender et al., 1999). Furthermore, the gene was shown to be up-regulated in Arabidopsis leaves by methyl jasmonate (MeJA) and wounding, and to require the COI1 gene for expression (Benedetti et al., 1998). When ATHCOR1 was identified, there were no hits to similar genes in the public databases, except that its predicted protein (CORI1) had a potential glycosylation site and a conserved motif found in various hydrolase enzymes, particularly Ser lipases (Benedetti et al., 1998).
ATHCOR1 (named AtCLH1) was shown recently to be related to a Chlase gene from Chenopodium album and to present Chlase activity in cell extracts of Escherichia coli transformed with the AtCLH1 cDNA (Tsuchiya et al., 1999). Here, we show that soluble recombinant CORI1, purified by affinity chromatography, has Chlase activity in vitro, but most importantly, we demonstrate that ATHCOR1 has Chlase activity in vivo, as revealed by the expression analysis of the gene correlated to the Chl and Chlide contents in the coi1 mutant, sense, and antisense Arabidopsis.
RESULTS
In Vitro Chlase Activity of Recombinant CORI1 Protein
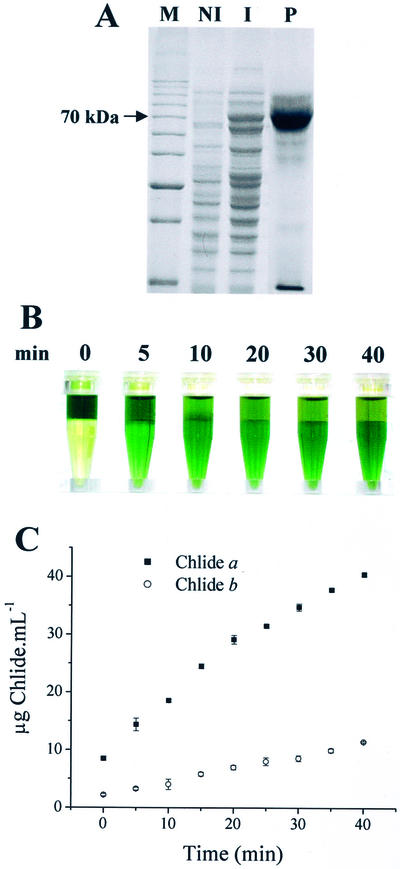
Recombinant CORI1 was expressed in bacteria as a fusion protein with the maltose-binding protein (MBP) in attempt to obtain soluble and functional Chlase. Although most of the recombinant fusion was retained into the insoluble fraction, a reasonable level of soluble CORI1-MBP was produced (Fig. 1A). The purified fusion protein was tested for in vitro Chlase activity, and when added to a solution containing Chl, the recombinant protein was capable of breaking down Chl into Chlide (Fig. 1B). Purified MBP without fusion did not show Chlase activity (not shown). The contents of Chlide a and b produced in the reaction were spectrophotometrically quantified. Figure 1C shows that the rate of Chlide formation was higher for Chlide a, suggesting an increased activity of CORI1 toward Chl a.
Figure 1.
Purification and in vitro activity of recombinant CORI1 protein. A, SDS-PAGE of soluble CORI1 produced as an MBP-fusion protein; M, 10-kD Mr ladder; whole E. coli protein extract before (NI) and after (I) induction with isopropylthio-β-galactoside; P, purified CORI1-MBP fusion. B, Aliquots of the purified CORI1-MBP fusion (approximately 50 μg) were incubated in 50 mm MOPS buffer, pH 7.0, containing Chl dissolved in acetone for different time periods. After incubation, 0.5 mL of the reaction was mixed to 0.4 mL of hexane plus 0.4 mL of acetone for partitioning of the remaining Chl to the hexane phase (top). C, Spectrophotometric measurements of the contents of Chlide a and b in the acetone phase from samples illustrated in B. Values are the mean of three independent measurements plus the sd.
Although Chlase appears to be a glycosylated protein (Terpstra, 1981; Tsuchiya et al., 1997), and its activity enhanced by divalent cations and some detergents (Terpstra, 1980), CORI1 produced in E. coli was functional and required no lipids or detergents to perform the cleavage of the Chl molecule. In addition, we observed that CORI1 was equally efficient at pH 6.0 to 7.5; it was stable at 4°C but less stable at 37°C. Activity was greatly reduced in the presence of 0.2% to 1.0% (v/v) Triton X-100 or in acetone above 40% (v/v). These properties are remarkably similar to what was found for the native rye Chlase (Tanaka et al., 1982).
The hydrolytic esterase activity of CORI1 appears to be very specific to Chls. The recombinant protein was not capable of hydrolyzing ρ-nitrophenyl-esters of fatty acid or tributyrin (not shown), as is the case of the bacterium oil-degrading enzyme HDE (Mizuguchi et al., 1999), although significant homology is found between CORI1 and HDE within their conserved lipase motifs.
In Vivo Chlase Activity of ATHCOR1 in Leaves
With the aim of studying the in vivo role of ATHCOR1, wild type and coi1 mutant were transformed with sense and antisense forms of the ATHCOR1 gene under the control of the cauliflower mosaic virus 35S promoter. After kanamycin selection, wild-type plants transformed with the sense (wtS1 and wtS4) and antisense (wtA1) constructs, and a coi1 mutant transformed with the sense construct (coi1S2) were isolated. The progeny seeds (T2 generation) obtained from wtS4 and wtA1 segregated for kanamycin resistance at a rate of 1:1, as opposed to the 3:1 ratio observed in the progeny of the wtS1 and coi1S2.
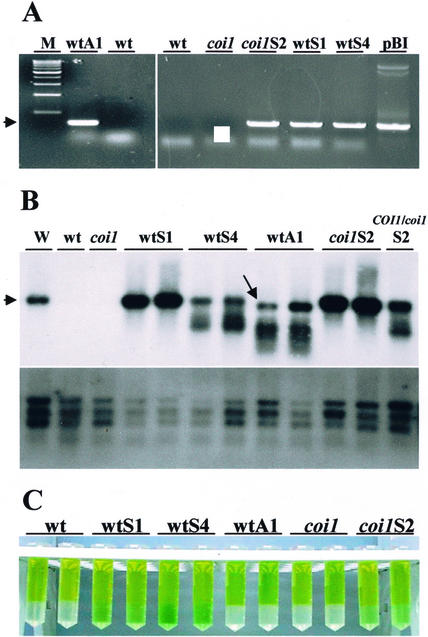
The phenotypes of the transgenic plants were similar and not distinguishable from nontransformed plants, by visual inspection. Genomic DNA from transgenic and control plants was isolated and used in PCR reactions to evaluate T-DNA insertions. Figure 2A shows that transgenic plants resistant to kanamycin (T2 plants) presented a PCR band corresponding to DNA fragments generated by internal 3′-end primers specific to ATHCOR1 and a 5′-end primer specific to the 35S promoter. The expected PCR bands were also detected in kanamycin resistant plants of the T3 generations (not shown).
Figure 2.
In vivo Chlase activity of CORI1 in wild-type, coi1, and transgenic plants. A, PCR products of genomic DNA extracted from Arabidopsis plants transformed with sense (wtS and coi1S) and antisense (wtA1) constructs of 35S::ATHCOR1, showing the amplification of DNA fragments of approximately 800 bp specific to the transgenes (arrowhead); pBI, control vector containing 35S::ATHCOR1 used for transformation of sense plants; M, 1-kb Mr ladder. B, Northern-blot analysis of total RNA extracted from leaves of wild type (wt), coi1 mutant, and two individuals of each transgenic line expressing the sense or antisense ATHCOR1, probed with ATHCOR1 or ubiquitin (below). An mRNA sample from wounded leaves of wild-type plants as positive control for ATHCOR1 induction is shown (W), and the arrowhead indicates the expected approximately 1.3-kb band; antisense plants showed a major approximately 1-kb band (arrow) plus smaller bands also observed in wtS4 and COI1/coi1S2, an F1 plant from the cross between wild type and homozygous coi1 transformed with the sense construct. C, Hexane/acetone partitioning of the Chl (top phase) and Chlide (acetone phase) extracted from leaves of wild-type, coi1 mutant, and transgenic plants.
As reported previously, expression of ATHCOR1 in Arabidopsis is dependent on the activity of the COI1 gene, and levels of ATHCOR1 mRNA are low in wild-type leaves but are rapidly increased after wounding or MeJA treatment (Benedetti et al., 1998). As expected, the levels of ATHCOR1 mRNA (approximately 1.3 kb) were significantly increased in leaves of sense plants wtS1, wtS4, and coi1S2, relative to nontransformed wild type and coi1 mutant (Fig. 2B). In contrast, leaves of the antisense plants showed high levels of an approximately 1-kb message (Fig. 2B), which probably corresponds to the antisense mRNA (an 800-bp construct plus a poly(A) tail). In addition, both wtS4 and wtA1 plants showed lower Mr bands with a “smear” suggestive of mRNA degradation. These lower Mr bands were also observed in the heterozygous COI1/coi1S2, an F1 plant from the cross between homozygous coi1 carrying the sense construct (coi1S2) versus the wild type.
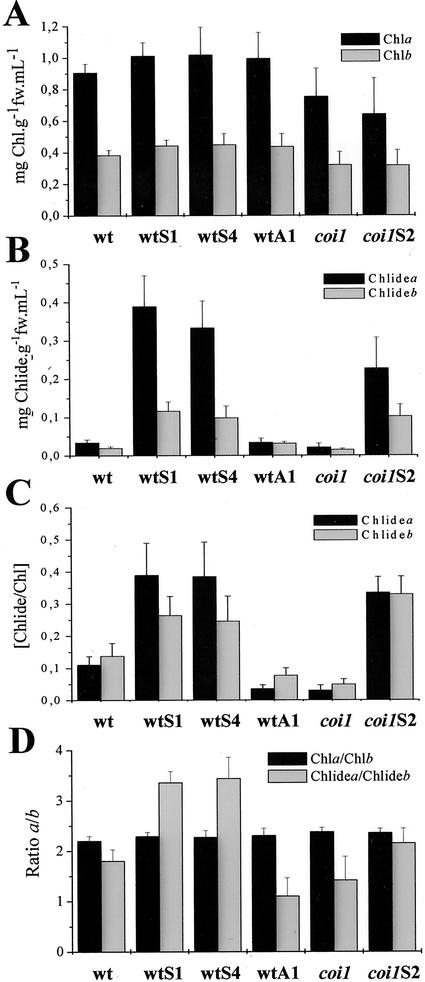
To examine the Chlase activity of ATHCOR1 in vivo, total Chl extracted from leaves of nontransformed and transgenic plants was fractionated into Chl/Chlide. Figure 2C shows that sense plants, including coi1S2, visually presented higher contents of Chlide relative to wild type, coi1 mutant, and antisense wtA1. The amount of extractable Chl/Chlide from leaves of normal and transgenic plants was quantified. Although the amount of Chls a and b did not vary substantially between nontransformed and transgenic plants or between wild type and coi1 mutant (Fig. 3A), the contents of Chlide a and b in leaves of sense plants were significantly increased relative to wild-type, coi1 mutant, and antisense plants (Fig. 3B). When the relative amount of Chlide per total Chl was measured as an estimate of the Chlase activity, it became clear that antisense and coi1 mutant had reduced Chlase activities compared with wild-type and sense plants (Fig. 3C). Moreover, the ratio of Chlide a/b was higher in sense than wild-type, coi1 mutant, and antisense plants, indicating that Chlase activity by ATHCOR1 is preferential toward Chl a (Fig. 3D).
Figure 3.
Relative contents of Chl and Chlide in wild-type, coi1, and transgenic plants. Contents of Chl and Chlide from leaves of wild-type (wt), coi1 mutant, sense (wtS, coi1S), and antisense (wtA) plants that were extracted in 1 mL of acetone. The amount of extractable Chl per gram of fresh weight did not vary significantly in wild-type, coi1, and transgenic plants (A). By contrast, Chide contents in leaves of sense plants were significantly higher than in wild-type, coi1, and wtA1 plants (B). C, In vivo Chlase activity (Chlide/Chl) was increased in sense plants, but diminished in the antisense wtA1 and coi1 mutant. D, The ratio of Chlide a to Chlide b was higher in sense than in wild-type, wtA1, and coi1 plants, indicating that CORI1 activity is increased toward Chl a. Values are the mean of 10 independent measurements plus the sd.
ATHCOR1 mRNA and Chlase Activity Are Increased in Flowers
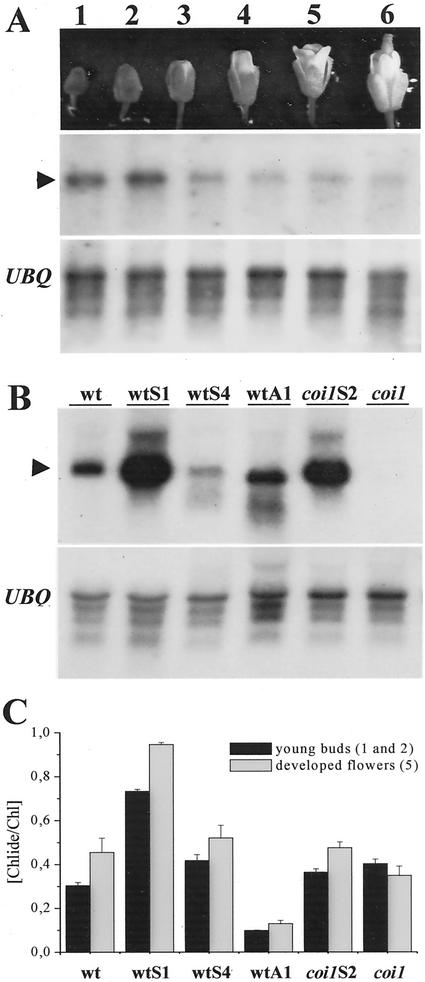
It has been shown that, whereas transcripts of ATHCOR1 are induced upon wounding, coronatine, or MeJA treatments in developing leaves, the gene appears to be constitutively expressed in flowers (Benedetti et al., 1998). Here, we show that ATHCOR1 transcripts are differentially expressed during flower development. Higher levels of ATHCOR1 transcripts were detected at the stage of buds (Fig. 4A), therefore, ATHCOR1 mRNA of young flowers (stages 1–3 shown in Fig. 4A) of nontransformed and transgenic plants were compared (Fig. 4B). The expression of ATHCOR1 messages in flowers of normal and transgenic plants was similar to the expression pattern observed in leaves, except that wtS4 flowers showed lower mRNA levels relative to wild type.
Figure 4.
ATHCOR1 mRNA and Chlide to Chl ratio during flower development. A, ATHCOR1 mRNA during wild-type flower development (stages 1–6) showing higher levels of transcripts (arrowhead) at the stage of buds, compared with ubiquitin (UBQ). B, Northern blot of ATHCOR1 in flower buds of wild-type (wt), coi1 mutant, sense (wtS, coi1S), and antisense (wtA) plants showing high expression of the sense (wtS1, coi1S2) and antisense (wtA1) mRNA relative to normal wild type. Lower levels of sense messages are observed in wtS4, wtA1, and coi1 plants, as compared with the major approximately 1.3-kb ATHCOR1 transcript (arrowhead). C, The ratio of Chlide a+b to Chl a+b in young (stages 1 and 2) and developed flowers (stage 5) of wild-type, coi1, sense, and antisense plants, indicating increased Chlase activities in mature flowers relative to buds, in all plants examined, excepted in coi1, where the difference between the means were not statistically significant. Values are the mean of five independent measurements plus the sd.
The a+b Chlide to Chl ratio from young and developed flowers was examined and, contrary to the mRNA accumulation, the estimated Chlase activity was slightly but significantly increased in developed rather than undeveloped flowers, except in the coi1 mutant, where the difference between the two activities was not statistically significant (Fig. 4C). The values of the a+b Chlide to Chl ratio in flowers were, on average, 2- to 5-fold higher than that of leaves (not shown), which correlates to the ATHCOR1 mRNA levels found in flowers. Chlase activity in flowers of sense wtS1 was twice of the normal flowers, whereas in the antisense it was significantly reduced (Fig. 4C). Chlase activity in coi1 flowers was unexpectedly comparable with that of the wild type (Fig. 4C), despite the fact that coi1 flowers showed no detectable mRNA bands on northerns (Fig. 4B). In addition, Chlase activity in developed flowers of sense wtS4 and coi1S2 was not statistically different from the wild type (Fig. 4C), and high levels of ATHCOR1 transcript in coi1S2 flowers (Fig. 4B) did not result in an increment on the Chlide to Chl ratio (Fig. 4C). These results suggested the presence of an additional Chlase activity in flowers not dependent on the COI1 gene. Therefore, the presence of transcripts of a second Arabidopsis Chlase gene (AtCLH2), which is expressed in leaves but is not induced by MeJA (Tsuchiya et al., 1999), was analyzed in normal and mutant flowers.
Transcripts of Chlase Gene AtCLH2 Is Present in coi1 Flowers
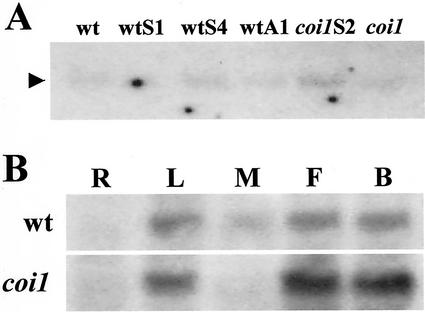
Transcripts of AtCLH2 were detected in very low abundance in Arabidopsis flowers; they were present in similar amounts in both wild-type and coi1 flowers and their relative levels were not altered between normal and transgenic plants (Fig. 5A). Transcripts of AtCLH2 were poorly detected in leaves of normal, coi1 mutant, and transgenic plants by northern blot (not shown). To further investigate the expression of AtCLH2 in different Arabidopsis organs, semiquantitative RT-PCR was employed. Figure 5B shows that AtCLH2 mRNA is present in leaves, flowers, and flower buds of both wild-type and coi1 plants, with an apparently higher level in coi1 flowers. Similar to ATHCOR1, AtCLH2 is not expressed in roots; however, treatment with MeJA did not induce its expression in leaves (Fig. 5).
Figure 5.
Transcripts of Chlase gene AtCLH2 is present in coi1 flowers. A, Northern-blot analyses of the same RNA samples shown in Figure 4B probed with the AtCLH2 cDNA. Transcripts of AtCLH2 are detected at low levels in wild-type (wt), coi1 mutant, transgenic sense (wtS, coi1S), and antisense (wtA) plants (arrowhead). B, Southern-blot analysis of AtCLH2 expression by reverse transcriptase (RT)-PCR. The amplified fragments were blotted, transferred to a nylon membrane, and hybridized to the AtCLH2 cDNA. AtCLH2 transcripts were detected in leaves (L), flowers (F), and flower buds (B) of both wild type and coi1 mutant. The gene was not detected in roots (R) and MeJA did not induce it in leaves (M).
DISCUSSION
In the present study, we provided evidence for the in vitro and in vivo function of the Arabidopsis ATHCOR1 gene as a Chlase. We showed that soluble CORI1 protein expressed in bacterial cells and purified by affinity chromatography has Chlase activity in vitro. This will help further studies on the molecular and biochemical properties of the enzyme.
Recombinant CORI1 has previously been shown to have in vitro Chlase activity, however, in experiments where whole bacterial cell fractions were used as the source of the enzyme (Tsuchiya et al., 1999). Similarly, recombinant and soluble Chlase from Citrus has recently been described, but further purification of the enzyme from bacterial fractions was not reported (Jacob-Wilk et al., 1999).
The in vivo activity of CORI1 as a Chlase was clearly characterized by the expression analysis of the gene associated to the Chlide to Chl ratios obtained from the coi1 mutant and transgenic Arabidopsis transformed with sense and antisense ATHCOR1. Wild-type Arabidopsis and coi1 mutant overexpressing ATHCOR1 presented higher Chlase activity in leaves, relative to nontransformed plants, whereas the opposite was observed for the antisense and normal coi1, which does not express ATHCOR1. In addition, the activity of CORI1 was increased toward Chl a, as revealed by the in vitro activities and by the changes in the Chlide a to b ratio in transgenic plants overexpressing ATHCOR1. This is consistent with the in vivo Chlase activities determined during leaf senescence in various plants. For instance, Chl a is degraded at a faster rate than Chl b in leaves of soybean (Glycine max; Jenkins et al., 1981), rice (Oryza sativa; Kura-Hotta et al., 1987), and wheat (Triticum aestivum; Patterson and Moss, 1979) and in the algae Phaeodactylum tricornutum (Schoch and Brown, 1987).
The Chlide to Chl ratio in flowers was higher than in leaves, which correlates with the constitutive expression of ATHCOR1 found in developing flowers. Although ATHCOR1 mRNA accumulates at the early stages of flower development, Chlase activity seems to increase as flowers are fully developed, suggesting that Chl breakdown may take place during sepal senescence. In developed flowers, Chlase activities found in the coi1 mutant, sense wtS4, and coi1S2 were similar to normal wild type. Therefore, although flowers of the antisense plant showed significantly reduced Chlide to Chl ratios, the results suggest that an additional COI1-independent Chlase is likely to be active in Arabidopsis flowers. The Arabidopsis gene AtCLH2, which is related to ATHCOR1, was shown to be constitutively expressed in rosette leaves at a low level and not to respond to MeJA (Tsuchiya et al., 1999). Here, evidence was presented indicating that AtCLH2 is expressed in coi1 flowers, therefore, supporting the hypothesis of a second Chlase activity in Arabidopsis not dependent on COI1. In addition, AtCLH2 transcript accumulation did not vary between normal and transgenic ATHCOR1 plants, indicating that the changes in the Chl to Chlide ratio observed in the transgenic plants were caused by the altered expression of the ATHCOR1 gene.
Interestingly, ATHCOR1 and AtCLH2 transcripts are apparently missing in roots, a strong indication of an association with chloroplast activity. This is consistent with the presence of a typical chloroplast transit peptide found in the amino acid sequence of AtCLH2 (Tsuchiya et al., 1999). Although the CORI1 protein presents several Ser and Thr residues in its N terminus, a common feature of chloroplast transit peptides, its sub cellular localization cannot be inferred from its sequence.
It was observed that both antisense wtA1 and sense wtS4 presented small Mr mRNA bands on northerns, which is suggestive of a post-transcriptional gene silencing (PTGS) phenomenon (Bass, 2000). In the case of the antisense plant, transcripts corresponding to the endogenous ATHCOR1 (approximately 1.3 kb) are apparently missing, particularly in flowers where the endogenous gene is normally detected. Interestingly, however, is that the suggested PTGS appears to be more pronounced in wtS4 flowers than in wtS4 leaves, perhaps because endogenous ATHCOR1 is expressed at a higher level in flowers. In accordance, Chlide to Chl ratio in wtS4 plants is significantly higher in leaves but not statistically different from the wild type in flowers. Similarly, heterozygous coi1 mutant transformed with sense ATHCOR1 (COI1/coi1S2) showed smaller Mr mRNA bands, which are not found in the homozygous coi1 transformed with sense ATHCOR1 (coi1S2). It is possible that in COI1/coi1S2, one copy of the COI1 gene is sufficient to up-regulate endogenous ATHCOR1 enhancing its transcript level and, thus, inducing PTGS.
The physiological role of Chlases in plants is not entirely clear. It has been shown for instance that Citrus Chlase1 is constitutively expressed through development, but its transcript level is not altered during fruit ripening, suggesting that CHLASE1 is not the regulating step of Chl breakdown during this process (Jacob-Wilk et al., 1999). In addition, maximum Chlase activities have been correlated to stages of increased Chl synthesis during olive development (Minguez-Mosquera and Gallardo-Guerrero, 1996). The fact that ATHCOR1 expression is rapidly induced upon wounding and coronatine treatment suggests that it may play a role in tissue repair or defense (Benedetti et al., 1998). Because of the photodynamic nature of Chl and its porphyrin breakdown products, these molecules can induce strong photooxidative damage by transferring electrons to oxygen species (Matile et al., 1996). Therefore, when tissues are damaged, Chlase would rapidly be activated to initiate Chl detoxification at the wounded sites. According to this, it has recently been proposed that accumulation of Chl breakdown products caused by a deficiency in the red Chl catabolite reductase enzyme is the cause of the cell death lesion phenotype of the Arabidopsis acd2 mutant (Mach et al., 2001). Moreover, light induced the acd2 phenotype, which could also be trigged by the chlorosis-inducing phytotoxin coronatine through a light-dependent process (Mach et al., 2001). Similarly, disease lesion mimic phenotype resembling those trigged during hypersensitive reactions induced by pathogens is observed in the maize mutant Les22, which is defective in uroporphyrinogen decarboxylase, an enzyme involved in Chl biosynthesis (Hu et al., 1998). This mutant also showed necrotic spots on leaves, which developed in a light-dependent manner (Hu et al., 1998). Therefore, if not properly detoxified, photodynamic porphyrins can enhance lesions at sites where they accumulate. During pathogen infection, this could substantially favor pathogens by causing increased plant cell death. It has been shown that in response to infection by a coronatine-producing strain of Pseudomonas syringae, transgenic plants overexpressing the ACD2 protein showed reduced disease symptoms (Mach et al., 2001).
Coronatine has been considered to play a critical role as a virulence factor during the early stages of bacterial infection (Mittal and Davis, 1995; Budde and Ullrich, 2000). A proposed mechanism for coronatine action suggests that the toxin suppresses the activation of defense-related genes (Mittal and Davis, 1995). However, coronatine is a mimic of MeJA (Feys et al., 1994), a signal for defense reactions. Thus, it appears that by inducing the expression of ATHCOR1, coronatine would increase Chl degradation provoking the formation of phototoxic porphyrins, predisposing the tissue to infection. It would be interesting to challenge the transgenic plants described here with coronatine-producing bacteria. Nevertheless, it has already been demonstrated that leaves of coi1 mutant showed reduced chlorosis after been infiltrated with P. syringae pv atropurpurea, which grew significantly less in coi1 than in wild-type leaves (Feys et al., 1994).
MATERIALS AND METHODS
Plant Growth
Wild-type Arabidopsis ecotype Columbia (Col-0) was obtained from the Nottingham Arabidopsis Stock Centre (UK), whereas the coi1 mutant was kindly provided by Dr. John G. Turner (University of East Anglia, UK). Seeds were germinated in Murashige and Skoog medium (Murashige and Skoog, 1962) and grown for 2 to 3 weeks before they were transferred to soil. coi1 seeds from an F2 population segregating for the Coi phenotype were first germinated in Murashige and Skoog medium containing 10 μm MeJA to select homozygous coi1 plants (Feys et al., 1994). Seedlings were grown under white light (70 μE m−2 s−1) with 16-h-day/8-h-night photoperiod at 20°C.
Plant Transformation
The entire coding sequence of ATHCOR1 was ligated downstream to the cauliflower mosaic virus 35S promoter, and the resulting 35S::ATHCORI construct was inserted into the SalI/SstI sites of pBI101 (CLONTECH, Palo Alto, CA), in which the GUS gene had been removed. For the antisense construct, an 800-bp fragment of the ATHCORI cDNA, starting from the ATG, was ligated in the inverted orientation into the XbaI/SstI sites of the pBI121 (CLONTECH), removing the GUS gene. Constructs were verified by sequencing and used to transform Agrobacterium tumefaciens LBA4404 cells. Arabidopsis wild-type and heterozygous coi1 plants were transformed via A. tumefaciens inoculation, essentially as described by Chang et al. (1994). Seeds of inoculated plants were sown in Murashige and Skoog medium containing 50 μg mL−1 kanamycin to select resistant plants. Seeds of transformed heterozygous coi1 plants were first germinated in MeJA plates to select homozygous coi1, which were then transferred to kanamycin plates. Genomic DNA from normal and transformed plants was isolated and used in PCR reactions to verify the presence of the T-DNA 35S-ATHCOR1 insertions using 35S promoter primers and ATHCOR1 internal primers.
Northern Blots
Total RNA from leaves, flowers, and roots was extracted with Trizol reagent (Invitrogen, Gaithersburg, MD). Aliquots of 10 μg of total RNA were fractionated on formaldehyde-agarose gels (Sambrook et al., 1989), transferred onto nylon membranes Hybond N+ (Amersham, Little Chalfont, UK) by capillary blot, and fixed by UV cross-linking according to the manufacturer's instructions. Blots were hybridized using the ATHCOR1, AtCLH2, and an ubiquitin cDNA as probes. Membranes were washed twice with 2.0× SSC containing 0.1% (w/v) SDS for 10 min at 42°C and twice with 0.2× SSC containing 0.1% (w/v) SDS for 10 min at 42°C.
Expression and Purification of Recombinant CORI1 Protein
A DNA fragment containing the coding region of ATHCOR1 cDNA was generated by PCR and subcloned into the EcoRI/SalI sites of pMalc2 vector, which allows the production of a MBP fusion (New England Biolabs, Beverly, MA). Recombinant constructs were verified by sequencing and used to transform Escherichia coli BL21-lysE cells. The recombinant MBP-CORI1 fusion was expressed in these cells in Luria-Bertani medium containing 1 mm isopropylthio-β-galactoside for 4 h and purified by affinity chromatography on an amylose resin, according to the manufacturer's protocol (New England Biolabs). Purified proteins were quantified by a Bradford-based method (Bio-Rad, Hercules, CA) and analyzed on SDS-PAGE (Laemmli, 1970).
Chlase Assay
Aliquots (0.1 mL) of the purified CORI1-MBP fusion (approximately 50 μg) were mixed to 2.0 mL of 0.1 m MOPS buffer, pH 7.0, and to 1.0 mL of purified Chl dissolved in acetone. The mixture was incubated at 25°C for different time periods and the reaction was stopped by transferring 0.5 mL of the reaction mixture to tubes containing to 0.5 mL of hexane plus 0.5 mL of acetone. The reaction was vortexed and centrifuged at 12,000g for 2 min for partitioning of the remaining Chl to the hexane phase. Leaves (approximately 0.2 g fresh weight) of wild-type, coi1 mutant, and transgenic plants were cut and immediately immersed in 6 mL of acetone and incubated at 4°C in the dark for 12 h. Aliquots of total Chl dissolved in acetone were mixed to hexane and 10 mm KOH at a ratio of 4:6:1 (v/v), as described by Jacob-Wilk et al. (1999). After vortexing and centrifugation (12,000g for 2 min), Chls a and b in the hexane phase and Chlides a and b in the acetone phase were quantified spectrophotometrically according to Arnon (1949): Chl a in mg mL−1 = 0.0127 A663 − 0.00269 A645; Chl b in mg mL−1 = 0.0229 A645 − 0.00468 A663. Total Chl and Chlide from flowers were extracted and quantified as above, except that flowers were carefully excised on the base of the sepals and frozen in liquid nitrogen before they were weighted and immersed in acetone.
Cloning of the AtCLH2 cDNA and RT-PCR Analysis
Ten micrograms of Dnase I-treated RNA from leaves, seedlings, and flowers of wild-type Arabidopsis was reversed transcribed using SuperScript reverse transcriptase (Invitrogen) and primer CATAAGCAACAAAAGCTGATG complementary to the 3′ end of the AtCLH2 gene. The cDNAs were used as templates in PCR reactions with primers 5′-ATGTCCTCTTCTTCATCAAGA-3′ and 5′-CATAAGCAACAAAAGCTGATG-3′, which amplified an approximately 1-kb fragment that was cloned in pGEM-t (Promega, Madison, WI). Three independent clones from each of the RT-PCR reactions were sequenced, and, in all cases, the sequence of the AtCLH2 cDNA was identified. The AtCLH2 cDNA was used as a probe in northern and Southern hybridizations.
RT-PCR reactions for expression analysis of the AtCLH2 gene were performed using Dnase I-treated RNA from roots, leaves, flowers, flower buds, and leaves of plants treated with MeJA for 4h. The PCR reactions were separated in standard agarose gels and the DNA fragments were transferred to a nylon membrane and hybridized with the AtCLH2 cDNA as probe. The membrane was washed several times at stringent conditions at 65°C, followed by the detection with x-ray films.
Footnotes
This work was supported by a grant (no. 95/06662–5) and a long-term fellowship (no. 97/0917–7 to C.E.B.) from the Fundação de Amparo à Pesquisa do Estado de São Paulo.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010813.
LITERATURE CITED
- Amir-Shapira D, Goldschmidt EE, Altman A. Chlorophyll catabolism in senescing plant tissues: in vitro breakdown intermediates suggest different degradative pathways for Citrus fruit and parsley leaves. Proc Natl Acad Sci USA. 1987;84:1901–1905. doi: 10.1073/pnas.84.7.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DT. Copper enzymes in isolated chloroplasts: polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti CE, Costa CL, Turcinelli SR, Arruda P. Differential expression of a novel gene in response to coronatine, methyl jasmonate, and wounding in the coi1 mutant of Arabidopsis. Plant Physiol. 1998;116:1037–1042. doi: 10.1104/pp.116.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandis A, Vainstein A, Goldschmidt EE. Distribution of chlorophyllase among components of chloroplast membranes in Citrus sinensis organs. Plant Physiol Biochem. 1996;34:49–54. [Google Scholar]
- Budde IP, Ullrich MS. Interactions of Pseudomonas syringae pv. glycinea with host and nonhost plants in relation to temperature and phytotoxin synthesis. Mol Plant-Microbe Interact. 2000;13:951–961. doi: 10.1094/MPMI.2000.13.9.951. [DOI] [PubMed] [Google Scholar]
- Chang SS, Park SK, Kim BC, Kang BJ, Kim DU, Nam HG. Stable genetic transformation of Arabidopsis thaliana by Agrobacterium inoculation in planta. Plant J. 1994;5:551–558. [Google Scholar]
- Engel N, Jenny TA, Mooser V, Gossauer A. Chlorophyll catabolism in Chlorella protothecoides: isolation and structure elucidation of a red bilin derivative. FEBS Lett. 1991;293:131–133. doi: 10.1016/0014-5793(91)81168-8. [DOI] [PubMed] [Google Scholar]
- Feys BJF, Benedetti CE, Penfold CN, Turner JG. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossauer A, Engel N. Chlorophyll catabolism: structures, mechanisms, conversions. J Photochem Photobiol. 1996;32:141–151. [Google Scholar]
- Hu G, Yalpani N, Briggs SP, Johal GS. A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell. 1998;10:1095–1105. doi: 10.1105/tpc.10.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y. Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 1999;20:653–661. doi: 10.1046/j.1365-313x.1999.00637.x. [DOI] [PubMed] [Google Scholar]
- Jenkins GI, Baker NR, Woolhouse HW. Changes in chlorophyll content and organization during senescence of the primary leaves of Phaseolus vulgaris L. in relation to photosynthetic electron transport. J Exp Bot. 1981;32:1009–1020. [Google Scholar]
- Kura-Hotta M, Satoh K, Katoh S. Relationship between photosynthesis and chlorophyll content during leaf senescence of rice seedlings. Plant Cell Physiol. 1987;28:1321–1329. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg J. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci USA. 2001;98:771–776. doi: 10.1073/pnas.021465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H. Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:67–95. doi: 10.1146/annurev.arplant.50.1.67. [DOI] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H, Kräutler B. Chlorophyll breakdown in senescent leaves. Plant Physiol. 1996;112:1403–1409. doi: 10.1104/pp.112.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Schellenberg M, Vicentini F. Localization of chlorophyllase in the chloroplast envelop. Planta. 1997;201:96–99. [Google Scholar]
- McFeeters RF. Substrate specificity of chlorophyllase. Plant Physiol. 1975;55:377–381. doi: 10.1104/pp.55.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguez-Mosquera MI, Gallardo-Guerrero L. Role of chlorophyllase in chlorophyll metabolism in olives cv. Gordal Phytochemistry. 1996;41:691–697. [Google Scholar]
- Mittal S, Davis K. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant-Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- Mizuguchi S, Amada K, Haruki M, Imanaka T, Morikawa M, Kanaya S. Identification of the gene encoding esterase, a homolog of hormone-sensitive lipase, from an oil-degrading bacterium, strain HD-1. J Biochem. 1999;126:731–737. doi: 10.1093/oxfordjournals.jbchem.a022510. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Patterson TG, Moss DN. Senescence in field grown wheat. Crop Sci. 1979;19:635–640. [Google Scholar]
- Rodríguez MT, González P, Linares JM. Degradation of chlorophyll and chlorophyllase activity in senescing barley leaves. J Plant Physiol. 1987;129:369–374. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schellenberg M, Matile P. Association of components of the chlorophyll catabolic system with pigment-protein complexes from solubilized chloroplast membranes. J Plant Physiol. 1995;146:604–608. [Google Scholar]
- Schoch S, Brown J. The action of chlorophyllase on chlorophyll-protein complexes. J Plant Physiol. 1987;126:483–494. [Google Scholar]
- Shioi Y, Tatsumi Y, Shimokawa K. Enzymatic degradation of chlorophyll in Chenopodium album. Plant Cell Physiol. 1991;32:87–93. [Google Scholar]
- Takamiya K, Tsuchiya T, Ohta H. Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci. 2000;5:426–431. doi: 10.1016/s1360-1385(00)01735-0. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kakuno T, Yamashita J, Horio T. Purification and properties of chlorophyllase from greened rye seedlings. J Biochem. 1982;92:1763–1773. doi: 10.1093/oxfordjournals.jbchem.a134106. [DOI] [PubMed] [Google Scholar]
- Terpstra W. Influence of lecithin liposomes on chlorophyllase-catalized chlorophyll hydrolysis: comparison of intramembraneous and solubilized PHAEODACTYLUM chlorophyllase. Biochim Biophys Acta. 1980;600:36–47. doi: 10.1016/0005-2736(80)90409-5. [DOI] [PubMed] [Google Scholar]
- Terpstra W. Identification of chlorophyllase as a glycoprotein. FEBS Lett. 1981;126:231–235. [Google Scholar]
- Trebitsh T, Goldschmidt EE, Riov J. Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc Natl Acad Sci USA. 1993;90:9441–9445. doi: 10.1073/pnas.90.20.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Masuda T, Mikami B, Kita N, Shioi Y, Takamiya K. Purification and characterization of two isozymes of chlorophyllase from mature leaves of Chenopodium album. Plant Cell Physiol. 1997;38:1026–1031. [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA. 1999;96:15362–15367. doi: 10.1073/pnas.96.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]