Abstract
We recently isolated two genes (OsGA3ox1 and OsGA3ox2) from rice (Oryza sativa) encoding 3β-hydroxylase, which catalyzes the final step of active gibberellin (GA) biosynthesis (H. Itoh, M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka, M. Kobayashi [2001] Proc Natl Acad Sci USA 98: 8909–8914). Using these cloned cDNAs, we analyzed the temporal and spatial expression patterns of the 3β-hydroxylase genes and also an α-amylase gene (RAmy1A) during rice seed germination to investigate the relationship between GA biosynthesis and α-amylase expression. Northern-blot analyses revealed that RAmy1A expression in the embryo occurs before the induction of 3β-hydroxylase expression, whereas in the endosperm, a high level of RAmy1A expression occurs 1 to 2 d after the peak of OsGA3ox2 expression and only in the absence of uniconazol. Based on the analysis of an OsGA3ox2 null mutant (d18-Akibare dwarf), we determined that 3β-hydroxylase produced by OsGA3ox2 is important for the induction of RAmy1A expression and that the OsGA3ox1 product is not essential for α-amylase induction. The expression of OsGA3ox2 was localized to the shoot region and epithelium of the embryo, strongly suggesting that active GA biosynthesis occurs in these two regions. The synthesis of active GA in the epithelium is important for α-amylase expression in the endosperm, because an embryonic mutant defective in shoot formation, but which developed epithelium cells, induced α-amylase expression in the endosperm, whereas a mutant defective in epithelium development did not.
During cereal seed germination, α-amylase in the aleurone layer plays an important role in hydrolyzing the endosperm starch into metabolizable sugars, which provide the energy for the growth of roots and shoots (Akazawa and Hara-Mishimura, 1985; Beck and Ziegler, 1989). Previous physiological and biochemical studies have revealed that α-amylase expression in the aleurone layer occurs as follows. First, active gibberellin (GA) biosynthesis commences in the embryo, and the GAs are transported from the embryo to the aleurone layer (Fincher, 1989). Active GAs trigger the expression of α-amylase at the transcriptional level through the induction of a positive transactivating factor for α-amylase transcription (Gubler et al., 1995). Then, α-amylase is secreted from the aleurone layer into the endosperm to catalyze the hydrating reaction of stored starch. Although the process of induction of α-amylase expression in the aleurone layer is well understood, the mechanism of GA biosynthesis in the embryo during cereal seed germination is still unclear. For example, quantitative analysis of GAs using GC-MS has revealed that the level of active GAs is increased in the shoot and scutella regions during the early stage of barley (Hordeum vulgare) seed germination (Lenton et al., 1994), but it is not known which region, shoot or scutella, is important for α-amylase induction.
A set of genes encoding enzymes involved in the GA biosynthetic pathway has been isolated from various plant species (Lange et al., 1994; Sun and Kamiya, 1994; Chiang et al., 1995; Yamaguchi et al., 1996; Lange, 1997; Helliwell et al., 1998; Thomas et al., 1999). We recently isolated two genes encoding GA 3β-hydroxylases, OsGA3ox1 (Oryza sativa GA 3β-hydroxylase 1) and OsGA3ox2, from rice (Oryza sativa; Itoh et al., 2001). Because GA 3β-hydroxylase catalyzes the final step of the GA biosynthetic pathway and produces bioactive GA1 and GA4 from the substrates GA20 and GA9, tissues or cells that express this enzyme produce the bioactive GAs if the substrates are supplied. OsGA3ox2 is preferentially expressed in young leaves of rice seedlings at the vegetative stage, and both genes are highly expressed in tapetum cells of the anther (Itoh et al., 2001). These tissues and organs are known to actively synthesize the active GAs (Kobayashi et al., 1988). Therefore, it should be possible to observe where and when active GAs are synthesized in the embryo during rice seed germination through the expression analysis of OsGA3ox1 and OsGA3ox2.
In this study, to elucidate the temporal and spatial pattern of GA synthesis during rice seed germination, we analyzed the expression kinetics and in situ localization of OsGA3ox1 and OsGA3ox2 in germinating seeds. Northern analyses showed that the OsGA3ox2 product is important for α-amylase expression, whereas the OsGA3ox1 product is not. Using mutants defective in shoot formation and/or scutella development, we found that the scutella is essential for α-amylase induction but that the shoot region is not involved in this process.
RESULTS
GA 3β-Hydroxylase Expression in the Embryo Occurs before α-Amylase Expression in Endosperm
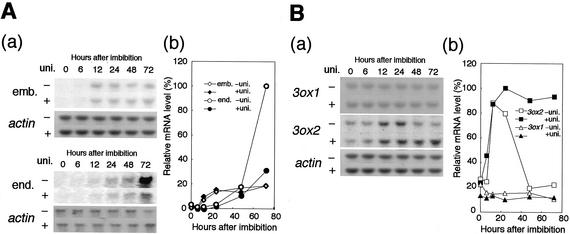
We compared the induction kinetics of the major GA-induced α-amylase gene, RAmy1A (O'Neil et al., 1990), to that of the two OsGA3ox genes in the embryo during the early germination stage. In the embryo, accumulation of RAmy1A mRNA started at 6 to 12 h after the beginning of imbibition with or without uniconazol, which is an inhibitor of GA biosynthesis, and then reached at plateau after 24 h (Fig. 1A, a and b). In contrast to the embryo, accumulation of RAmy1A mRNA in the endosperm was hardly detected until 12 h after imbibition with or without the uniconazol treatment (Fig. 1A, a). Up to 48 h after imbibition, RAmy1A expression in treated or untreated endosperm reached a similar level to that in the embryo at 24 h after imbibition (Fig. 1A, b). Then, the level of RAmy1A mRNA increased more than 5-fold during the following 24 h in the untreated endosperm, whereas it increased by only about 2-fold during the same period in the treated endosperm (Fig. 1A, a and b). This inhibitory effect of uniconazol on α-amylase induction indicates that α-amylase expression in the endosperm is dependent on de novo-synthesized GA during seed germination.
Figure 1.
Expression of RAmy1A (A), OsGA3ox1, and OsGA3ox2 (B) in germinating rice seeds. A (a), Northern analysis using the RAmy1A probe. Seeds were germinated for the indicated hours after imbibition with or without uniconazol (uni. + or −), 5 μg of total RNA was prepared from the treated and untreated embryo, and 1 μg of total RNA was prepared from the treated and untreated endosperm. A (b), Relative amount of RAmy1A transcript in the embryo (diamonds) and endosperm (circles) in the presence (black symbols) or absence (white symbols) of uniconazol. The level of the RAmy1A transcript was normalized against the level of the actin transcript. The normalized values shown here are relative to the expression levels detected in endosperm of untreated seeds at 72 h (100%). B (a), Northern analysis of OsGA3ox2 and OsGA3ox1 in embryo. Five micrograms of total RNA was prepared to detect OsGA3ox2, and 20 μg of total RNA was prepared to detect OsGA3ox1. B (b), Relative level of the OsGA3ox1 (triangles) and OsGA3ox2 (squares) transcripts in the germinating rice embryo treated with (black symbols) or without (white symbols) uniconazol. Normalization of OsGA3ox expression is the same as in A.
To further investigate the relationship between α-amylase expression in the endosperm and GA biosynthesis in the embryo, we studied the expression kinetics of the two GA 3β-hydroxylase genes, OsGA3ox1 and OsGA3ox2, in the embryo during seed germination (Fig. 1B). The expression of OsGA3ox1 was constitutively seen at low level in this period with or without uniconazol. On the other hand, OsGA3ox2 expression rapidly increased during the first 12 h to reach a level several times higher than that of dry seeds, or 10 times higher than that of OsGA3ox1. During the next 24 h, OsGA3ox2 expression was reduced to the basal level in untreated seeds but was maintained at a high level in uniconazol-treated seeds (Fig. 1B, a and b). OsGA3ox2 mRNA levels at 0 and 6 h were not increased by uniconazol. This delayed response is probably because the time needed for uniconazol to inhibit GA biosynthesis after diffusion into the embryos and for the GA catabolism to reduce the amount of bioactive GAs (Fig. 1B, a). The observed rapid increase and high-level expression of OsGA3ox2 after imbibition suggest that the OsGA3ox2 product may have a major role in the de novo GA synthesis to induce RAmy1A expression. The induction kinetics of OsGA3ox2 in the embryo seems to be consistent with the GA-dependent induction of RAmy1A-expression in the endosperm; the peak of OsGA3ox2 expression occurred about 50 h before the rapid increase in RAmy1A expression. The rapid decrease in OsGA3ox2 expression in the untreated seeds can be explained by a negative feedback repression of OsGA3ox2 by GA (see “Discussion”).
The OsGA3ox2 (D18) Product Is Necessary for RAmy1A Expression in the Endosperm
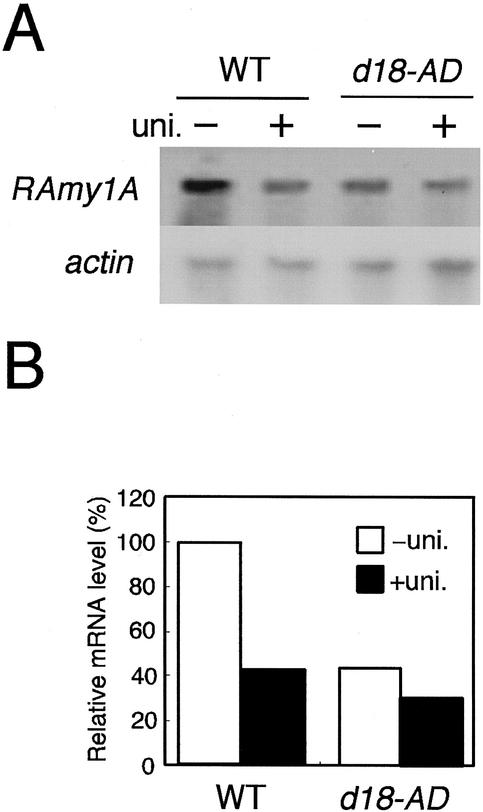
The comparative study of the induction kinetics of GA 3β-hydroxylase and α-amylase suggested that 3β-hydroxylase produced by OsGA3ox2 may be important for the induction of α-amylase expression in the endosperm, whereas the product of OsGA3ox1 may not importantly contribute to α-amylase induction. To confirm this hypothesis, we compared the pattern of RAmy1A expression in the endosperm between the wild-type rice plant, Akibare, and the loss-of-function mutant of OsGA3ox2, d18-Akibare dwarf (d18-AD; Fig. 2). The d18-AD mutant was derived from the wild-type cultivar by chemical treatment and has a null allele of OsGA3ox2 caused by complete deletion of all exon and intron sequences (Itoh et al., 2001).
Figure 2.
Comparison of the level of RAmy1A expression in wild-type (Akibare) and d18-AD mutant plants. A, After 72 h of imbibition with or without uniconazol (uni. + or −), total RNA was extracted from the endosperm of wild-type and d18-AD plants; 1 μg of total RNA was used for the northern analysis. The blot was re-probed with a radiolabeled actin probe as a loading control. B, Relative amount of RAmy1A mRNA. The RAmy1A mRNA level was normalized to the corresponding actin transcript level. The values shown are relative to that of the wild type without uniconazol treatment (100%).
In the wild-type plant, accumulation of the RAmy1A transcript in the endosperm occurred at a level about 2.5 times higher than that in the seeds treated with uniconazol (Fig. 2B). RAmy1A mRNA did not accumulate to this extent in the d18-AD mutant and the uniconazol treatment had no significant effect on the mutant.
GA Biosynthesis in the Scutella Epithelium Is Essential for the Induction of α-Amylase Expression
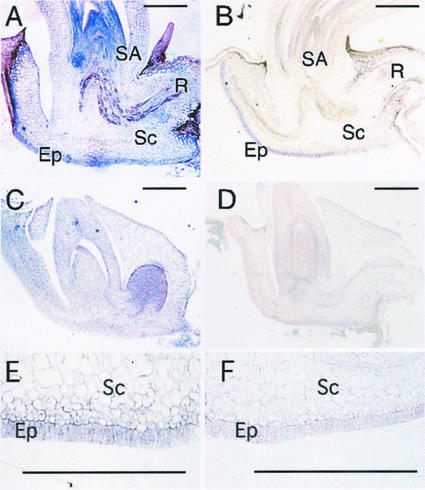
To identify the site of GA biosynthesis, we performed an in situ hybridization study of the OsGA3ox2 transcript and also of the OsGA3ox1 in germinating seeds. A strong signal for the OsGA3ox2 transcript was detected in the shoot apical region, including young leaf primodia and in the epithelial cells facing the endosperm (Fig. 3, A and E). The OsGA3ox1 transcript was also localized in the epithelium but not in the shoot apical region (Fig. 3, B and F). When we used the sense strands as probes, such specific signal was not seen at all (Fig. 3, C and D). The localized expression of OsGA3ox2 indicates that there are two regions where the active GA is actively synthesized in germinating rice seeds, namely the shoot apical region and the epithelium.
Figure 3.
In situ hybridization of OsGA3ox1 and OsGA3ox2 transcripts in germinating embryo. Longitudinal sections through rice embryos fixed after 24 h of imbibition were hybridized with digoxigenin-UTP-labeled OsGA3ox1 (B and F) and OsGA3ox2 (A and E) antisense probes. E and F, Higher magnification views of epithelium cell layers. The section was hybridized with the sense probe for OsGA3ox1 (D) or OsGA3ox2 (C) as control experiment. SA, Shoot apex; R, root; Sc, scutellum; Ep, epithelium. Bar indicates 500 μm.
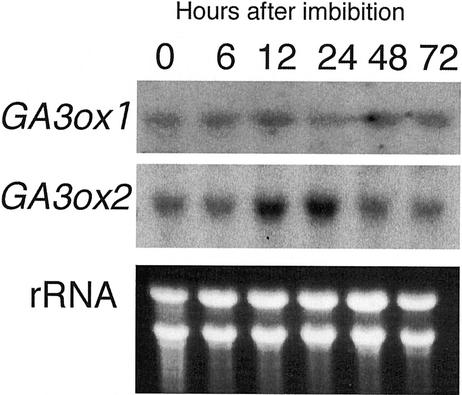
We also examined which gene, OsGA3ox1 or OsGA3ox2, was dominantly expressed in epithelium by northern analysis (Fig. 4), because the in situ analysis demonstrated that both genes expressed in epithelium. Total RNA was isolated from the embryo removed from the shoot region. The expression pattern of each gene agreed well with the previous results using embryo including the shoot region as shown in Figure 1B, and the amount of OsGA3ox2 was much higher than that of OsGA3ox1 at 0.5 or 1 d after imbibition.
Figure 4.
Level of OsGA3ox1 and OsGA3ox2 expression in the shoot-removed embryo. Total RNAs were hybridized with OsGA3ox1 (upper panel) and OsGA3ox2 (lower panel) probes. Seeds were germinated for the indicated hours after imbibition. Ten micrograms of total RNA from shoot-removed embryo was loaded on the gel and stained with ethidium bromide (rRNA).
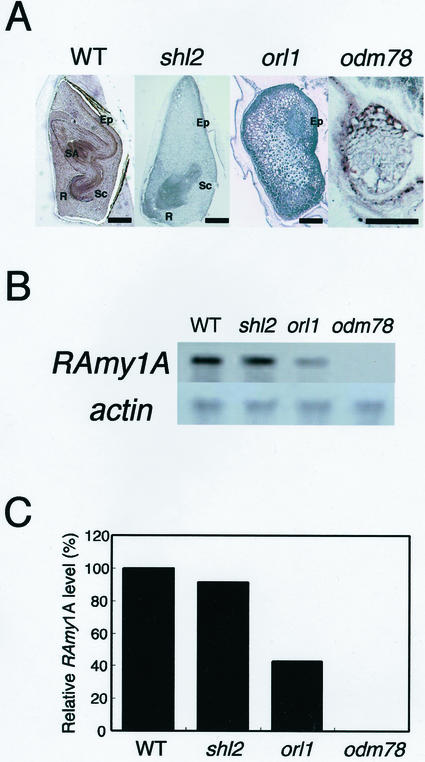
The above results indicate that the shoot apex and epithelial layer where OsGA3ox2 expression occurs is responsible for the induction of α-amylase in endosperm. Nagato et al. (1989) isolated many rice embryonic mutants, and some of these mutants cannot form shoot organs. By using these embryonic mutants, we investigated whether both organ and tissue are required for the α-amylase induction in endosperm. We used three mutants defective in the formation of shoots (Fig. 5A). shootless2 (shl2) develops almost all of the embryonic organs, such as scutellum with epithelium, root, and vascular tissues, but does not form shoot-related tissues, such as the shoot apical meristem, coleoptile, epiblast, and leaf primodia (Satoh et al., 1999). organless1 (orl1) forms a scutellum-like organ with the palisade cells facing toward the endosperm, but lacks almost all other organs. The expression of RAmy1A in the palisade cells in orl1 occurs the same as in the wild-type epithelium (Nagato et al., 1989), indicating that the orl1 palisade cells have epithelium-like characteristics at least in terms of α-amylase expression. The organ deficient mutant 78, odm78, cannot develop any organized organs and only forms small globular embryo with vacuolated cells, although the development of the endosperm occurs normally (Hong et al., 1995). RAmy1A expression in the endosperm was observed in the shl2 and orl1 mutants but not in odm78 (Fig. 5, B and C). The level of the RAmy1A transcript detected in shl2 was similar to that found in the wild-type plant, but in orl1 it was about 40% of the level in the wild type. The similar level of RAmy1A expression in the wild-type and shl2 plants demonstrates that the shoot apical region plays only a small role in α-amylase induction in the endosperm. In contrast to the shoot apical region, the epithelium is essential for α-amylase induction. odm78 seeds defective in epithelium formation did not induce RAmy1A expression at all.
Figure 5.
RAmy1A expression in three types of embryonic organ-deficient mutants. A, Phenotypes of the mature mutant embryos. Each embryo is shown with the embryo-epithelium boundary to the right side. The characteristic phenotype of each mutant is described in the text. SA, Shoot apex; R, root; Sc, scutellum; Ep, epithelium. Bar indicates 500 μm. B, RAmy1A expression in the mutant endosperm. After 96 h of imbibition, total RNA was extracted from the endosperm; 1 μg of total RNA was used for the northern analysis. The lower panel shows a loading control probed with actin cDNA. C, Relative levels of RAmy1A expression in B. The RAmy1A expression levels were normalized against the level of expression of the actin gene. The RAmy1A level in the wild type was set at 100%.
DISCUSSION
The high-level expression of RAmy1A occurred in endosperm without treatment of uniconazol, whereas such high-level expression was not observed in endosperm treated with uniconazol or in the embryo (Fig. 1A). The fact that the uniconazol treatment had no effect on the RAmy1A expression in the embryo (Fig. 1A) indicates that α-amylase expression in the embryo does not depend on de novo-synthesized GAs but may depend on pre-existing GAs synthesized during seed maturation, as occurs in wheat (Triticum aestivum) seeds (Lenton et al., 1994). In contrast to this situation, the induction of RAmy1A expression in endosperm was caused by the de novo-synthesized active GA (Fig. 1A). Based on the following evidence, we have concluded that the GA 3β-hydroxylase encoded by OsGA3ox2 mainly catalyzes the active GA synthesis to induce the RAmy1A expression in endosperm. First, the dramatic increase in RAmy1A expression in endosperm occurred after the rapid induction of the OsGA3ox2 expression in embryo (Fig. 1). Second, the level of RAmy1A expression in the loss-of-function mutant of OsGA3ox2 (d18-AD) was almost the same as that of uniconazol-treated seeds, whereas the level in the nontreated wild-type endosperm was about 2.5 times higher than that of treated endosperm (Fig. 2).
The expression kinetics of OsGA3ox2 is also consistent with a previous finding that the increase in the level of GA1 during rice seed germination starts 1 d after imbibition (Choi et al., 1996), whereas the peak of the OsGA3ox2 expression was from 12 to 24 h after imbibition. The correlation between the OsGA3ox2 expression pattern reported here and the previous observation confirms that the OsGA3ox2 product mainly contributes to the de novo GA synthesis in germinating seeds. In contrast to the rapid increment of OsGA3ox2 expression at the early time course, the expression of OsGA3ox2 was then suppressed to basal level during next 24 h (Fig. 1B). This OsGA3ox2 suppression may be regulated by the negative feedback mechanism by GA. It is known that the expression of some 3β-hydroxylase genes in various plants is negatively regulated by bioactive GAs at the transcriptional level (Chiang et al., 1995; Lester et al., 1997; Martin et al., 1997; Ait-Ali et al., 1999). In fact, OsGA3ox2 expression is suppressed in seedlings by treatment with GA3, whereas the expression of OsGA3ox1 is not affected by this treatment (Itoh et al., 2001). The maintenance of a high level of OsGA3ox2 in the uniconazol-treated seeds can be also explained by the same mechanism, that is, the treatment with uniconazol inhibits active GA biosynthesis even under the high level of expression of OsGA3ox2.
Although the OsGA3ox2 product is important for the RAmy1A expression in endosperm, the fact that some RAmy1A mRNA was detected in d18-AD also indicates that some other factor(s) contributes to the RAmy1A mRNA induction. There are two possible explanations for this pattern of RAmy1A mRNA accumulation. One is that the product of OsGA3ox1, which is constitutively expressed at a low level in the embryo, functions to induce RAmy1A expression. However, this hypothesis does not explain why the wild-type and d18-AD seeds treated with uniconazol accumulated the RAmy1A transcript at a similar level to that in the untreated d18-AD seeds (Fig. 2). Uniconazol blocks early GA biosynthesis by inhibiting ent-kaurene oxidase. If the OsGA3ox1 product functions to produce active GAs in the mutant, the d18-AD seeds treated with uniconazol should show a low level of α-amylase expression relative to that in the untreated seeds. The other possibility is that pre-existing active GAs trigger the accumulation of the RAmy1A transcript. This hypothesis seems more plausible than the former because it would also explain why some RAmy1A expression was detected in the uniconazol-treated seeds.
In situ hybridization analysis revealed that the OsGA3ox2 transcript was localized in the shoot apical region and in the epithelial cells (epithelium; Fig. 3). The OsGA3ox2 expression in young leaf primordia is consistent with our previous finding that OsGA3ox2 expression occurs around the shoot apex of vegetative seedlings (Sakamoto et al., 2001), and also with the quantitative analysis that the active GA content is preferentially higher in young leaves (Choi et al., 1995). Based on their biochemical and molecular studies, Appleford and Lenton (1997) proposed that the scutellum tissue may be important for de novo GA biosynthesis in wheat seeds. Our observations confirm the importance of the scutellum tissue for induction of α-amylase expression in the endosperm.
Our other approach using rice embryonic organ-deficient mutants has clearly demonstrated that the epithelial cells are essential to induce the RAmy1A expression in endosperm, whereas the shoot apex region, which is the other site for the OsGA3ox2 expression, is not important for the RAmy1A expression (Fig. 5). The biological activity of the scutellum or epithelium may directly influence the level of expression of α-amylase, because the orl1 mutant with a partially developed scutellum induced about one-half the level of RAmy1A transcript relative to that of the wild-type plant.
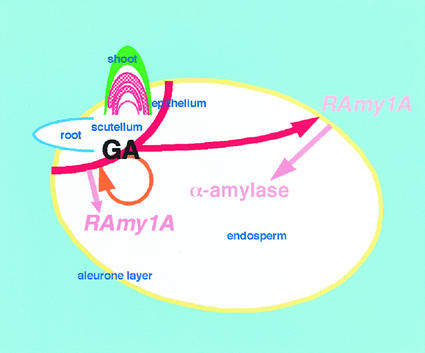
Taking all of the above observations into consideration, we have modeled the relationship between GA biosynthesis in the embryo and the α-amylase expression in the endosperm during rice seed germination as shown in Figure 6. At the early stage of imbibition, pre-existing GA in the embryo triggers the expression of α-amylase in the embryo without de novo synthesis of GA (orange arrow). Then, OsGA3ox2 expression is induced in the epithelium cells, and the product synthesizes the active GA (red bold line at the epithelium layer). OsGA3ox2 expression also occurs in the shoot apical region (red hatch) but the product in this region does not contribute to α-amylase induction. The GAs produced in the epithelium layer are mainly transported to the aleurone (red arrow), where they induce the production of α-amylase. Endosperm starch is gradually hydrolyzed by α-amylase to supply energy for germination.
Figure 6.
Schematic illustration of the sites of GA synthesis and RAmy1A expression during germination. The sites of GA biosynthesis, the shoot apical region and epithelium, during seed germination are shown in red, and the transport of GA for RAmy1A induction is indicated by the red arrow. The expression of RAmy1A in the aleurone layer (yellow) is triggered by the transported GA and induces the α-amylase activity in the endosperm. The pre-existing GA in the embryo induced the expression of RAmy1A in the epithelium (orange circular arrow).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Several types of rice (Oryza sativa) seeds were used in this study. Taichung 65 (T65) is the original wild-type line for the shl2, orl1, and odm78 mutants, and Akibare is the original wild-type line for d18-AD. Mature dry seeds were sterilized with 30% (v/v) sodium hypochloride and then rinsed several times with sterilized water. The seeds were incubated on filter paper wetted with water in the presence or absence of 10 μm uniconazol in 0.2% (v/v) ethanol for odm for 72 h at 30°C. Total RNA was extracted separately from the embryo and endosperm. For in situ hybridization, the sterilized seeds were incubated on 1% (w/v) agar for 24 h at 30°C.
In Situ Hybridization
Plant materials were fixed in 4% (w/v) paraformaldehyde and 0.25% (v/v) glutaraldehyde in 0.1 m sodium phosphate buffer, pH 7.4, overnight at 4°C, dehydrated through a graded ethanol series followed by a t-butanol series (Sass, 1958), and finally embedded in Paraplast Plus (Sherwood Medical, St. Louis). Microtome sections (8–10 μm thick) were mounted on glass slides treated with silane. Digoxygenin-labeled RNA probes were prepared from the 3′-terminal halves of cDNA clones of OsGA3ox1 and OsGA3ox2 (Itoh et al., 2001). Hybridization and immunological detection of the hybridized probes were performed according to the method of Kouchi and Hata (1993).
Northern-Blot Analysis
Total RNA from the embryo and endosperm was extracted by the standard method (Sambrook et al., 1989). Five micrograms of total RNA from the embryo and 1 μg of total RNA from the endosperm were electrophoresed in a 1% (w/v) agarose/formaldehyde gel and then transferred to Hybond N+ membrane (Amersham, Buckinghamshire, UK) with 20× SSC. Twenty micrograms of total RNA from the embryo was electrophoresed to detect OsGA3ox1. Hybridization was performed at 65°C in sodium chloride/sodium phosphate/EDTA, 5× Denhardt's solution, 0.5% (w/v) SDS, 10% (w/v) dextran sulfate, and 0.1 mg mL−1 denatured salmon sperm DNA. Filters were washed with 2× SSC and 0.1% (w/v) SDS for 20 min at 65°C and 0.2× SSC and 0.1% (w/v) SDS for 5 min at room temperature.
Quantification of mRNA
To determine the level of hybridization, the membranes were exposed to a Imaging Plate (Fuji Photo Film, Tokyo) and analyzed on a BAS2000 by using Image Gauge (version 3.41) software (Fuji Photo Film). After autoradiography, the membranes were washed and reprobed with a radiolabeled actin fragment from rice. The highest level of the ratio of target gene to actin mRNA was set at 100.
ACKNOWLEDGMENTS
We thank Dr. Junji Yamaguchi (Hokkaido University, Sapporo, Japan) for providing the RAmy1A clone and also thank Dr. Hidemi Kitano (Nagoya University) and Dr. Yasuo Nagato (University of Tokyo) for providing the embryonic mutant seeds.
Footnotes
This work was supported in part by a grant-in-aid from the Program for Promotion of Basic Research Activities for Innovative Biosciences and by the Special Coordination Fund of the Science and Technology Agency (to M.M.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010785.
LITERATURE CITED
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y. Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol. 1999;121:783–791. doi: 10.1104/pp.121.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa T, Hara-Mishimura I. Topographic aspects of biosynthesis, extracellular section and intracellular storage of proteins in plant cells. Annu Rev Plant Physiol. 1985;70:441–472. [Google Scholar]
- Appleford NEJ, Lenton JR. Hormonal regulation of α-amylase gene expression in germinating wheat (Triticum aestivum) grains. Physiol Plant. 1997;100:534–542. [Google Scholar]
- Beck E, Ziegler P. Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:95–117. [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y-H, Kobayashi M, Fujioka S, Matsuno T, Hirosawa T, Sakurai A. Fluctuation of endogenous gibberellin levels in the early development of rice. Biosci Biotech Biochem. 1995;59:285–288. [Google Scholar]
- Choi Y-H, Kobayashi M, Sakurai A. Endogenous gibberellin A1 level and a-amylase activity in germinating rice seeds. J Plant Growth Regul. 1996;15:147–151. [Google Scholar]
- Fincher GB. Molecular and cellular biology associated with endosperm mobilization in germinating cereal grains. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:305–345. [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.1105/tpc.7.11.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JA, Peacock WJ, Dennis ES. Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA. 1998;95:9019–9024. doi: 10.1073/pnas.95.15.9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-T, Aoki T, Kitano H, Satoh H, Nagato Y. Phenotypic diversity of 188 rice embryo mutants. Dev Genet. 1995;16:298–310. [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Two differently expressed gibberellin 3β-hydroxylase genes are involved in the regulation of vegetative and reproductive growth of rice. Proc Natl Acad Sci USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N. Fluctuation and localization of endogenous gibberellins in rice. Agric Biol Chem. 1988;52:1189–1194. [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol Gen Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenton JR, Appleford NEL, Croker SJ. Gibberellins and α-amylase gene expression in germinating wheat grains. Plant Growth Regul. 1994;15:261–270. [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Proebsting WM, Hedden P. Mendel's dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci USA. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagato Y, Kitano H, Kamijima O, Kikuchi S, Satoh H. Developmental mutants showing abnormal organ differentiation in rice embryo. Theor Appl Genet. 1989;78:11–15. doi: 10.1007/BF00299746. [DOI] [PubMed] [Google Scholar]
- O'Neil S, Kumagai MH, Majumdar A, Hunang N, Sutliff TD, Rodriguez RL. The alpha-amylase genes in Oryza sativa: characterization of cDNA clones and mRNA expression during seed development. Mol Gen Genet. 1990;221:235–244. doi: 10.1007/BF00261726. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 2001;15:581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cell: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sass AE. Botanical Microtechnique. 3rd Ed. Ames, IA: Iowa State University Press; 1958. [Google Scholar]
- Satoh N, Hong S-T, Nishimura A, Matsuoka M, Kitano H, Nagato Y. Initiation of shoot apical meristem in rice: characterization of four SHOOTLESS genes. Development. 1999;126:3629–3636. doi: 10.1242/dev.126.16.3629. [DOI] [PubMed] [Google Scholar]
- Sun T-P, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene syntheses B from pumpkin (Cucurbita maxima L.) Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]