Abstract
The most abundant soluble tuber protein from the Andean crop oca (Oxalis tuberosa Mol.), named ocatin, has been purified and characterized. Ocatin accounts for 40% to 60% of the total soluble oca tuber proteins, has an apparent molecular mass of 18 kD and an isoelectric point of 4.8. This protein appears to be found only in tubers and is accumulated only within the cells of the pith and peridermis layers (peel) of the tuber as it develops. Ocatin inhibits the growth of several phytopathogenic bacteria (Agrobacterium tumefaciens, Agrobacterium radiobacter, Serratia marcescens, and Pseudomonas aureofaciens) and fungi (Phytophthora cinnamomi, Fusarium oxysporum, Rhizoctonia solani, and Nectria hematococcus). Ocatin displays substantial amino acid sequence similarity with a widely distributed group of intracellular pathogenesis-related proteins with a hitherto unknown biological function. Our results showed that ocatin serves as a storage protein, has antimicrobial properties, and belongs to the Betv 1/PR-10/MLP protein family. Our findings suggest that an ancient scaffolding protein was recruited in the oca tuber to serve a storage function and that proteins from the Betv 1/PR-10/MLP family might play a role in natural resistance to pathogens.
The Andean root and tuber crops constitute a unique reservoir of germplasm biodiversity in the world. They grow at altitudes of 2,400 to 4,000 m in the Andes; therefore, they have great potential for introduction into other highland areas where crops from the Old World are not well adapted (King and Gershoff, 1987). However, the Andean root and tuber crops have been largely overlooked and poorly studied at the biological and biochemical level (Tapia, 1984; National Research Council, 1989; Flores and Flores, 1997).
One of the Andean tuber crops, oca (Oxalis tuberosa Mol.), grows in greatest diversity in the highlands of Ecuador, Peru, and Bolivia, although it is found as far north as Venezuela and as far south as Chiloe Island in Chile. Oca and potato (Solanum tuberosum) are cultivated in the same areas and the same range of altitude, but oca is more resistant to cold temperatures than potato (Pulgar, 1981). Oca is tolerant to temperatures as low as 5°C and grows in moderately cool climates and in very poor soils with pH of 5.3 to 7.8, where other crops cannot survive (Leon, 1964). Oca tubers have a nutritional value as good as, or better than, potato (National Research Council, 1989), and they vary in nutritional value depending on the variety (Cortes, 1977). In a study by Janick and Simon (1988), the authors identified oca as an excellent source of carbohydrates that should be highly digestible by monogastric animals because of the low content of alpha-galactosidase and fiber and the high content of digestible sugars. Compared with other Andean tuber crops, including potato, some oca morphotypes have considerably higher levels of essential amino acids (Hodge, 1957). For example, high concentrations of Val and Lys (4.0 and 4.5 mg g−1 fresh weight, respectively) were found in some of the oca morphotypes. Amounts of riboflavin and thiamine present in oca tubers (0.07 and 0.05 mg g−1 fresh weight, respectively) were lower than those obtained in some potato varieties (1.0 and 0.065 mg g−1 fresh weight, respectively); however, the amounts of iron and phosphorus in oca (0.8 and 34.0 mg g−1 fresh weight, respectively) were considerably higher than those in potato (0.6 and 25 mg g−1 fresh weight, respectively; Hodge, 1957; Seminario, 1988). Studies by Cortes (1977) on different oca tuber morphotypes showed that oca tubers can be a potential commercial source of flour, starch, and alcohol. According to Ortega (1992), oca is an excellent source of carbohydrates, alcohol, sugar, phosphorus, calcium, iron, and vitamins.
Oca is now cultivated in New Zealand, Australia, and Mexico (National Research Council, 1989), but its basic biology and biochemistry are virtually unknown (Stegemann and Schmiediche, 1981; Stegemann et al., 1988). Little is known about the biochemistry of most vegetative storage organs except for potato, sweet potato (Ipomoea batata), and yam (Dioscorea cayensis; Racussen and Foote, 1980; Maeshima, et al., 1985; Coursey, 1991).
Vegetative storage organs contain a large amount of storage proteins (constituting 30%–80% of total soluble proteins), which serve as a nutritional resource to support sprouting and regrowth (Stegemann and Schmiediche, 1981). Vegetative storage proteins are synthesized with a signal peptide, which targets them to the vacuole (Matsuoka and Nakamura, 1991). In addition to their storage function, they could play additional roles in the plant: sporamin, from sweet potato, has an antiprotease activity and is thought to play a role in defense against insects (Yeh et al., 1997). Patatin, from potato tubers, has phospholipase A2 activity, inhibits worm larval growth (Strickland et al., 1995), and is suggested to play a role in the hypersensitivity reaction (Senda et al., 1996).
In this study, we found that the protein patterns of the soluble proteins from 36 different oca tuber morphotypes are very similar when separated by SDS-PAGE. The major tuber soluble protein (ocatin) was characterized. The amino acid sequence of ocatin showed homology with proteins that accumulate in response to elicitation, pathogens, or wounding in a wide variety of angiosperms. Ocatin was found to have antibacterial and antifungal activities against several soil-borne microbes. Thus, ocatin serves as a storage protein and might play a role in natural resistance to pathogens.
RESULTS
Ocatin, an Acidic Protein of 18 kD, Is the Major Soluble Protein of Oca Tubers
The moisture content and protein concentration (dry weight basis) of 36 different oca tuber morphotypes were determined. The moisture content was similar among all morphotypes, ranging from 62% to 84%, whereas major differences were observed in protein concentration varying from 67 mg g−1 (morphotype cc) to 24 mg g−1 (morphotype t). The protein content of the tuber increases at earlier stages of development until the tuber is ready to harvest (3–4 months old), but decreases thereafter (data not shown).
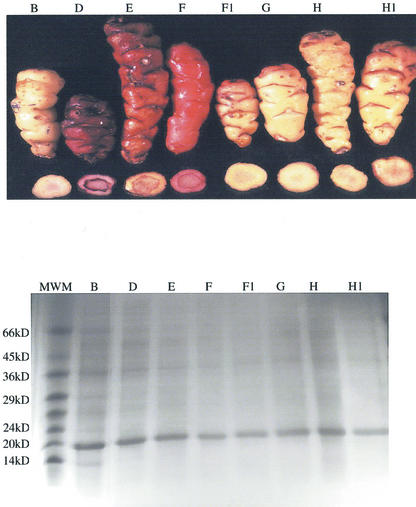
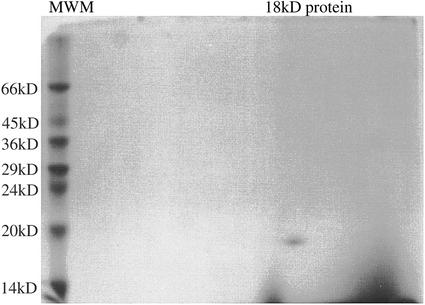
When the oca tuber-soluble proteins were separated by SDS-PAGE under reducing conditions, all 36 morphotypes examined displayed a very similar protein pattern (Fig. 1, a–c). The most prominent band, which showed an apparent molecular mass of 18 kD accounts for 40% to 60% of the total soluble tuber proteins, as determined by densitometric analysis. This protein was isolated by a two-step procedure. In the first step, the total oca tuber-soluble proteins were separated in five fractions by anion-exchange chromatography and subsequently analyzed by SDS-PAGE. In the second step, the proteins contained in fraction 3 were further fractionated by anion-exchange chromatography using a less steep gradient. After purification, a single protein band with a molecular mass of 18 kD was separated by SDS-PAGE. A two-dimensional gel electrophoresis showed a single protein spot (Fig. 2), thus corresponding to a homogeneous protein with a pI point of 4.8; this protein was named ocatin. Amino acid composition of ocatin showed that it has high amounts of Val, Thr, Tyr, Gln, Gly, and Ala when compared with those amino acids of patatin and sporamin (Table I).
Figure 1.
a through c, Pattern of tuber-soluble proteins from 24 different oca morphotypes. The tuber-soluble proteins were extracted as described in “Materials and Methods,” separated in SDS-PAGE under reducing conditions, and stained with Coomassie Blue. Fifteen micrograms of protein was loaded per lane. The right lane shows the migration of Mr markers.
Figure 2.
Two-dimensional isoelectric focusing SDS-polyacrylamide gel (13.5% [w/v]). Twenty micrograms of purified ocatin was loaded on the isoelectro-focusing gel. The gel was stained with Coomassie Blue. Only one spot was detected in the gel. Mr markers (MWM) are indicated on the left.
Table I.
Comparison of the amino acid composition (milligrams per gram of ocatin with patatin; Kapoor et al., 1975) and sporamin A and B (Maeshima et al., 1985)
| Amino Acid | Ocatin | Patatin | Sporamin A | Sporamin B |
|---|---|---|---|---|
| mg g−1 protein | ||||
| Val | 50 | 41 | 25 | 29 |
| Met | 10 | 19 | 5 | 3 |
| Thr | 20 | 7 | 7 | 15 |
| Ile | 25 | 35 | 48 | 50 |
| Leu | 60 | 92 | 70 | 56 |
| Phe | 45 | 52 | 53 | 52 |
| Tyr | 60 | 52 | 36 | 38 |
| Trp | 4 | 10 | 9 | 7 |
| Lys | 55 | 66 | 38 | 41 |
| Asn | 120 | 95 | 131 | 132 |
| Ser | 62 | 49 | 63 | 68 |
| Gln | 165 | 128 | 130 | 141 |
| Gly | 38 | 28 | 26 | 23 |
| Pro | 35 | 32 | 36 | 33 |
| Ala | 70 | 51 | 60 | 61 |
| His | 9 | 3 | 10 | 7 |
| Cys | 4 | – | 2 | 2 |
| Arg | 38 | 29 | 36 | 45 |
Ocatin Is a Tuber Storage Protein and Is Developmentally Regulated
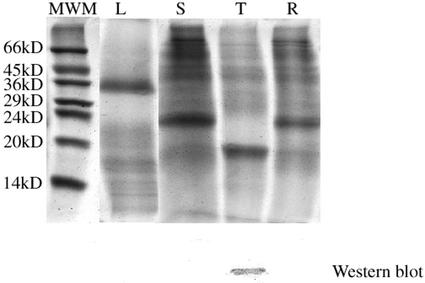
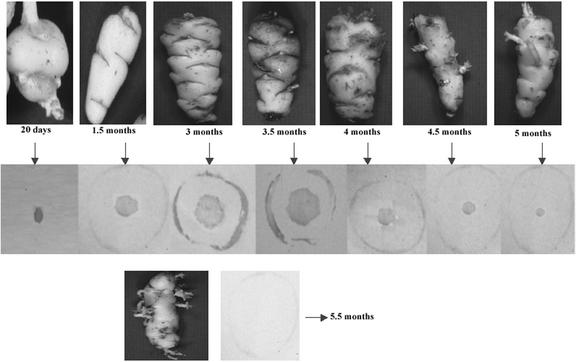
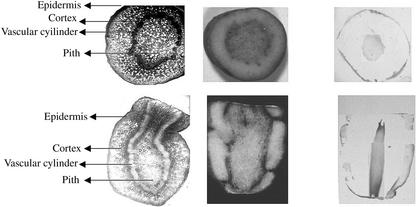
Western-blot analysis of protein extracts from tubers, roots, leaves, and stems of oca tuber (morphotype cc) at different developmental stages suggested that ocatin is a tuber protein (Fig. 3). Tissue print analysis revealed that ocatin is restricted to the pith, subepidermal, and epidermal regions of the tuber (Fig. 4). The content of ocatin gradually increases from 20-d-old to 3.5-month-old tubers, but decreases at older stages and especially under storage and upon sprouting. In 20-d-old tubers, ocatin is already accumulating but it is present only in the pith. In 1.5-month-old tubers, ocatin began to accumulate also in the subepidermal and epidermal layers and its accumulation in these tissues increases during tuber development until the tubers are 3.5 months old (approximate harvest time for this morphotype). Thereafter, the content of ocatin decreases steadily, becoming very low in 5.5-month-old tubers (Fig. 4). The sharp decrease in ocatin is correlated with the appearance of sprouts, which in this morphotype occurs shortly after harvest. Tissue prints also show the spatial distribution of ocatin in the tuber is restricted to the epidermal and the pith tissue; ocatin was absent in the cortical region of the tuber (Fig. 5). The tissue specificity of ocatin, its presence in high amounts in the oca tuber, and its developmental regulation suggested by western blot and tissue print analysis strongly suggest that ocatin plays a major role as a tuber storage protein.
Figure 3.
Pattern of soluble proteins from different organs of the oca plant, and a western-blot analysis using antibodies against ocatin. The soluble proteins were extracted from leaves (L), stems (S), roots (R), and tubers (T) as described in “Materials and Methods,” separated in SDS-PAGE under reducing condition,s and stained with Coomassie Blue. Twenty micrograms of protein was loaded per lane. The most right lane shows the migration of molecular markers. Immunoblots showing the reactivity of the proteins from each organ with specific antibodies against ocatin are shown below the SDS-PAGE profile of the corresponding extract.
Figure 4.
Reactivity of tissue prints from cross sections of oca tuber (morphotype cc) at different developmental stages with antibodies against ocatin. Prints were prepared from cross sections of 20-d, 1.5-, 2.0-, 2.5-, 3.0-, 3.5-, 4.0-, 4.5-, 5.0-, and 5.5-month-old oca tubers and probed with antibodies against ocatin as described in “Materials and Methods.” The size of the image was adjusted for illustration purposes.
Figure 5.
Tissue distribution of ocatin within the oca tuber (morphotype cc). Cross (upper) and longitudinal (lower) sections showing the epidermis, cortex, vascular cylinder, and the pith of oca tuber (40×). Reactivity of prints from cross and longitudinal sections of oca tuber with antibodies against ocatin.
Ocatin Has Antibacterial and Antifungal Activities
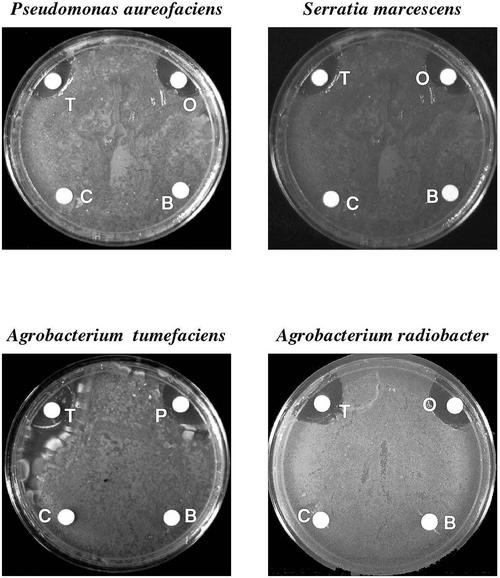
The effect of total soluble oca tuber proteins, the basic oca tuber-soluble proteins, and purified ocatin on the in vitro growth of pathogenic bacteria and fungi was determined. The total soluble oca tuber proteins as well as purified ocatin inhibited the growth of Pseudomonas aureofaciens, Serratia marcescens, Agrobacterium tumefaciens, and Agrobacterium radiobacter. In contrast, the basic oca tuber protein fraction did not affect the growth of any of the bacteria tested (Fig. 6). The effect of different amounts of ocatin on bacterial growth was tested. Table II shows the quantification of antibacterial activity by ocatin.
Figure 6.
Antibacterial activity of ocatin. The effect of filter discs containing total oca tuber soluble proteins (T), basic soluble proteins (B), purified ocatin (O), or buffer as a control (C) on the in vitro growth of A. tumefaciens, A. radiobacter, P. aureofaciens, and S. marcescens.
Table II.
Effect of ocatin on the in vitro growth of different bacterial species
| Bacterial Pathogen | Area of Inhibition |
|---|---|
| mm2 | |
| A. radiobacter | 4.6 |
| A. tumefaciens | 4.9 |
| Agrobacterium rhizogenes R100nal | 0 |
| Bacillus cepacea | 0 |
| Bacillus subtilis | 0 |
| Bacillus thuringiensis | 0 |
| Clavibacter nebraskensis | 0 |
| Erwinia carotovora | 0 |
| Erwinia amylovora | 0 |
| Erwinia herbicola | 0 |
| Escherichia coli ESS | 0 |
| Pseudomonas syringae “B” | 0 |
| P. syringae pv tomato | 0 |
| Pseudomonas phaseolicola | 0 |
| Pseudomonas solanacearum | 0 |
| P. aureofaciens gacA | 0 |
| P. aureofaciens 30–84 | 4.6 |
| P. aureofaciens 30–84 I | 0 |
| Pseudomonas fluorescens 2–79 | 0 |
| Rhizobium leguminosarum | 0 |
| Streptomyces griseovirides “mycostop” | 0 |
| S. marcescens | 4.3 |
| Xanthomonas campestris pv vesicatoria | 0 |
| X. campestris pv peligonia | 0 |
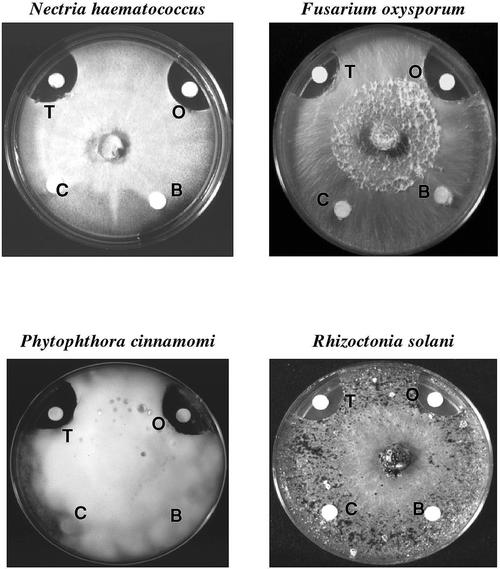
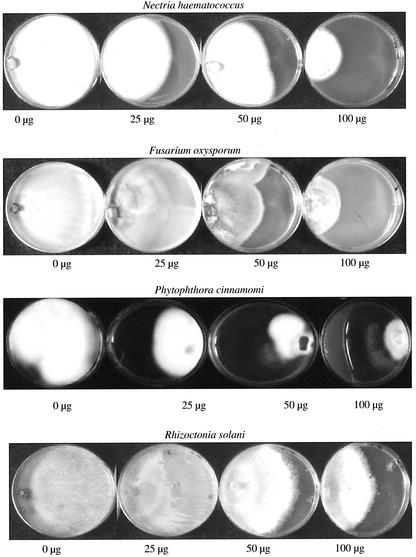
The total soluble oca tuber proteins retarded the growth of ascomycetes (Nectria hematococcus and Fusarium oxysporum), oomycetes (Phytophthora cinnamomi), and basidiomycetes (Rhizoctonia solani; Fig. 7). The growth of these fungi was was inhibited by ocatin in a dose-dependent manner but not affected by the basic tuber protein fraction (Table III; Fig. 8). Hyphal growth inhibition of the slow-growing P. cinnamomi was observed even at the lowest ocatin dose tested (25 μg), whereas the inhibition of the fast-growing R. solani and F. oxysporum required higher ocatin doses (50 and 100 μg).
Figure 7.
Antifungal activity of ocatin. Effect of filter discs containing total oca tuber soluble proteins in (T), basic soluble proteins (B), purified ocatin (O), or buffer as a control (C), in the in vitro growth of F. oxysporum, N. hematococcus, P. cinnamomi, and R. solani.
Table III.
Effect of different amounts of ocatin on the in vitro growth of different fungal species
| Fungus | 25 μg Ocatin | 50 μg Ocatin | 100 μg Ocatin | 200 μg Ocatin |
|---|---|---|---|---|
| Fusarium momordicae | 100 | 100 | 100 | 100 |
| F. oxysporum | 100 | 65 | 60 | 20 |
| Nectria haematococcus | 63 | 57 | 43 | 22 |
| Phytium aphanidermatum | 100 | 100 | 100 | 100 |
| Phytium irregulare | 100 | 100 | 100 | 100 |
| Phytium ultimum | 100 | 100 | 100 | 100 |
| P. cinnamomi | 44 | 23 | 15 | 12 |
| Phytophthora cryptogea | 100 | 100 | 100 | 100 |
| Phytophthora drechsleri | 100 | 100 | 100 | 100 |
| Phytophthora griseae | 100 | 100 | 100 | 100 |
| Phytophthora lateralis | 100 | 100 | 100 | 100 |
| Phytophthora megasperma | 100 | 100 | 100 | 100 |
| P. megasperma glycinea | 100 | 100 | 100 | 100 |
| R. solani | 100 | 66 | 50 | 20 |
| Verticillium dahliae | 100 | 100 | 100 | 100 |
Inhibition of hyphal growth on different fungal species by ocatin (as percentage of total area covered by fungal hyphae).
Figure 8.
Effect of different amounts of purified ocatin (25, 50, 100, and 200 μg) on growth of P. cinnamomi, R. solani, F. oxysporum, and N. hematococcus.
Ocatin Shows Sequence Similarity with Intracellular Pathogenesis-Related Proteins
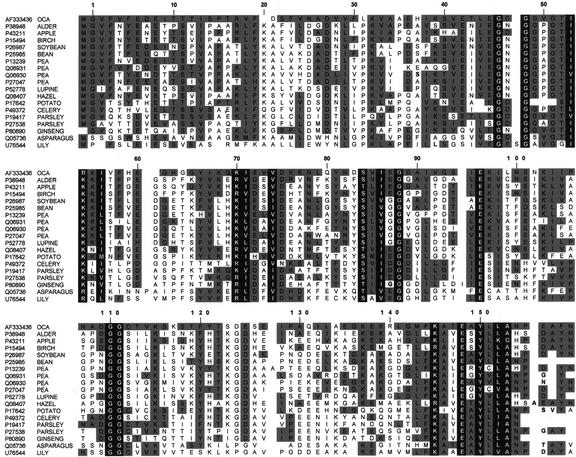
Amino acid sequence analysis of five ocatin tryptic peptides purified after in-gel digestion after two-dimensional gel electrophoresis revealed substantial similarity to proteins of the Bet v 1/PR-10/MLP family of proteins found in a wide variety of species (Fig. 9). This protein family includes several intracellular pathogenesis related proteins (of the PR-10 group; Moiseyev et al., 1997), ribonucleases (Breiteneder et al., 1992, 1993, 1994, 1995), tree pollen, and food allergens related to Bet v 1 (Breiteneder et al., 1992) and major latex proteins (MLP; Osmark, et al., 1998).
Figure 9.
Sequence alignment of ocatin with selected members of the PR-10/Bet v 1/MLP protein family. Accession numbers in the Swissprot or GenBank databases are given on the left. Residues conserved within the family are given against a black background, and residues similar between ocatin and other family members are given against a gray background. Numbers at the top refer to the sequence of Bet v 1, the only member of this family whose three-dimensional structure is known. Segments of ocatin sequence underlined correspond to the five tryptic peptides isolated after in-gel digestion. They correspond to the following proteins and species: P38948, major pollen allergen ALN G 1, Alnus glutinosa; P43211, major allergen MALD1, Malus domestica; P15494, major pollen allergen BET V 1-A, Betula verrucosa; P26987, stress-induced protein Sam22, Glycine max; P25985, pathogenesis-related protein 1, Phaseolus vulgaris; P13239, disease resistance response protein 176, Pisum sativum; Q06931, abscisic acid-responsive protein ABR17, P. sativum; Q06930, abscisic acid-responsive protein ABR18, P. sativum; P27047, disease resistance response protein DRRG49-C, P. sativum; P52778, intracellular pathogenesis-related protein L1R18A, Lupinus luteus; Q08407, major pollen allergen COR A 1, Corylus avellana; P17642, pathogenesis-related protein STH-2, potato; P49372, major allergen API G 1, Apium graveolens; P19417, pathogenesis-related protein A (PR1-1), Petroselinum crispum; P27538, pathogenesis-related protein 2, P. crispum; P80890, ribonuclease 2, Panax ginseng; Q05736, pathogenesis-related protein 1 (AOPR1), Asparagus officinalis; and U76544, tapetal protein L1PR2, Lilium longiflorum.
Ocatin cDNA was amplified by reverse transcription (RT)-PCR as described in '93Materials and Methods'94 and its complete nucleotide sequence was established and deposited at GenBank (accession no. AF333436). The deduced amino acid sequence of ocatin (Fig. 9) corresponds to a protein with a predicted pI of 4.75 and a molecular mass of 17.29 kD, which consists of 153 residues and shows 36% to 44% pair-wise residue identity with those of members of the Bet v 1/PR-10/MLP protein family. The ocatin peptides analyzed by sequence analysis were identical to the deduced sequence at positions 1 through 15, 18 through 32, 71 through 85, 118 through 137, and 146 through 159 in the alignment showed in Figure 8. It is remarkable that ocatin shares the same residue conservation pattern of the Bet v 1/PR-10/MLP protein family, including the nine strictly conserved residues (Fig. 9). Furthermore, it shows a hydropathy curve extensively similar to those of other proteins of this family (not shown). Thus, the pI, molecular size, amino acid sequence, and hydropathy profile of ocatin suggests that it is a new member of the Bet v 1/PR-10/MLP protein family and has sequence similarity with intracellular pathogenesis-related proteins. The proteins of this family are acidic, consist of 145 to 162 amino acid residues, and although many exhibit residue identities of only 15% to 30%, they display similar hydropathy profiles and show a residue conservation pattern (with several invariant glycines separated by highly diverse sequences), typical of distantly related proteins with a conserved three dimensional structure (Osmark et al., 1998).
DISCUSSION
Ocatin is the name we gave to the major storage protein purified and characterized from oca tubers, with an apparent molecular mass of approximately 18 kD. This protein was isolated by anion-exchange chromatography and is present in all oca tuber morphotypes studied. Ocatin expression seems to be highly regulated because it is found initially only in the pith of the primordial tubers swelling at the tip of the stolons. It further accumulates in both pith and epidermal tissues as tuberization proceeds, reaching maximum levels when the tubers are ready to harvest. Thereafter, ocatin levels decrease as the tubers age in storage and start to sprout. Similar expression patterns have been reported for patatin and sporamin, the tuber storage proteins of potato and sweet potato (Hannapel, 1990, 1991). Consistent with its proposed role as a vegetative storage protein, ocatin appears to be synthesized in large amounts in tubers at precise stages of development. This is also the case for other storage proteins such as patatin and sporamin, which account for 40% to 45% and 70% to 80% of the total tuber-soluble proteins, respectively (Racussen and Foote, 1980; Maeshima et al., 1985).
In addition to being a storage protein, ocatin also shows in vitro activity as an antifugal and antibacterial protein. In most cases, storage proteins lack biological activity, although they may be related to metabolically active proteins. However, in a small number of cases, the storage protein itself has been shown to exhibit biological activity. Patatin has phospholipase C activity (Andrews et al., 1988) and sporamin acts as an enzyme inhibitor (Hattori et al., 1985). Ocatin shows little homology with other storage proteins from different species. Storage tuber proteins such as patatin, dioscorin, and sporamin are encoded by multigene families that fall into two subfamilies (Shewry, 1995), but this is not the case for ocatin. In addition, patatin and sporamin are not tuber or root specific, respectively, but can be synthesized in other organs under appropriate conditions and they show differential expression in the vegetative storage organs.
The deduced amino acid sequence of ocatin is similar to proteins of the PR-10/Bet v 1/MLP family and ocatin has the residues conserved within this protein family. Thus, the protein recruited to serve a storage function in the oca tuber is a member of an ancient family of widespread occurrence within the plant kingdom. The proteins in this family are encoded by polymorphic genes and apparently have diverse biological functions. Some of them display RNAse activity (Swodoba et al., 1994) and others are expressed in response to phytohormones or development (Carpin et al., 1998), whereas others accumulate in response to wounding, elicitation, or the presence of pathogenic fungi and bacteria (Walter et al., 1990; Chiang and Hadwiger, 1996). Because of their evolutionary conservation and the rapid transcriptional activation of their genes in response to physical interaction with pathogens, it has been suggested that they may have a role in plant disease resistance. However, until now the data supporting such a role were purely correlative and thus ocatin is the first member of the PR-10/Bet v 1/MLP protein family shown to have clear antibacterial and antifungal activities. These findings suggest that ocatin may play a role in the resistance of tubers to pathogens and raise the possibility that other family members also have antimicrobial activities and could play a role in host-pathogen interactions.
Further mutagenesis studies of ocatin will allow us to identify the structural determinants responsible for its antimicrobial activities. The cloning and expression of ocatin could lead to the production of transgenic crops with an increased resistance to pathogens. The precise pathway of ocatin synthesis, processing, and deposition remains unclear and thus is an important target for future research.
MATERIALS AND METHODS
Extraction of the Oca (Oxalis tuberosa Mol.) Tuber-Soluble Proteins
Thirty-six different oca tuber morphotypes were obtained from the Peruvian Andean regions of Cuzco and Ayacucho. Oca nodal cuttings from morphotypes cc and j were also grown in the greenhouse under hydroponic conditions. Three tubers per morphotype were harvested at 20 d, 1.5, 2.0, 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, and 6.0 months after tuber initiation. Because tubers started to sprout at 4.5 months, sprouts were removed upon harvesting old tubers. All tubers were weighed, frozen in liquid nitrogen, and stored at −80°C.
Three samples (1 g sample−1) of tubers from each morphotype were homogenized in 10 mL of ice-cold extraction buffer (50 mm Tris [pH 6.5], 10 mm thiourea, 10 mm dithiothreitol [DTT], 1.5% [w/v] polyvinylpyrroline, and 2 mm phenyl-methyl-sulfonyl-fluoride). Homogenates were centrifuged at 39,200g for 30 min at 4°C, the supernatants filtrated through glass microfiber filters, and the proteins precipitated by adding 6 mL of cold acetone (−17°C). The samples were then centrifuged and the pellets dissolved in 2 mL of suspension buffer (50 mm Tris [pH 6.5], 20 mm EDTA, and 5 mm DTT). Protein concentration was determined as described by Bradford (1976), using bovine serum albumin (Pierce, Rockford, IL) as standard.
Electrophoresis and Densitometric Analysis
Extracts containing more than 100 μg protein μL−1 were mixed with an equal volume of sample buffer (Laemmli, 1970). Proteins from more diluted extracts were precipitated by the method of Peterson (1977). In brief, 1 mL of the extract was mixed with 100 μg of solid sodium deoxycholate, incubated on ice for 15 min, and centrifuged (4,350g, 10 min). The protein pellets were washed three times with ice-cold 80% (v/v) acetone, and dissolved in sample buffer. Fifteen to 20 μg of protein sample was loaded to each lane and electrophoretic separations in polyacrylamide gels (13.5% [w/v]) were done under reducing conditions on SDS-PAGE (Laemmli, 1970). The gels were run in a double-slab electrophoresis cell (Mini-Protein II, Bio-Rad, Richmond, CA) for 1 h at 150 V. The gels were stained with Coomassie Brilliant Blue R-250 or silver stain (Sigma, St. Louis), and the intensity of the bands quantified after scanning with an LKB Ultrascan XL laser densitometer (Pharmacia Biotech, Piscataway, NJ).
Purification of Ocatin and Antiserum Production
A protein extract from tubers was ultrafiltrated through 10 NMWL membranes (Millipore, Medford, MA) and concentrated to 2 μg protein mL−1. Proteins were precipitated with (NH4)2SO4 (80% [w/v] final saturation) and dialysed against 25 mm Tris (pH 6.5). Batches of 250 mg of protein were fractionated on a 4.6- × 100-mm HPLC column packed with Poros HQ50 (PE Applied Biosystems, Foster City, CA) with a linear gradient from 60 to 600 mm NaCl in 20 min at a flow rate of 5 mL min−1 using a 600E HPLC instrument (Waters, Milford, MA). Five fractions of 1 mL were collected and analyzed by SDS-PAGE. Material corresponding to fraction 3 from different fractionations were pooled, diluted, applied to the same column, and eluted with a linear gradient of 150 to 200 mm NaCl in 20 min. The material eluting at 160 mm NaCl was collected and its homogeneity was verified by two-dimensional PAGE (O'Farrel, 1975).
Polyclonal antibodies to ocatin were prepared in a New Zealand white rabbit at the Laboratory of Animal Resources (Centralized Biological Laboratory, Penn State University) by subcutaneous injection of native protein (0.5 mg) emulsified in Freund's complete adjuvant. Subsequent booster injections with 0.5 to 0.8 mg of native protein were given after 14, 21, and 28 d in Freund's incomplete adjuvant. The antiserum was collected by heart puncture after 38 d and stored at −20°C.
Western Blotting and Tissue Printing Studies
Total soluble proteins (20 μg lane−1) from oca roots, stems, or tubers were submitted to electrophoresis under reducing conditions in an SDS polyacrylamide gel (13.5% [w/v]) and separated proteins were electroblotted on to an Immobilon-P polyvinylidene difluoride (PVDF) membrane (Millipore) as described (LeGendre and Matsudaira, 1989) using a Bio-Rad Mini-Trans electrotransfer cell. The membrane was then saturated with blocking solution (0.25% [w/v] bovine serum albumin, 0.25% [w/v] gelatine, and 0.3% [v/v] Tween 20 in Tris-buffered saline solution [20 mm Tris-HCL and 0.9% {w/v} NaCl {pH 74}]) for 1 to 3 h at room temperature with constant shaking before a 1- to 3-h incubation with antibodies against ocatin diluted in blocking solution (1:5,000 [v/v]). Unbound antibodies were removed by washing three times (30 min each) with Tris-buffered saline solution containing 0.3% (v/v) Tween 20 before incubation with an anti-rabbit IgG-alkaline phosphatase conjugate (Promega, Madison, WI). The membrane was washed as described above and incubated with nitroblue tetrazolium (250 μg mL−1) and 5-bromo-4chloro-3-indolyl phosphate (56 g mL−1) (Promega) in 50 mm Tris-HCL, pH 9.8, containing 1 mm MgCl2 until the bands appeared. The reaction was stopped by quickly washing the membrane with distilled water.
Tissue printing was performed as described by Reid et al. (1992). In brief, tubers were cut in radial or longitudinal sections of 1 to 2 mm thick and preblotted on a 3-mm filter paper. The sections were then transferred with forceps to a 0.45 m nitrocellulose membrane (Schleicher and Schuell) previously soaked in 0.2 m CaCl2 for 30 min and quickly dried on a 3-mm filter paper. Using a gloved fingertip, the sections were pressed onto the membrane for 15 to 30 s and the print was dried with a hair drier. The membranes were then treated as described for western blotting. Tissue print replicates were performed three times for each developmental stage.
Antimicrobial Activity Assays
The effect of total soluble oca tuber proteins, the basic oca tuber-soluble proteins, or purified ocatin in the in vitro growth of 24 bacterial species and 15 fungal species (Table IV) was tested. Bacterial cultures were grown overnight at 24°C with shaking in Luria-Bertani broth (Sambrook, et al., 1989) to an optical density reading of 0.2 at 600 nm. A 100-μL aliquot of this culture was spread on 80-mm plates containing Luria-Bertani agar media.
Table IV.
List of bacterial and fungal species tested against ocatin
| Bacterial Pathogen | Source |
|---|---|
| A. radiobacter | Penn State University |
| A. tumefaciens | U.S. Department of Agriculture (USDA; Harrisburg, PA) |
| A. rhizogenis R100nal | USDA |
| B. cepacea | Penn State University |
| B. subtilis | Penn State University |
| B. thuringenesis | Louisana State University (Baton Rouge) |
| C. nebraskensis | American Type Culture Collection (ATCC) |
| E. carotovora | Penn State University |
| E. amylovora | Louisiana State University |
| E. herbicola | Louisiana State University |
| E. coli ESS | Louisiana State University |
| P. syringae “B” | Penn State University |
| P. syringae pv tomato | Louisiana State University |
| P. phaseolicola | Louisiana State University |
| P. solanacearum | ATCC |
| P. aureofaciens gacA | Louisiana State University |
| P. aureofaciens 30–84 | Penn State University |
| P. aureofaciens 30–84 I | Louisiana State University |
| P. fluorescens 2–79 | Louisiana State University |
| R. leguminosarium | Penn State University |
| S. griseovivides “mycostop” | USDA |
| S. marcescens | Penn State University |
| X. campestris pv vesicatoria | USDA |
| X. campestris pv peligonia | Louisiana State University |
| Fungi Species | |
| F. momoridicae | ATCC |
| F. oxysporum | Penn State University |
| N. haematococcus | USDA |
| P. aphanidermatumm | Penn State University |
| P. irregulare | Penn State University |
| P. ultimum | Louisiana State University |
| P. cinnamomi | USDA |
| P. cryptogea | USDA |
| P. drechsleri | Louisiana State University |
| P. griseae | Louisiana State University |
| P. lateralis | ATCC |
| P. megasperma | Louisiana State University |
| P. megasperma glycenea | Louisiana State University |
| R. solani | Louisiana State University |
| V. dahiae | USDA |
The in vitro growth of pathogenic fungi was determined on agar plates as described (Roberts and Selitrennikoff, 1990). A sterile filter paper disc saturated with 50 mm Tris buffer (pH 6.5) containing either 50 μg of total crude extract of soluble oca tuber protein, 50 μg of the basic oca proteins, or 50 μg of ocatin was placed in the inoculated plates. As a control, we used a sterile filter paper disc saturated with a 50 mm Tris buffer solution (pH 6.5) without protein. Protein solutions were filter sterilized using Ultrafree-MC Durapore 0.2-μm filters (Millipore). The plates were incubated in the dark at 24°C and inhibition was defined as a bacteria- or mycelium-free zone surrounding a filter disc 24 h after inoculation. Three replicates per treatment were included, and each experiment was done at least twice. In the case of antibacterial activity, this was quantified by measuring the area of bacteria-free zone in the plates (Table II) upon exposure to 50 μg of ocatin.
A dose response experiment was performed with the fungi in which the growth was inhibited by ocatin. Ocatin amounts of 25, 50, 100, and 200 μg was placed directly in 40-mm petri dishes containing potato dextrose agar medium. Ocatin was added to the medium just before it solidified. A 3-mm plug of fungal hyphae was placed on one side of the plate and hyphal length was measured at 24-h intervals. Solid potato dextrose medium containing only Tris buffer (pH 6.5) was used as a control.
Structural Analysis
The amino acid composition of total soluble protein extracts from the oca morphotypes cc and j, as well as that from ocatin, were determined using a Pharmacia-LKB Alpha Plus 4151 ninhydrin-based analyzer (Pharmacia Biotech, Piscataway, NJ) after hydrolysis at 110°C for 24 h in evacuated tubes with 6 m HCl containing 0.5% (w/v) phenol.
The Coomassie-stained ocatin band was cut from a two-dimensional polyacrylamide gel and the piece was placed in an Eppendorf tube for in-gel digestion. In brief, washing was carried out in 0.2 m ammonium bicarbonate containing 50% (v/v) acetonitrile. The protein was reduced with DTT and alkylated with idioacetamide followed by in-gel digestion with 0.5 to 3 μg of trypsin (Promega) in 0.2 m ammonium bicarbonate overnight at 37°C. The tryptic peptides were extracted using acetonitrile in 0.1% (v/v) trifluoroacetic acid, first at 60%, then 40% (v/v). The peptide extract was separated and fragments isolated on a PVDF membrane for sequence analysis using a Microblotter system (Perkin-Elmer/Applied Biosystems Division, Foster City, CA). Edman degradation of PVDF bound peptides was carried out with a Procise CLC sequencer (PE/Applied Biosystems).
RNA Isolation, RT-PCR Amplification, and Sequence Analysis of Ocatin cDNA
Total RNAs from 3.5-month-old ocatin tubers were isolated and RNA concentration was determined spectrophotometrically as A260. RT was performed as follows: 1.5 μg of the purified RNA was incubated with 2 μL of oligo(dT) (50 μm; the volume was adjusted to 10 μL with distilled water) at 70°C for 5 min and then chilled on ice. Then, 1 μL of avian myeoloblastosis virus-RT (24 units μL−1), 4 μL of dNTPs (2.5 mm), 0.5 units of ribonuclease inhibitor (40 units μL−1), and 2 μL of 10× RT-first strand buffer were added, and the volume was adjusted to 20 μL with distilled water. After heating the reaction mixture to 95°C for 5 min and incubation for 60 min at 40°C, the resulting cDNAs were stored at −20°C until use. Primers were initially designed according to the sequences of the tryptic fragments of the N- and C-terminal regions of ocatin (5′-ATGGGTGTTTTCGTATTCGAGG-3′ and 5′-GATTAGTTG-TAATCGGGATGGG-3) to amplify a 479-bp fragment. PCR was performed with 3 μL of cDNA, 4 μL of dNTPs (2.5 mm), 3 μL of each primer (10 μm), 2.5 μL of Dynazyme (2 units μL−1; Finnzymes Oy, Espoo, Finland), and 5 μL of 10× Dynazyme buffer, the volume being adjusted to 50 μL with distilled water. After 3 min at 95°C, the mixture was incubated for 27 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 2 min. The fragment was run on a 1% (w/v) low-Tm agarose gel, stained with ethidium bromide, excised under UV transillumination, and purified using the BandPrep kit (Pharmacia Biotech Inc.). The purified fragment was submitted to sequence analysis using the GeneAmp PCR system 9600 and the Applied Biosystems Prism dye terminator cycle sequencing kit (PE/Applied Biosystems) according to the manufacturer's instructions. To complete the full sequence, two primers were designed based on the sequence of 5′- and 3′- flanking regions of the soybean stress-induced gene GMSAM, which showed maximal homology with the sequence of ocatin at the protein level. The primers used were 5′-CTCAAACTAGTAG-TATTATTCTTCC-3′ and 5′-GATGACAAGTAAGTTGAA-GAGG-3′. Sequence analysis was performed twice on both strands.
Sequence Alignment and Structural Predictions
Hydropathy plots were calculated using a sliding six-residue window as described (Kyte and Doolittle, 1978). Similarity searches were done at Swiss Prot using FASTA (Pearson and Lipman, 1988) and sequences were aligned using the program CLUSTAL W (Higgins et al., 1996) with manual adjustments.
ACKNOWLEDGMENTS
We thank Dr. Monica Thelestam (Karolinska Institute, Stockholm) for laboratory facilities. We also thank Dr. Brett Savary (USDA-ARS, ERRC, Wyndmoor, PA) for technical advice, Mitchelle Peipher for help with immunoelectron microscopy analysis, and Dr. Jorge Vivanco (Colorado State University, Fort Collins) and Paula Michaels for critical reading of the manuscript. Special thanks to the farmers from Picol and San Jose de Arizona (Cuzco and Ayacucho, Peru, respectively) and to Dr. Marleni Ramirez (U.S. State Department, Washington, DC) for their help with the oca collection. Amino acid composition and sequence analysis was performed at the Protein Analysis Center (Karolinska Institute).
Footnotes
This work was supported by a grant from the McKnight Foundation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010541.
LITERATURE CITED
- Andrews DL, Beames B, Summers M, Park WD. Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J. 1988;252:199–226. doi: 10.1042/bj2520199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Ferreira F, Hoffmann-Sommergruber K, Ebner C, Breitenbach M, Rumpold H, Kraft D, Scheiner O. Four recombinant isoforms of Cor I, the major allergen of hazel pollen, show different IgE-binding properties. Eur J Biochem. 1993;212:355–362. doi: 10.1111/j.1432-1033.1993.tb17669.x. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Ferreira F, Reikerstorfer A, Duchene MK, Valenta R, Hoffmann-Sommergruber K, Ebner C, Breitenbach M, Kraft D, Scheiner O. Complementary DNA cloning and expression in Escherichia coli of alng I, the major allergen in pollen of alder (Alnus glutinosa) J Allergy Clin Immunol. 1992;90:909–917. doi: 10.1016/0091-6749(92)90463-c. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Hoffmann-Sommergruber K, O'riordan G, Susani M, Ahorn H, Ebner C, Kraft D, Scheiner O. Molecular characterization of Api g I the major allergen of celery (Apium graveolens), and its immunological and structural relationships to a group of 17 kDa tree pollen allergens. Eur J Biochem. 1995;233:484–489. doi: 10.1111/j.1432-1033.1995.484_2.x. [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, Scheiner O, Breitenbach M. The gene coding for the major birch pollen allergen Betvl, is highly homologous to a pea disease resistance response gene. EMBO J. 1994;8:1935–1938. doi: 10.1002/j.1460-2075.1989.tb03597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpin S, Laffer S, Schoentgen F, Valenta R, Chenieux J, Rideau M, Hamdi S. Molecular characterization of a cytokinin-inducible periwinkle protein showing sequence homology with pathogenesis-related proteins and the Betv1 allergen family. Plant Mol Biol. 1998;36:791–798. doi: 10.1023/a:1005951208815. [DOI] [PubMed] [Google Scholar]
- Chiang CC, Hadwiger Cloning and characterization of a disease resistance response gene in pea inducible by Fusarium solani. Mol Plant-Microbe Interact. 1996;3:78–85. doi: 10.1094/mpmi-3-078. [DOI] [PubMed] [Google Scholar]
- Cortes H. I Congreso Internacional de Cultivos Andinos, Ayacucho: Serie de Cursos y Conferencias y Reuniones No. 178. Lima, Peru: Instituto Interamericano de Cooperacion para la Agricultura; 1977. Avances en la investigación de la oca; pp. 45–48. [Google Scholar]
- Coursey DG. Yams: an account of the nature, origins, cultivation and utilization of the useful members of the Discoreaceae. London: Tropical Products Institute; 1991. pp. 32–39. [Google Scholar]
- Flores H, Flores T. Biochemistry of plant storage organs. In: Romeo JT, editor. Recent Advances in Phytochemistry. 31: Functionality of Food Phytochemicals. New York: Plenum Publishing Corporation; 1997. pp. 113–132. [Google Scholar]
- Hannapel D. Differential expression of potato tuber protein genes. Plant Physiol. 1990;94:919–925. doi: 10.1104/pp.94.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel D. Characterization of early events of potato tuber development. Physiol Plant. 1991;83:568–573. [Google Scholar]
- Hattori T, Nakagawa T, Maeshima M, Nakumara K, Asahi M. Molecular cloning and nucleotide sequence of cDNA for sporamin, the major soluble protein of sweet potato tuberous roots. Plant Mol Biol. 1985;5:313–320. doi: 10.1007/BF00020629. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gidson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hodge WH. Three native tubers of the high Andes. Econ Bot. 1957;5:185–201. [Google Scholar]
- Janick J, Simon J. Proceedings of the First National Symposium New Crops: Research Development, Economics: Indianapolis, Indiana. Portland, OR: Timber Press; 1988. Advances in New Crops; pp. 422–435. [Google Scholar]
- King S, Gershoff S. Nutritional evaluation of three underexploited Andean tubers: Oxalis tuberosa (Oxalidaceae), Ullucus tuberosus (Basellaceae), and Tropaeolum tuberosum (Tropaeolaceae) Econ Bot. 1987;4:503–511. [Google Scholar]
- Kyte J, Doolittle I. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1978;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeGendre N, Matsudaira PT. Purification of proteins and peptides by SDS-PAGE. In: Matsudaira PT, editor. A Practical Guide to Protein and Peptide Purification for Microsequencing. New York: Academic Press; 1989. pp. 49–69. [Google Scholar]
- Leon J. Plantas Alimenticias Andinas. Instituto Interamericano de Cooperacion para la Agricultura. Bol. Tec. 6. Lima, Peru. 1964. pp. 10–14. [Google Scholar]
- Maeshima M, Sasaki T, Asahi T. Characterization of the major proteins in sweet potato tuberous root. Phytochemistry. 1985;24:1899–1902. [Google Scholar]
- Matsuoka K, Nakamura L. Propeptide of a precursor to a plant vacuolar protein required for vacuolar targeting. Proc Natl Acad Sci USA. 1991;88:834–838. doi: 10.1073/pnas.88.3.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseyev GP, Fedoreyeva LI, Zhuravlev YN, Yasnetskaya E, Jekel Pa, Beintema JJ. Primary structure of two ribonucleases from ginseng calluses new members of the PR-10 family of intracellular pathogenesis-related plant proteins. FEBS Lett. 1997;407:207–210. doi: 10.1016/s0014-5793(97)00337-2. [DOI] [PubMed] [Google Scholar]
- National Research Council. Lost Crops of the Incas. Washington, DC: National Academy Press; 1989. [Google Scholar]
- Osmark P, Boyle B, Brisson N. Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol Biol. 1998;38:1243–1246. doi: 10.1023/a:1006060224012. [DOI] [PubMed] [Google Scholar]
- O'Farrel PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Ortega L. Usos y Valor Nutritivo de los Cultivos Andinos. Programa de Investigación de Cultivos Andinos, Puno, Peru. 1992. pp. 250–265. [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson GL. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Annal Biochem. 1977;83:281–285. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Pulgar-Vidal J. Geografía del Perú: Las Ocho Regiones Naturales del Perú. Ed 7. Lima, Peru: Editorial Universo S.A.; 1981. [Google Scholar]
- Racusssen D, Foote M. A major soluble glycoprotein of potato. J Food Biochem. 1980;4:43–52. [Google Scholar]
- Reid P, Pont-Lezica RF, Del Campillo E, Taylor R, editors. Tissue Printing. Tools for the Study of Anatomy, Histochemistry and Gene Expression. San Diego: Academic Press, Inc.; 1992. [Google Scholar]
- Roberts WK, Selitrennikoff CP. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol. 1990;136:1771–1778. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. A Laboratory Manual. Ed 2. Cold Spring, Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Seminario C. Cultivos Andinos. Universidad Nacional de Cajamarca, Peru. 1988. pp. 45–55. [Google Scholar]
- Senda K, Yoshioka H, Doke N, Kawakita K. A cytosolic phospholipase A2 from potato tissues appears to be patatin. Plant Cell Physiol. 1996;37:347–353. doi: 10.1093/oxfordjournals.pcp.a028952. [DOI] [PubMed] [Google Scholar]
- Shewry PR. Plant storage proteins. Biol Rev. 1995;70:375–426. doi: 10.1111/j.1469-185x.1995.tb01195.x. [DOI] [PubMed] [Google Scholar]
- Stegemann H, Majino S, Scmiediche P. Biochemical differentiation of clones of oca (Oxalis tuberosa) by their tuber proteins and the properties of these proteins. Econ Bot. 1988;42:37–44. [Google Scholar]
- Stegemann H, Schmiediche P. Proteinmuster von Sauerklee (Oxalis tuberosa) Jahresber Biol Bundesanst. 1981;4:11–114. [Google Scholar]
- Strickland J, Orr L, Alsh T. Inhibition of Diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiol. 1995;109:667–674. doi: 10.1104/pp.109.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swodoba L, Scheiner O, Kraft D, Breitenbach M, Hederle-Bors E, Vicente O. A birch gene family encoding pollen allergens and pathogenesi-related proteins. Biochem Biophys Acta. 1994;1219:457–464. doi: 10.1016/0167-4781(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Tapia ME. Agricultura Andina, Proyecto PISCA IIICA/CIID. Lima, Peru. 1984. Avances en las investigaciones sobre tuberculos alimenticios de los Andes; pp. 35–49. [Google Scholar]
- Walter HM, Liu JW, Grand C, Lamb CJ, Hess D. Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol Gen Genet. 1990;222:353–360. doi: 10.1007/BF00633840. [DOI] [PubMed] [Google Scholar]
- Yeh KW, Chen J, Lin M, Chen Y, Lin Functional activity of sporamin from sweet potato (Ipomoea batatas Lam.): a tuber storage protein with trypsin inhibitory activity. Plant Mol Biol. 1997;33:565–570. doi: 10.1023/a:1005764702510. [DOI] [PubMed] [Google Scholar]