Abstract
Ethylene-responsive element binding factors (ERF) proteins are plant-specific transcription factors, many of which have been linked to stress responses. We have identified four Arabidopsis ERF genes whose expression was specifically induced by avirulent and virulent strains of the bacterial pathogen Pseudomonas syringae pv tomato, with overlapping but distinct induction kinetics. However, a delay in ERF mRNA accumulation after infection with the virulent strain was observed when compared with the avirulent strain. The induction of ERF gene expression in most cases preceded the mRNA accumulation of a basic chitinase gene, a potential downstream target for one or more of these ERFs. The expression of the ERF genes was examined among different Arabidopsis tissues, in response to the signaling molecules ethylene, methyl jasmonate, and salicylic acid (SA), and in Arabidopsis mutants with decreased or enhanced susceptibility to pathogens, and significant differences were observed. For example, in seedlings, some of the ERF genes were not induced by SA in the wild-type but were SA responsive in the pad4-1 mutant, suggesting that PAD4-1, which acts upstream of SA accumulation, is also involved in repressing the SA-induced expression of specific ERF genes. The four ERF proteins were shown to contain transcriptional activation domains. These results suggest that transcriptional activation cascades involving ERF proteins may be important for plant defense to pathogen attack and that some ERF family members could be involved in the cross-talk between SA- and jasmonic acid-signaling pathways.
Many plant genes are transcriptionally regulated in response to pathogen attack or environmental stresses. Plant signals, like salicylic acid (SA), ethylene, and jasmonic acid (JA), which accumulate in plants during pathogen infection, are involved in the regulatory pathways mediating these responses (Glazebrook, 2001). These regulatory pathways require the coordination of highly specific DNA-protein and protein-protein interactions, most of which are not fully understood. A number of plant promoter elements that can respond to diverse environmental stimuli have been identified including the GCC box, an ethylene-responsive element initially found in several pathogenesis-related (PR) gene promoters (Hart et al., 1993; Ohme-Takagi and Shinshi, 1995; Sessa et al., 1995; Sato et al., 1996). Proteins that specifically bind to the GCC box were initially discovered in tobacco (Nicotiana tabacum) and are called ERFs (ethylene-responsive element binding factors; Ohme-Takagi and Shinshi, 1995; Suzuki et al., 1998). The tobacco ERFs share a well-conserved 58- to 59-amino acid domain called the ERF domain (Hao et al., 1998), which has only been found in plants. The ERF domain has a novel structure consisting of a β-sheet and an α-helix (Allen et al., 1998), which binds to DNA as a monomer (Hao et al., 1998).
There are numerous ERF proteins in plants (Riechmann et al., 2000), and the similarity is primarily confined to the ERF domain. ERF proteins play important roles in plant responses to various hormones or environmental cues. In Arabidopsis, ERF proteins are involved in mediating responses to dehydration, salt, and cold stress (Stockinger et al., 1997; Liu et al., 1998; Fujimoto et al., 2000; Park et al., 2001), abscisic acid (Finkelstein et al., 1998), ethylene (Büttner and Singh, 1997; Solano et al., 1998; Fujimoto et al., 2000), and pathogen infection (Solano et al., 1998; Maleck et al., 2000; Schenk et al., 2000; Park et al., 2001). ERF proteins have also been found to be involved in defense responses in other plants. In periwinkle, elicitor and/or jasmonate-inducible ERF genes have been identified (Menke et al., 1999; van der Fits and Memelink, 2000), whereas tobacco and tomato ERF genes are induced after infection by Pseudomonas syringae (Zhou et al., 1997; Thara et al., 1999), tobacco mosaic virus (Horvath et al., 1998), or Cladosporium fulvum (Durrant et al., 2000). Some of the tomato ERFs can interact specifically with the PTO protein, which confers resistance to P. syringae (Zhou et al., 1997). Overexpression of a tobacco ERF enhances resistance against pathogen attack and osmotic stress (Park et al., 2001).
To further analyze the role that ERFs play in plant defense responses, we tried to identify ERF genes in Arabidopsis whose expression was specifically induced after pathogen attack. We identified four Arabidopsis ERF genes that are specifically induced by infection with either an avirulent or virulent P. syringae strain, with the induction in most cases occurring earlier with the avirulent pathogen. The four ERF genes studied here displayed overlapping but distinct induction kinetics after pathogen attack and all contained transcriptional activation domains. Further characterization of the Arabidopsis ERF genes revealed that there were interesting differences in their expression in response to defense signaling molecules and in Arabidopsis mutants altered in their defense responses. These results suggest that the ERF proteins may form part of a transcriptional cascade that regulates the temporal response of plant gene expression in response to pathogen attack.
RESULTS
Identification of Arabidopsis ERF Genes Induced after Pathogen Attack
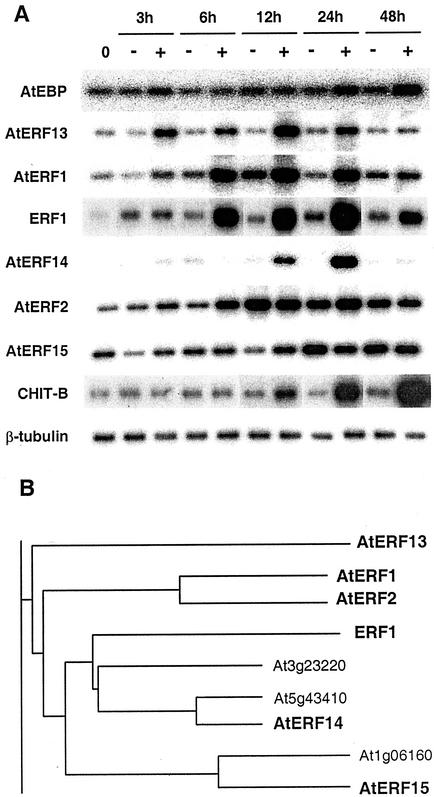
Previously we had isolated an Arabidopsis ERF-like protein called AtEBP by virtue of its interaction with an ocs-element binding protein (Büttner and Singh, 1997). Because AtEBP (At3g16770) was induced by ethylene and the encoded protein was able to bind to the GCC box, we tested whether AtEBP expression could also be induced by pathogen attack. We infiltrated leaves of Arabidopsis plants containing the RPS2 resistance gene with either a mock solution or a suspension containing the bacterial pathogen Pseudomonas syringae pv tomato (Pst) strain DC3000 carrying the avirulence gene avrRpt2 (Kunkel et al., 1993) and isolated RNA from the infiltrated leaves at different time points. The reverse transcriptase (RT)-PCR analysis shown in Figure 1A, demonstrated that AtEBP mRNA was not significantly induced at any of the time points analyzed although a small induction appeared 24 to 48 h after the inoculation. In contrast, mRNA levels of a basic chitinase (CHIT-B; Samac et al., 1990) started to accumulate between 6 and 12 h after inoculation, and by 24 h, a large induction had occurred that continued to increase at the 48-h time point. CHIT-B (also called PR3) had been shown previously to be inducible by pathogen infection (Thomma et al., 1998).
Figure 1.
RNA expression after pathogen infection and dendrogram of Arabidopsis ERF1-related proteins. A, Induction of Arabidopsis ERF genes after pathogen attack. Arabidopsis leaves were infiltrated with a mock solution as a control (−) or with the same solution containing approximately 107 colony forming units (cfu)/mL of the avirulent pathogen Pst DC3000(avrRpt2) (+). Infiltrated leaves were harvested at the indicated time points, and the RNAs were isolated and subjected to RT-PCR using primers specific for each gene. An Arabidopsis basic chitinase (CHIT-B) and a constitutively expressed β-tubulin (Snustad et al., 1992) were used as controls in RT-PCR analysis. B, Fragment of the phylogenetic tree produced after 107 Arabidopsis ERF protein sequences were aligned. The Multiple Sequence Alignment application (AlignX) of the Vector NTI Suite program (InforMax, Inc., North Bethesda, MD), based on the Clustal W algorithm, was used. The genes in bold were available in the Arabidopsis database at the time this work was initiated and were chosen for further analysis. Protein identification numbers are AtERF13, AAK48967; AtERF1, BAA32418; AtERF2, BAA32419; ERF1, AAD03545; At3g23220, BAA95733; At5g43410, BAA97420; AtERF14, AAB70439; At1g06160, AAF80213; and AtERF15, AAD20668.
Because overexpression of ERF1 (At3g23240), another Arabidopsis ERF protein, resulted in enhanced expression of CHIT-B (Solano et al., 1998), we tested whether ERF1 expression was induced by Pst DC3000(avrRpt2) infection. As shown in Figure 1A, a substantial induction in ERF1 expression was observed, which first appeared between 3 and 6 h after inoculation and peaked around 24 h. We then searched the Arabidopsis database to identify other ERFs that were closely related to ERF1. We focused on the five most closely related proteins available at the time these studies were initiated, and these are shown in bold in Figure 1B. An alignment of the amino acid sequences for all of the ERF proteins shown in Figure 1B is available as supplemental data at www.plantphysiol.org. The extensive amino acid similarity among these ERF proteins is primarily confined to the ERF domain. In addition, there are stretches of amino acid similarity outside the ERF domain encoded by AtERF1 (At4g17500; Fujimoto et al., 2000) and AtERF2 (At5g47220; Fujimoto et al., 2000), AtERF14 (At1g04370) and At5g43410, and AtERF15 (At2g31230) and At1g06160, respectively (see supplemental data available at www.plantphysiol.org). Three of the ERF genes shown in bold in Figure 1B have not previously been characterized, and we named them AtERF13 (At2g44840), AtERF14, and AtERF15 after finding in the database that AtERF12 (At1g28360; Ohta et al., 2001) was the last published member of the AtERF series.
As shown in Figure 1A, three of the ERF genes were specifically induced by Pst DC3000(avrRpt2) infection with induction patterns distinct from ERF1. AtERF13 and AtERF1 had quite similar induction patterns, with both showing a small increase in mRNA levels within 3 h that peaked at 12 h, although AtERF1 also showed a small induction in expression after the mock treatment. In contrast, AtERF14 first showed a response at 12 h that peaked at 24 h. The other two clones tested, AtERF2 and AtERF15, were induced by Pst DC3000(avrRpt2) infection but were also induced by the mock treatment, although in the case of AtERF2, the response to Pst DC3000(avrRpt2) started earlier than AtERF15. Other ERF genes have also been shown to be induced by a mock treatment (Thara et al., 1999). For our subsequent studies we focused on the four ERF genes that showed a significant and specific increase in expression after infection by Pst DC3000(avrRpt2).
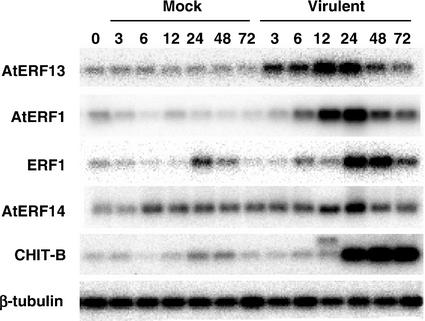
We tested whether the expression of the ERF genes changes after infection with the virulent pathogen Pseudomonas syringae pv tomato strain DC3000, which does not contain the avrRpt2 gene. As shown in Figure 2, all four ERF genes tested were induced with distinct induction kinetics after infiltration with the virulent strain, similar to what was seen with the avirulent strain (Fig. 1A). Thus, AtERF13 and AtERF1 were the first to be induced followed by ERF1 and AtERF14. AtERF1, ERF1, and, to a lesser extent, CHIT-B showed a delay in mRNA accumulation patterns when compared with the induction elicited by the avirulent pathogen (Fig. 1A), whereas AtERF14 displayed lower levels of induction in the compatible interaction (Fig. 2).
Figure 2.
RNA expression of the Arabidopsis ERF genes in response to a virulent pathogen. Arabidopsis leaves were infiltrated with a mock solution (Mock) or with the same solution containing approximately 107cfu mL−1 of the virulent pathogen Pst DC3000 (Virulent). RNAs isolated from leaves at the indicated time points were subjected to RT-PCR.
mRNA Expression Patterns of the ERF Genes
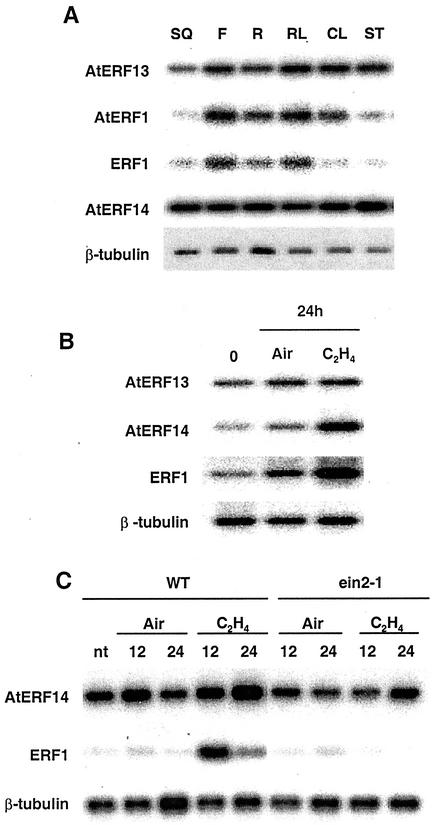
The mRNA expression patterns of the ERF genes were examined by RT-PCR in different Arabidopsis tissues. As shown in Figure 3A, the four ERF mRNAs were detected in all the tissues analyzed, although in some cases there were differences in the level of expression in specific tissues. Although AtERF14 was equally expressed in all the tissues tested, AtERF13, AtERF1, and ERF1 had the highest level of mRNA expression in flowers and rosette leaves compared with other tissues.
Figure 3.
Expression of the ERF genes in Arabidopsis tissues and in response to ethylene and in an ethylene insensitive mutant. A, RNAs isolated from Arabidopsis siliques (SQ), flowers (F), roots (R), rosette leaves (RL), cauline leaves (CL), and stems (ST) were analyzed by RT-PCR. B, Arabidopsis plants were placed in a glass chamber and flushed with ethylene (C2H4) or air. Plants were harvested after 24 h for RNA extraction and RT-PCR analysis. ERF1, which is up-regulated by C2H4 (Solano et al., 1998), was used as a control for the ethylene treatment. C, As in B but including the Arabidopsis ein2-1 mutant. Plant samples were collected after 12 and 24 h of treatment.
Ethylene has been shown to play important roles in a number of plant stress responses including responses to pathogens and in the expression of some ERF genes, including ERF1 and AtERF1 (Solano et al., 1998; Fujimoto et al., 2000). To determine whether AtERF13 or AtERF14 expression is regulated by ethylene, we treated 3-week-old Arabidopsis plants with ethylene. For this experiment we used the ERF1 gene as a positive control for the ethylene treatment. Figure 3B shows that AtERF14 and ERF1 are up-regulated by ethylene, whereas AtERF13 is not induced after 24 h of treatment. We also did not observe any change in AtERF13 expression after ethylene treatment for 12 h (data not shown). AtERF14 expression in ein2-1, an ethylene insensitive mutant, was also examined. As shown in Figure 3C, the ethylene induction of AtERF14 is reduced in ein2-1 as was also the case for ERF1.
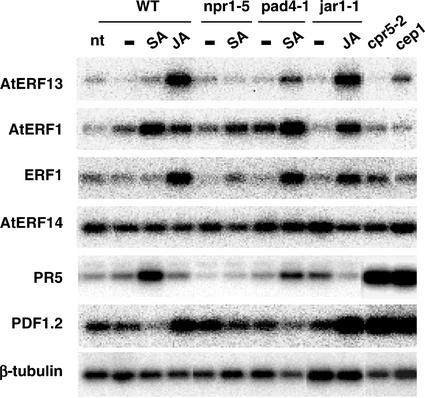
Like ethylene, SA and JA are important phytohormones involved in signaling in response to pathogen infection. To test the possible involvement of the ERF genes in SA or JA signaling pathways, we examined their mRNA expression after treatment of 2-week-old Arabidopsis seedlings with these hormones. As shown in Figure 4, AtERF13 and ERF1 were induced by methyl jasmonate (MeJA) but not by SA whereas AtERF1 was induced by both hormones. AtERF14 expression was not affected by either SA or MeJA. For these experiments, the SA-regulated PR5 gene (Ward et al., 1991) and the MeJA-regulated PDF1.2 gene (Penninckx et al., 1996) were used as controls and were induced by SA and MeJA, respectively.
Figure 4.
Analysis of ERF RNA levels in mutants with altered responses to defense signaling molecules and/or pathogens. RT-PCR analysis of RNA samples from Arabidopsis (WT) and the npr1-5, pad4-1, jar1-1, cpr5-2, and cep1 mutants. Seedlings were treated for 6 h with 0.1% (v/v) ethanol (−), 1 mm SA (SA), or 100 μm MeJA (JA) or were not treated (nt). PR5 and PDF1.2, which are up-regulated by SA (Ward et al., 1991) and MeJA (Penninckx et al., 1996), respectively, were used as controls. For the WT, results for the Columbia ecotype are shown, but similar results were obtained with the Nossen ecotype.
To further characterize the role of SA and MeJA in the expression of the ERF genes, we investigated their mRNA levels in different Arabidopsis mutants with enhanced (cpr5-2, Bowling et al., 1997; cep1, Silva et al., 1999) or reduced (npr1-5, Cao et al., 1994; pad4-1, Glazebrook et al., 1996; jar1-1, Staswick et al., 1998) disease resistance to pathogens and/or altered responses to SA (npr1-5, pad4-1, cpr5-2, and cep1) or MeJA (jar1-1 and cpr5-2). AtERF13, AtERF1, ERF1, and PDF1.2 were induced by MeJA in jar1-1 to levels similar to those observed in the wild-type (WT; Fig. 4). AtERF1 was induced by SA in the WT, in the pad4-1 mutant, and, to a lesser extent, in the npr1-5 mutant. Although AtERF13 and ERF1 were not induced by SA in the WT, they showed an increase in mRNA accumulation after SA treatment in the pad4-1 mutant. In contrast, PDF1.2 exhibited reduced expression in all samples treated with SA. In the two mutants cpr5-2 and cep1, in which the plant defense response is enhanced, the levels of PDF1.2 were high. However, none of the ERF genes were significantly induced in cpr5-2 or cep1, suggesting that they were not involved in the enhanced expression of PDF1.2 in these mutant backgrounds. The small increase in AtERF13 expression in cep1 was not reproducibly seen in other experiments. The mRNA levels of AtERF14 did not show significant changes after SA or MeJA treatments or in any of the mutant backgrounds.
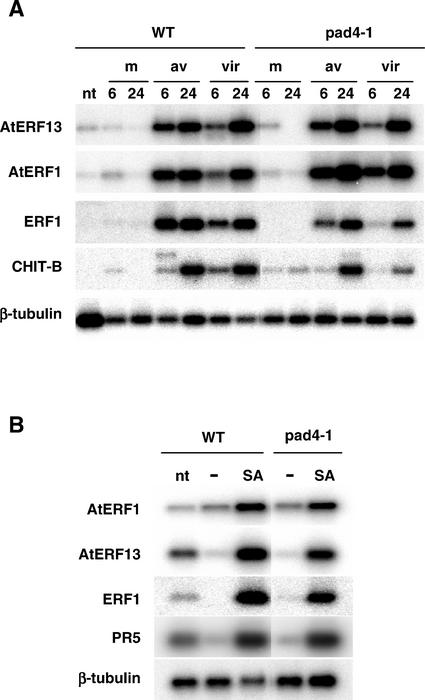
We also looked to see whether the expression patterns of the ERF genes after pathogen infection were altered in the pad4-1 mutant. As shown in Figure 5A, AtERF13, AtERF1, and ERF1 were induced in a similar fashion after infection with either the avirulent or virulent P. syringae strains in the pad4-1 mutant compared with WT. These experiments used 4-week-old plants grown in soil in contrast with the experiments presented in Figure 4, which used 2-week-old seedlings grown in Murashige and Skoog agar. Therefore, we repeated the SA treatment with 4-week-old plants grown in soil. As shown in Figure 5B, AtERF13 and ERF1 were induced by SA in the WT and pad4-1, in contrast to the results observed with the 2-week-old seedlings. These results suggest that the regulation of AtERF13 and ERF1 by PAD4 is complex and may be controlled by developmental and/or growth conditions.
Figure 5.
RNA expression of the Arabidopsis ERF genes in response to pathogen infection and SA treatment in the pad4-1 mutant. A, Infection of WT and pad4-1 with P. syringae. Arabidopsis leaves from 4-week-old plants were infiltrated with a mock solution (m) or with the same solution containing approximately 107cfu mL−1 of the avirulent pathogen Pst DC3000(avrRpt2) (av) or approximately 107cfu mL−1 of the virulent pathogen Pst DC3000 (vir). RNAs isolated from leaves at the indicated time points were subjected to RT-PCR. B, Treatment of WT and pad4-1 with SA. Four-week-old Arabidopsis plants were sprayed with 0.1% (v/v) ethanol (−) or 1 mm SA or were not treated (nt). RNAs isolated from leaves after 6 h of treatment were analyzed by RT-PCR.
Transcriptional Properties of the ERFs
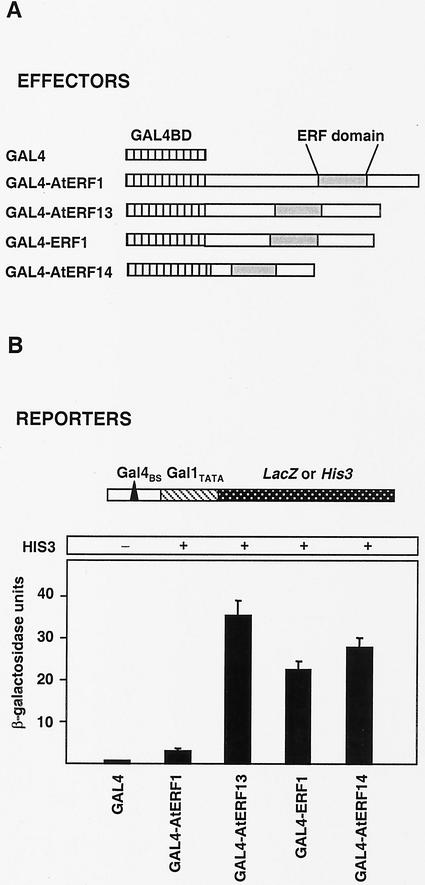
ERF proteins have been shown to function as either transcriptional activators or repressors (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Solano et al., 1998; Fujimoto et al., 2000; van der Fits and Memelink, 2000; Ohta et al., 2001; Park et al., 2001). Of the four ERF proteins examined in this study, AtERF1 has been analyzed with regard to its transcriptional properties and shown to be a transcriptional activator (Fujimoto et al., 2000), whereas overexpression of ERF1 resulted in enhanced expression of PDF1.2 and CHIT-B (Solano et al., 1998). We used a yeast one-hybrid system to examine the transcriptional properties of the Arabidopsis ERF proteins. As shown in Figure 6A, we generated effector plasmids containing translational fusions between the ERF and the GAL4-binding domain coding regions. Two reporter constructs integrated in the genome of two different strains of the yeast Saccharomyces cerevisiae were used. In both cases, the Gal4-binding site (Gal4BS) was fused to a minimal promoter (Gal1TATA) to control the expression of either the LacZ or the His3 reporter genes. The effector constructs expressing the ERF proteins were introduced into the two yeast strains specified above. As shown in Figure 6B, all the ERFs were able to activate transcription, with AtERF13, ERF1, and AtERF14 showing significantly stronger activation compared with AtERF1, which was used as a positive control. ERF1, AtERF13, and AtERF14 contain acidic rich regions that may function as transcriptional activation domains, as has been reported for AtERF1 (Fujimoto et al., 2000).
Figure 6.
Transcriptional properties of the ERF proteins. A, Schematic diagram of the effector and reporter constructs used in the yeast assays. The effectors contained the GAL4 DNA-binding domain coding region (GAL4BD) fused to each of the ERFs (GAL4-AtERF1, GAL4-AtERF13, GAL4-ERF1, and GAL4-AtERF14). The reporter genes were the LacZ or His3 gene under the control of a minimal promoter (Gal1TATA) plus a GAL4 binding site (Gal4BS). B, β-Galactosidase activity and growth in the absence of His induced by the effectors shown in A. β-Galactosidase values are from at least three independent replicates. Error bars represent ses.
DISCUSSION
ERF proteins comprise one of the largest families of transcription factors in plants with 124 family members present in Arabidopsis (Riechmann et al., 2000). We have identified four Arabidopsis ERF genes that are specifically induced after inoculation with avirulent or virulent Pst DC3000 strains. One possibility for why a number of ERF factors are enhanced in response to a specific pathogen may be to help orchestrate the correct temporal response in defense gene expression. Support for this possibility comes from our results showing distinct induction patterns among the four Arabidopsis ERF genes in response to P. syringae infection. The same pattern of induction kinetics was seen with both the avirulent and virulent strains of P. syringae, although, in most cases, the induction in ERF expression was delayed and/or reduced after inoculation with the virulent strain. These results suggest that the ERF proteins analyzed here play roles in both compatible and incompatible interactions.
Our results and, in the case of AtERF1, the results of Fujimoto et al. (2000), show that all four ERF proteins contain transcriptional activation domains. One possibility is that these ERF proteins form part of a transcriptional activation cascade whereby ERF proteins induced early in response to P. syringae infection, such as AtERF1 and AtERF13, are directly involved in regulating the expression of ERF members induced later in the infection, such as AtERF14. Although detailed analysis of the promoters of the ERF genes will be required to fully test this possibility, their promoters do not contain any obvious GCC box sequences. An alternative possibility is that the different ERF proteins induced in response to P. syringae infection regulate the expression of specific sets of plant defense genes. Support for this possibility comes from studies with AtERF1 and ERF1. AtERF1, one of the ERF genes that shows the earliest response to P. syringae inoculation, is a positive regulator of Hookless-1, an Arabidopsis gene containing a GCC box motif in its promoter (Fujimoto et al., 2000). In contrast, overexpression of ERF1, which is induced later than AtERF1 after P. syringae inoculation, did not cause any change in Hookless-1 expression. However, the expression of two other genes that contain potential GCC boxes in their promoters, PDF1.2 and CHIT-B, was induced (Solano et al., 1998).
Our analysis of Arabidopsis ERF proteins induced after P. syringae inoculation has not been exhaustive. First, the cut-off for the genes initially chosen on the basis of similarity to ERF1 was arbitrary. Moreover, as shown in Figure 1B, there are other Arabidopsis ERF proteins that are as similar to ERF1 as the ones used in this study, but these were not present in the databases at the time that we initiated these studies. Some of these genes may also be induced by P. syringae. Because some ERF proteins have been shown to act as transcriptional repressors (Fujimoto et al., 2000; Ohta et al., 2001), it would be interesting to know whether any Arabidopsis ERF members that belong to this category are inducible by P. syringae, and, if so, what are their temporal accumulation patterns.
Although the mRNA levels of three of the ERF genes, AtEBP, AtERF2, and AtERF15, did not show specific and/or significant changes in response to P. syringae (avirulent) inoculation, these proteins may still play roles in plant defense gene expression, possibly in response to other pathogens and/or through post-transcriptional regulation. For instance, Hermsmeier et al. (2000) have found that AtEBP RNA levels are locally down-regulated at the syncytium in a compatible cyst nematode infection and have proposed that the nematode may actively suppress the plant defense response. Moreover, Schenk et al. (2000) have shown using a genomic approach that AtEBP expression, also called RAP2.3 (Okamuro et al., 1997), was increased 4.3 times in response to infection by the incompatible fungal pathogen Alternaria brassicicola. Post-translational control has been observed for some ERF proteins, for example the tomato Pti4 protein is specifically phosphorylated by the product of the disease resistance gene Pto, and this phosphorylation enhances the binding of Pti4 to the GCC box (Gu et al., 2000). In periwinkle, ORCA3 regulates the JA-mediated expression of several terpenoid indole alkaloids biosynthetic genes (van der Fits and Memelink, 2001). The regulation by ORCA3 does not depend on de novo protein synthesis, and the JA-inducible expression of at least two of these genes is sensitive to protein kinase inhibitors (Menke et al., 1999).
The large number of ERF proteins involved in defense responses may also be to help orchestrate the spatial response in defense gene expression to specific pathogens. Although we found that the four ERF genes examined here were expressed in all of the plant tissues analyzed, there were differences in the levels of expression in specific tissues for some of the genes. We also found interesting differences in the expression of the four ERF genes in response to defense signaling molecules. Although ERF1 (Solano et al., 1998), AtERF1 (Fujimoto et al., 2000), and AtERF14 were responsive to ethylene, only AtERF1 was responsive to SA in 2-week-old seedlings. Treatment with MeJA resulted in enhanced expression for ERF1, AtERF1, and AtERF13, although the induction of AtERF1 was less pronounced. Interestingly, whereas AtERF1 expression is enhanced within 3 h after P. syringae inoculation, the response to ethylene treatment is significantly slower and starts between 6 and 12 h (Fujimoto et al., 2000). In contrast, ERF1 expression is enhanced between 3 and 6 h after P. syringae infection but as soon as 15 min after ethylene treatment (Solano et al., 1998). These results demonstrate differences between the responses to a pathogen and a defense signal and are consistent with those of Thara et al. (1999), who showed that the induction of Pti4 and Pti5 by P. syringae was independent of ethylene, SA, and JA. Fujimoto et al. (2000) also found that the induction of AtERF1 by wounding and cycloheximide treatment was both faster than and independent of the induction caused by ethylene.
There were also interesting differences in the expression of the ERF genes in Arabidopsis mutants altered in their responses to defense signaling molecules and/or pathogens. The jar1-1 mutant has decreased sensitivity to JA inhibition of root elongation (Staswick et al., 1992). JAR1 has been linked to plant defense responses, because jar1-1 mutants suppress resistance responses of cpr5-2 and cpr6 (Clarke et al., 2000) and are more susceptible to the soil fungus Pythium irregulare (Staswick et al., 1998). However, the expression of the JA-responsive ERF genes and the PDF1.2 gene, commonly used as a marker for JA-mediated defense responses, were still inducible by MeJA in the jar1-1 mutant. Because the MeJA induction of PDF1.2 expression has been shown to be blocked in another JA signaling mutant called coi1 (Penninckx et al., 1998), our results are consistent with multiple JA signaling pathways operating in the plant defense response.
Plant defense responses are constitutively expressed in the cpr5-2 and cep1 mutants. Thus, cpr5-2 has elevated levels of SA, enhanced levels of PR, increased PDF1.2 gene expression thought to reflect an activated JA and/or ethylene signaling pathways, and enhanced resistance to virulent strains of P. syringae and Peronospora parasitica (Bowling et al., 1997). The cep1 mutant has higher levels of SA and PR gene expression (Silva et al., 1999), and our results show that PDF1.2 expression is increased (Fig. 4). However, the SA- and/or JA-responsive ERF genes were not induced in either of these mutant backgrounds. Although the PDF1.2 promoter contains a GCC box (Manners et al., 1998), our results make it unlikely that the ERF genes analyzed here are regulating PDF1.2 expression in the cep1 and cpr5-2 mutants, although a post-transcriptional role cannot be ruled out. Whether PDF1.2 is being induced in cep1 or cpr5-2 by other ERF proteins or through some other transcription factor/promoter system remains to be investigated.
Two mutants, npr1-5 and pad4-1, that are altered in SA responses were also tested. NPR1 acts downstream of SA to promote the expression of genes like PR-1, although some SA responses are NPR1-independent (for review, see Glazebrook, 2001). The induction of AtERF1 expression by SA seemed to be, in part, NPR1-independent because it was reduced but not eliminated in the npr1-5 mutant, whereas PR5 induction was abolished in this mutant background. The PAD4-1 gene, encoding a lipase-like protein, is inducible by SA and virulent P. syringae infection (Jirage et al., 1999), and pad4-1 Arabidopsis plants are defective in camalexin production, SA synthesis, and PR gene expression after infection with a virulent strain of P. syringae (Zhou et al., 1998). Our results demonstrating that in 2-week-old seedlings AtERF13 and ERF1 are induced by SA in pad4-1 but not in the WT, suggest a new role for PAD4, whereby it regulates the expression of specific ERF genes by preventing their induction by SA. These results further support the evidence for cross-talk between the SA and JA signaling pathways.
ERF members clearly have significant differences in their RNA expression patterns and transcriptional properties. Superimposed upon these differences may be differences in DNA binding site preference (Park et al., 2001), post-translational control (Gu et al., 2000), and/or specific interactions with other proteins (Büttner and Singh, 1997; Xu et al., 1998) that may help to modulate the specificity of plant defense/stress gene expression in response to different signal transduction pathways. Given the prominent role ERF proteins play in plant stress responses and the large size of the ERF family, it will be an important task to determine the function of each member of this large family of transcription factors.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis mutants cep1, cpr5-2, jar1-1, and pad4-1 were obtained from the Nottingham Arabidopsis Stock Centre (University of Nottingham, Nottingham, UK), and the Arabidopsis mutants ein2-1 and npr1-5 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus) and from Dr. Dan Klessig (Bruce Thompson Institute for Plant Research, Ithaca, NY), respectively. All the mutants were in the Columbia ecotype except for npr1-5 (Nossen) and cep1 (Wassilewskija). Seeds from these mutants and from Columbia ecotype (WT) were stratified for 2 d at 4°C and grown on Murashige and Skoog agar plates under 16-h-light/8-h-dark cycle at 22°C unless otherwise noted. Plant samples were harvested by freezing whole seedlings or plant tissues in liquid nitrogen, which were then stored at −80°C until RNA was isolated.
Pathogen Infection and Chemical Treatments
Pseudomonas syringae pv tomato (Pst) strain DC3000 and Pst DC3000 expressing the avirulence gene avrRpt2 were a gift from B. Staskawicz (University of California, Berkeley). Leaves from 4- to 5-week-old plants containing the RPS2 resistance gene that were grown on soil under a 12-h-light/12-h-dark regime, were infiltrated with a mock (10 mm MgSO4) solution as a control or with the same solution containing approximately 107cfu mL−1 of the plant pathogens Pst DC3000(avrRpt2) or Pst DC3000. Infiltrated leaves were harvested at the indicated time points and RNA was isolated. For the ethylene treatment, 3- to 4-week-old Arabidopsis plants that had been grown in pots were placed in a glass chamber, and ethylene was injected to a final concentration of 100 μL L−1. Control plants were handled in the same way and flushed with air. For the SA and MeJA treatments, 2-week-old seedlings that had been grown on Murashige and Skoog plates in vertical position were used, unless otherwise stated. Murashige and Skoog solutions containing 0.1% (v/v) ethanol plus 1 mm SA or 100 μm MeJA were poured onto Murashige and Skoog plates so the liquid covered the roots but not the aerial tissues. No treatment or 0.1% (v/v) ethanol were used as controls. Whole seedlings were harvested after 6 h of treatment, and RNA was isolated.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from seedlings or leaves using the Purescript reagent (Gentra Systems, Minneapolis), treated with 1 unit of RNase-free DNase (Promega, Madison, WI) at 37°C for 1 h and repurified with Purescript. One microgram of total RNA was used for the first-strand cDNA synthesis after incubation at 65°C for 10 min. cDNA was synthesized in a volume of 25 μL that contained 50 mm Tris-HCl (pH 8.3), 50 mm KCl, 10 mm MgCl2, 0.5 mm spermidine, 10 mm dithiothreitol, 4 μm poly(dT) primer, 0.5 mm dNTPs, 2 units of avian myeloblastosis virus RT (Promega), and 12.5 units of RNasin (Promega) at 37°C for 1 h. All PCR reactions were performed with 0.5 unit of Taq polymerase (Invitrogen, Carlsbad, CA), the buffer provided by the supplier, 0.2 mm dNTPs, and a pair of primers (0.1 μm each) in a final volume of 20 μL. The gene-specific primer pairs used for the RT-PCR are listed: CHIT-B, 5′-CGGTGGTACTCCTCCTGGACCCACCGGC-3′ and 5′-CGGCGGCACG GTCGGCGTCTGAAGGCTG-3′; AtEBP, 5′-GCCATGGATCCGAATTCAGCGGCG AAGCAG-3′ and 5′-TACACTCTAGACTCGAGACATCATCAGCAG-3′; β-tubulin, 5′-CGTGGATCACAGCAATACAGAGCC-3′ and 5′-CCTCCTGCACTTCCACTT CGTCTTC-3′; AtERF1, 5′-CAATCTTGTGTAACCGGTCAGAGC-3′ and 5′-CACCGTCAATCCCTTATCCATTCC-3′; AtERF2, 5′-TGTACGGACAGTGCAATA TAGAATCCG-3′ and 5′-CACCTCCGACGTCAGATTCTCTGC-3′; ERF1, 5′-ATTCTATCGGATCTTCTCCAGATTC-3′ and 5′-CCTAATCTTTCACCAAGT CCCACT-3′; AtERF13, 5′-CCATTATGAGCTCATCTGATTCCG-3′ and 5′-CAGAATAAGAACATTCTGATTGGTCC-3′; AtERF14, 5′-GGATCAAGGAGGT CGTAGCAGTGG-3′ and 5′-TTATTGCCTCTTGCCCATGTTG-3′; AtERF15, 5′-TCATTTGAGTCACCGGAGATGATG-3′ and 5′-CCACAAGTGTACCACTTTCT TGC-3′; PDF1.2, 5′-AATGGATCCATGGCTAAGTTTGCTTCCATC-3′ and 5′-AATGAATTCAATACACACGATTTAGCACC-3′; PR5, 5′-ATGGCAAATATCTC CAGTATTCACA-3′ and 5′-ATGTCGGGGCAAGCCGCGTTGAGG-3′.
When the PCR reactions were in the exponential phase of amplification, typically after 20 to 25 cycles, the products were run on a 1.5% (w/v) agarose gel, transferred onto Zeta-Probe GT Genomic blotting membranes (Bio-Rad, Hercules, CA), and hybridized with the appropriate randomly primed 32P-labeled probes following standard procedures (Sambrook et al., 1989). Chronex #4 x-ray films (AGFA-Gevaert, Nunawading, Australia) or a Cyclone phosphor imager (Packard, Meriden, CT) were used to image the hybridized membranes.
Yeast Strains and LacZ Assays
The effector plasmid pGBT9 (CLONTECH, Palo Alto, CA) was used to generate in-frame C-terminal fusions of the complete coding sequences of AtERF1, AtERF13, ERF1, and AtERF14 with the GAL4-DNA binding domain (GAL4BD). The constructs were generated after PCR amplification of the ERFs using the Pfu turbo DNA polymerase (Stratagene, La Jolla, CA) and appropriate oligonucleotides with engineered restriction sites for cloning. These constructs were introduced into two haploid strains of Saccharomyces cerevisiae; HF7c and SFY526 contained the His3 and LacZ reporter genes, respectively, under the control of a minimal Gal1 promoter (Gal1TATA) containing an upstream Gal4-binding site (Gal4BS). The bacterial LacZ gene encodes a β-galactosidase, and the His3 gene encodes a S. cerevisiae imidazole glycerol-phosphate dehydratase, which catalyzes one of the enzymatic steps in His biosynthesis. Growth of HF7c transformants on minimal media without His indicated activation of transcription (+), which was quantified by measuring β-galactosidase activity in SFY526 cells as described by Ausubel et al. (1990). His media was supplemented with 2.5 mm of 3-amino-1,2,4-triazole (Sigma, St. Louis) to inhibit the HIS3 protein derived from the leaky expression of the His3 gene.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this paper that would limit their use in noncommercial research purposes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Brian Staskawicz for the P. syringae DC3000 strains, Dr. Dan Klessig for the npr1-5 seeds, Dr. Hong-Gu Kang for helpful advice on the RT-PCR analysis, and Dr. Isabel Aguilar and Dr. César Poza for helpful advice on the plant infection experiments. We thank Elaine Smith and Louise Thatcher for technical support, members of the Singh laboratory for helpful discussion, and Dr. Carol Andersson and Dr. John Klingler for helpful comments on the manuscript. We also thank the Nottingham Arabidopsis Stock Centre for providing the cep1, cpr5-2, jar1-1 and pad4-1 seeds and the Arabidopsis Biological Resource Center for the ein2-1 seeds.
Footnotes
This work was supported in part by a postdoctoral fellowship (to L.O.-S.) from Fundación Séneca (Murcia, Spain).
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010862.
LITERATURE CITED
- Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JM, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1990. [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5-2 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Volko SM, Ledford H, Ausubel FM, Dong X. Roles of salicylic acid, jasmonic acid, and ethylene in cpr-induced resistance in Arabidopsis. Plant Cell. 2000;12:2175–2190. doi: 10.1105/tpc.12.11.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell. 1998;10:1043–1054. doi: 10.1105/tpc.10.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis-2001 status. Curr Opin Plant Biol. 2001;4:301–308. doi: 10.1016/s1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–786. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins F., Jr A 61 bp enhancer element of the tobacco β-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol Biol. 1993;21:121–131. doi: 10.1007/BF00039623. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Hart JK, Byzova M, Rodermel SR, Baum TJ. Changes in mRNA abundance within Heterodera schachtii-infected roots of Arabidopsis thaliana. Mol Plant-Microbe Interact. 2000;13:309–315. doi: 10.1094/MPMI.2000.13.3.309. [DOI] [PubMed] [Google Scholar]
- Horvath DM, Huang DJ, Chua NH. Four classes of salicylate-induced tobacco genes. Mol Plant-Microbe Interact. 1998;11:895–905. doi: 10.1094/MPMI.1998.11.9.895. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton K, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BP, Broekaert WF. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol. 1998;38:1071–1080. doi: 10.1023/a:1006070413843. [DOI] [PubMed] [Google Scholar]
- Menke FL, Champion A, Kijne JW, Memelink JA. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999;18:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Eggermont K, Terras FR, Thomma BP, De Samblanx GW, Buchala A, Metraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallay PE, Shah DM. Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sato F, Kitajima S, Koyama T, Yamada Y. Ethylene-induced gene expression of osmotin-like protein, a neutral isoform of tobacco PR-5, is mediated by the AGCCGCC cis-sequence. Plant Cell Physiol. 1996;37:249–255. doi: 10.1093/oxfordjournals.pcp.a028939. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–11660. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Meller Y, Fluhr R. A GCC element and a G-box motif participate in ethylene-induced expression of the PRB-1b gene. Plant Mol Biol. 1995;28:145–153. doi: 10.1007/BF00042046. [DOI] [PubMed] [Google Scholar]
- Silva H, Yoshioka K, Dooner HK, Klessig DF. Characterization of a new Arabidopsis mutant exhibiting enhanced disease resistance. Mol Plant-Microbe Interact. 1999;12:1053–1063. doi: 10.1094/MPMI.1999.12.12.1053. [DOI] [PubMed] [Google Scholar]
- Snustad DP, Haas NA, Kopczak SD, Silflow CD. The small genome of Arabidopsis contains at least nine expressed β-tubulin genes. Plant Cell. 1992;4:549–556. doi: 10.1105/tpc.4.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell H. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Yuen GY, Lehman CC. Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 1998;15:747–754. doi: 10.1046/j.1365-313x.1998.00265.x. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Suzuki N, Ohme-Takagi M, Shinshi H. Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 1998;15:657–665. doi: 10.1046/j.1365-313x.1998.00243.x. [DOI] [PubMed] [Google Scholar]
- Thara VK, Tang X, Gu Y-Q, Martin G, Zhou J-M. Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 1999;20:475–483. doi: 10.1046/j.1365-313x.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001;25:43–53. doi: 10.1046/j.1365-313x.2001.00932.x. [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Narasimhan ML, Samson T, Coca MA, Huh GH, Zhou J, Martin GB, Hasegawa PM, Bressan RA. A nitrilase-like protein interacts with GCC box DNA-binding proteins involved in ethylene and defense responses. Plant Physiol. 1998;118:867–874. doi: 10.1104/pp.118.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4-1 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.