Abstract
We developed a method for expression in Arabidopsis of a transgene encoding a cleavable chimeric polyprotein. The polyprotein precursor consists of a leader peptide and two different antimicrobial proteins (AMPs), DmAMP1 originating from Dahlia merckii seeds and RsAFP2 originating from Raphanus sativus seeds, which are linked by an intervening sequence (“linker peptide”) originating from a natural polyprotein occurring in seed of Impatiens balsamina. The chimeric polyprotein was found to be cleaved in transgenic Arabidopsis plants and the individual AMPs were secreted into the extracellular space. Both AMPs were found to exert antifungal activity in vitro. It is surprising that the amount of AMPs produced in plants transformed with some of the polyprotein transgene constructs was significantly higher compared with the amount in plants transformed with a transgene encoding a single AMP, indicating that the polyprotein expression strategy may be a way to boost expression levels of small proteins.
Genetic modification of plants often requires co-expression of two or more transgenes in addition to a selectable marker gene. For instance, to achieve resistance against a broader range of pathogens in plants, co-expression of transgenes encoding antimicrobial proteins (AMPs) with different biochemical targets is an attractive approach. Other examples include the production of a particular metabolite in plants by co-expressing multiple transgenes that are involved in the biosynthetic pathway of that metabolite.
There are different ways to obtain transgenic plants expressing multiple transgenes. One frequently chosen option is to introduce each transgene individually via separate transformation events and to cross the different single transgene-expressing lines (Zhu et al., 1994; Bizily et al., 2000). A drawback of this method is that the different transgenes in the resulting progeny are inserted at different loci, which complicates the subsequent breeding process. Moreover, this method is not applicable to crops that are propagated vegetatively; for instance, in potato (Solanum tuberosum) and many ornamental and fruit tree species.
A second possibility is to introduce the different transgenes via sequential single-gene transformation steps. The drawback of this method is that a different selectable marker has to be used for every transformation step, which is complicated by the fact that there is only a limited choice of selectable markers.
A third possible way is the introduction of the different transgenes as linked expression cassettes, each with a promoter and terminator, within a single transformation vector (Slater et al., 1999). It has been observed, however, that the presence of multiple copies of the same promoter within a transgenic plant often results in transcriptional silencing of the transgenes (Matzke and Matzke, 1998). In an attempt to introduce a vector containing four linked transgenes each driven by the cauliflower mosaic virus 35S (CaMV35S) promoter in tobacco (Nicotiana tabacum), it was observed that none of the analyzed transgenic lines expressed all four transgenes at a reasonably high level (Van den Elzen et al., 1993). To avoid this problem, different promoters for each of the expression cassettes in the construct could be used. However, there is only a very limited choice of promoter sets that have comparable characteristics in terms of expression levels, cell type and developmental specificity, and response to environmental factors.
A fourth option would be to produce multiple proteins from one transcription unit by separating the distinct coding regions by so-called internal ribosomal entry sites, which allow ribosomes to reiterate translation at internal positions within an mRNA species. Although internal ribosomal entry sites are well documented in animal systems (Kaminski et al., 1994), it is not known at present whether such sites are also functional in nuclear-encoded genes from plants. Polycistronic genes can be expressed when inserted in plant chloroplastic genomes (Daniell et al., 1998), but the gene products in this case are confined to the chloroplast, which is not always the preferred site of deposition for foreign proteins.
A fifth strategy is based on the production of multiple proteins by proteolytic cleavage of a polyprotein precursor encoded by a single transcription unit. Potyviruses, for example, translate their genomic RNA into a single polyprotein precursor that encompasses domains with proteolytic activity able to cleave the polyprotein precursor in cis and to release the individual proteins (Carrington and Dougherty, 1988). The potyviral system has been exploited to co-produce different proteins in plants (Marcos and Beachy, 1994; Beck von Bodman et al., 1995). In the construct developed by Beck von Bodman et al. (1995), two coding sequences for enzymes involved in mannopine biosynthesis were fused within one open reading frame together with a coding sequence for a protease derived from a potyviral polyprotein precursor. The adjoining regions were separated by sequences that represent, after translation, specific cleavage sites for the viral protease. The tobacco plants transformed with this construct synthesized mannopine, suggesting that the two enzymes had somehow been produced in a form that was at least partially functional, although direct evidence for the presumed cleavage in planta was not presented. A disadvantage of this system is that a viral protease needs to be produced, together with the proteins of interest, that may cause cleavage of endogenous proteins and hence entail undesired side effects. An alternative approach was followed by Urwin et al. (1998). A polyprotein precursor consisting of a Cys protease inhibitor (oryzacystatin from rice [Oryza sativa]), a protease-sensitive propeptide from a pea (Pisum sativum) metallothionein-like protein and a Ser protease inhibitor (cowpea [Vigna unguiculata] trypsin inhibitor), was found to be cleaved in Arabidopsis plants. The cleavage, however, was only partial because uncleaved polyprotein precursor could also be detected in the transgenic plants. Still another approach was followed by Halpin et al. (1999), who linked the coding regions of two different reporter genes by a viral sequence. This viral sequence encodes a 20-amino acid-long sequence, called 2A sequence. The 2A sequence originates from the foot-and-mouth disease virus, where it mediates cotranslational cleavage at its own carboxy terminus by an apparantly enzyme-independent type of reaction. A plasmid was constructed in which the 2A sequence is inserted between the reporter genes chloramphenicol acetyltransferase (CAT) and β-glucuronidase (GUS) while maintaining a single open reading frame. After expression in tobacco, the CAT-2A-GUS polyprotein underwent cleavage to yield CAT-2A and GUS. The limitation of this method is the attachment of 19 amino acids of the 2A sequence to the carboxy terminus of the first protein of interest, which can disturb or even abolish the action of that protein of interest. Moreover, the 20th amino acid of the 2A sequence remains attached to the amino-terminus of the second protein of the polyprotein precursor.
We have reported previously on a unique natural polyprotein, IbAMP, occurring in seeds of the plant Impatiens balsamina (Tailor et al., 1997). The predicted IbAMP polyprotein precursor consists of a leader peptide, followed by six 20-amino acid-long mature peptide domains, each flanked at either side by propeptide domains ranging from 16 to 35 amino acids in length. In this paper, we have addressed the question of whether one of the IbAMP propeptides can be used as a linker peptide for the production of a cleavable chimeric polyprotein precursor leading to the delivery of different AMPs in Arabidopsis.
RESULTS
Characterization of Transgenic Plants Expressing Polyprotein Constructs with IbAMP-Based Linker Peptides
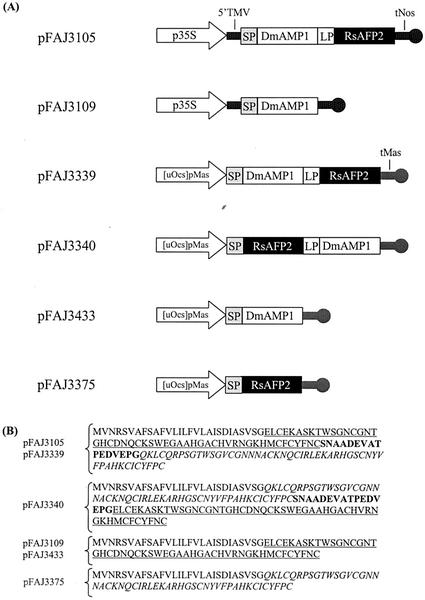
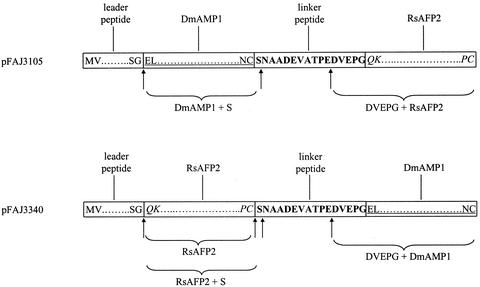
To explore the possibility of exploiting the IbAMP polyprotein system for co-expression of multiple AMPs, a plant transformation vector (pFAJ3105) was constructed as depicted in Figure 1. In the predicted expression product of this construct, the fifth propeptide (16 amino acids) of the IbAMP polyprotein is flanked at either side by the mature protein domain of the AMPs DmAMP1 and RsAFP2 (50 and 51 amino acids, respectively). Moreover, the expression product has a leader peptide (28 amino acids) at its amino-terminus derived from the DmAMP1 precursor. RsAFP2 and DmAMP1 belong to the family of the plant defensins (Broekaert et al., 1995) and originate from Raphanus sativus and Dahlia merckii seeds, respectively (Terras et al., 1992; Osborn et al., 1995). For comparative purposes, a single-protein construct was made of which the predicted expression product consisted of the DmAMP1 leader peptide and the DmAMP1 mature protein domain (pFAJ3109, Fig. 1).
Figure 1.
A, Diagram of the expression cassettes in plant transformation vectors pFAJ3105, pFAJ3109, pFAJ3339, pFAJ3340, pFAJ3433, and pFAJ3375. p35S, Enhanced CaMV35S promoter (Kay et al., 1987); TMV, 5′ leader sequence of tobacco mosaic virus; SP, coding region of the leader peptide derived from the DmAMP1 precursor; DmAMP1, coding region of the mature DmAMP1; LP, coding region of the linker peptide; RsAFP2, coding region of the mature RsAFP2; tNos, 3′-untranslated terminator region of the Agrobacterium tumefaciens nopaline synthase gene (Bevan et al., 1983); [uOcs]pMas, chimeric promoter consisting of the enhancer of the A. tumefaciens octopine synthase gene and the promoter of the A. tumefaciens mannopine synthase gene (M.F.C. De Bolle, unpublished data); tMas, 3′-untranslated terminator region of the A. tumefaciens mannopine synthase gene (Barker et al., 1983). B, Amino acid sequence of the (poly)proteins encoded by constructs pFAJ3105, pFAJ3109, pFAJ3339, pFAJ3340, pFAJ3433, and pFAJ3375. The leader peptide of DmAMP1 is in normal font. The mature DmAMP1 domain is underlined. The linker peptide domain is in bold. The mature RsAFP2 domain is italicized.
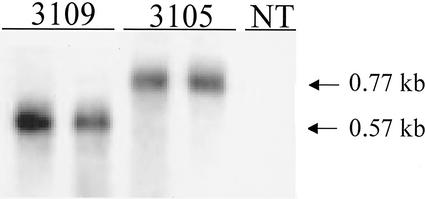
Different transgenic T1 generation lines transformed with constructs pFAJ3105 and pFAJ3109, respectively, were analyzed for expression of the transgene both at the mRNA and protein level. Analysis by northern blotting, using the coding region of DmAMP1 as a probe, indicated the presence of transcripts in leaves of lines transformed with either pFAJ3105 or pFAJ3109, but not in untransformed lines (Fig. 2). A clear difference in size of the DmAMP1-specific mRNA in plants transformed with the double protein construct as compared with plants transformed with the single-protein construct could be observed. This size difference is because of the fact that the double protein construct is transcribed as an mRNA coding for the leader peptide, DmAMP1, the linker peptide, and RsAFP2, whereas the single-protein construct is transcribed as an mRNA only coding for the leader peptide and DmAMP1. Quantification of band intensity in nine lines transformed with pFAJ3105 and eight lines transformed with pFAJ3109 indicated that the amounts of transcripts did not differ significantly between both types of transgenic plants (data not shown).
Figure 2.
Northern-blot analysis of DmAMP1 expression in leaves of transgenic T1 generation lines transformed with constructs pFAJ3109 and pFAJ3105. All analyzed samples represent 15 μg of total RNA. NT, Non-transformed Arabidopsis plants; 3109, Arabidopsis plants transformed with the single-protein construct pFAJ3109 (transgenic line 1 and 2); 3105, Arabidopsis plants transformed with the double-protein construct pFAJ3105 (transgenic line 1 and 2). Similar northern-blot analyses were obtained for another eight lines of Arabidopsis plants transformed with construct pFAJ3105 and six lines of Arabidopsis plants transformed with construct pFAJ3109.
The amount of DmAMP1 and RsAFP2 in leaves from a series of T1 Arabidopsis plants transformed with pFAJ3105 was determined using ELISA assays (Table I). Most of the lines transformed with the polyprotein construct expressed both DmAMP1-cross-reactive proteins (CRPs) and RsAFP2-CRPs. In general, there was a good correlation between DmAMP1-CRP and RsAFP2-CRP levels. However, RsAFP2-CRP levels were generally 2- to 5-fold lower than the DmAMP1-CRP levels. The ELISA assays for measuring the RsAFP2-CRP levels in the extracts were, however, less reliable than those for the DmAMP1-CRPs. In the RsAFP2 ELISA assays, dilutions of extracts of transgenic plants yielded dose-response curves that deviated from those obtained for dilutions of standard solutions containing native RsAFP2, indicating that the majority of the RsAFP2-CRPs in the extracts was not immunologically identical to the native RsAFP2 itself.
Table I.
Expression levels of DmAMP1 and RsAFP2 in leaves of transgenic Arabidopsis transformed with the constructs pFAJ3105 and pFAJ3109
| Construct | Line | Expression Level of DmAMP1 | Expression Level of RsAFP2 |
|---|---|---|---|
| % | |||
| pFAJ3105 | 1 | 0.77 | 0.29 |
| 2 | 1.13 | 0.22 | |
| 3 | 0.48 | 0.20 | |
| 4 | 0.005 | <0.001 | |
| 5 | 0.36 | 0.05 | |
| 6 | 0.99 | 0.25 | |
| 7 | 0.60 | 0.09 | |
| 8 | 0.13 | <0.001 | |
| 9 | 0.25 | 0.08 | |
| 10 | 1.35 | 0.35 | |
| 11 | 0.24 | 0.07 | |
| 12 | 1.18 | 0.24 | |
| 13 | 0.68 | 0.17 | |
| 14 | 0.49 | 0.07 | |
| Average | 0.62 | 0.15 | |
| pFAJ3109 | 1 | 0.19 | nda |
| 2 | 0.05 | nd | |
| 3 | 0.02 | nd | |
| 4 | 0.20 | nd | |
| 5 | 0.10 | nd | |
| 6 | 0.06 | nd | |
| 7 | 0.07 | nd | |
| 8 | 0.003 | nd | |
| Average | 0.09 | nd | |
Expression levels of DmAMP1 and RsAFP2 are expressed as the ratio of the amount of DmAMP1-CRP or RsAFP2-CRP to the amount of total soluble protein in crude extracts from leaves of transgenic Arabidopsis plants. The Arabidopsis plants are transformed with either the double protein construct (pFAJ3105) coding for DmAMP1 and RsAFP2 or the single protein construct (pFAJ3109) coding only for DmAMP1.
nd, Not determined.
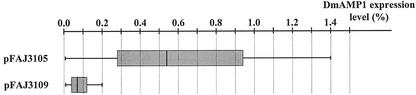
The average DmAMP1-CRP expression level of 14 lines transformed with pFAJ3105 was 0.62% of total soluble protein content versus only 0.09% for eight lines transformed with pFAJ3109, indicating that the polyprotein construct resulted in a dramatic increase in expression level as compared with the single-protein construct (Fig. 3). Statistical analysis demonstrated that this difference in expression level is significantly different (P < 0.05).
Figure 3.
Box plots of the DmAMP1 expression levels of T1 generation transgenic Arabidopsis plants transformed with constructs pFAJ3105 and pFAJ3109, respectively. DmAMP1 expression level is expressed as the ratio of the amount of DmAMP1 to the amount of total soluble protein in crude extracts from leaves of transgenic T1 Arabidopsis plants. Gridlines depicted as vertical dotted lines represent the DmAMP1 expression level scale. Vertical lines in the box plots correspond to the 0th, 25th, 50th (or median), 75th, and 100th percentile, respectively.
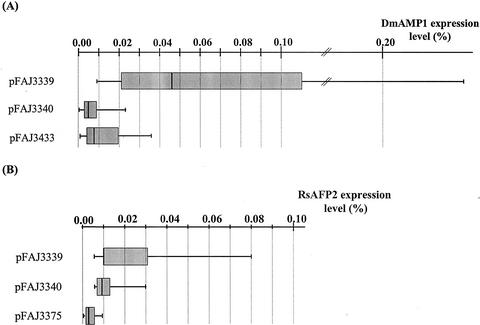
To test whether enhanced expression observed for the polyprotein construct pFAJ3105 is restricted to the particular configuration of the expression unit, four other constructs (pFAJ3339, pFAJ3340, pFAJ3433, and pFAJ3375) were made (Fig. 1). The promoter used in these constructs was the chimeric promoter [uOcs]pMas, consisting of the A. tumefaciens octopine synthase enhancer linked to the A. tumefaciens mannopine synthase promoter instead of the CaMV35S promoter used in construct pFAJ3105. The terminator was the mannopine synthase terminator instead of the nopaline synthase terminator used in construct pFAJ3105. Constructs pFAJ3339 and pFAJ3340 are both polyprotein-encoding constructs, with the polyprotein encoded by pFAJ3339 being exactly the same as the polyprotein encoded by pFAJ3105 and the one encoded by pFAJ3340 having a reversed order of the DmAMP1 and RsAFP2 coding regions relative to that of pFAJ3105 and pFAJ3339. Constructs pFAJ3433 and pFAJ3375 are both single-protein constructs, with pFAJ3433 encoding mature DmAMP1 preceded by the DmAMP1 leader peptide at its amino terminus and pFAJ3375 encoding mature RsAFP2 with the DmAMP1 leader peptide.
Comparison of the expression levels in the different transgenic plant populations (Table II) revealed that DmAMP1-CRP and RsAFP2-CRP levels in the polyprotein construct pFAJ3339 plants were on average 9- and 8-fold higher relative to the levels in the single-protein constructs pFAJ3433 and pFAJ3375 plants, respectively (Fig. 4). The RsAFP2-CRP levels in plants carrying the polyprotein construct pFAJ3340 were on average 3-fold higher than those in pFAJ3375 plants expressing only RsAFP2. There was, however, no significant difference in DmAMP1-CRP levels between polyprotein construct pFAJ3340 plants and pFAJ3433 plants expressing only DmAMP1.
Table II.
Expression levels of DmAMP1 and RsAFP2 in leaves of transgenic Arabidopsis transformed with the constructs pFAJ3339, pFAJ3340, pFAJ3433, and pFAJ3375
| Construct | na | Average Expression Level
|
Pb | Pc | |
|---|---|---|---|---|---|
| DmAMP1 | RsAFP2 | ||||
| % | |||||
| pFAJ3339 | 13 | 0.085 | 0.024 | a | a |
| pFAJ3340 | 19 | 0.006 | 0.01 | b | a |
| pFAJ3433 | 20 | 0.009 | ndd | b | nd |
| pFAJ3375 | 13 | nd | 0.003 | nd | b |
Expression levels are expressed as described in Table I.
n is the no. of independent transformants for which expression was measured.
Comparison of the average of the ln-transformed DmAMP1 expression levels at significance level 0.05. Identical letters indicate that the averages are not significantly different according to Tukey's studentized range test.
Comparison of the average of the ln-transformed RsAFP2 expression levels at significance level 0.05. Identical letters indicate that the averages are not significantly different according to Tukey's studentized range test.
Not determined.
Figure 4.
A, Box plots of the DmAMP1 expression levels of T1 generation Arabidopsis plants transformed with the polyprotein constructs pFAJ3339 and pFAJ3340 and the single-protein construct pFAJ3433, respectively. B, Box plots of the RsAFP2 expression levels of T1 generation Arabidopsis plants transformed with the polyprotein constructs pFAJ3339 and pFAJ3340 and the single-protein construct pFAJ33775, respectively. Legend for both box plots as in Figure 3.
Analysis of the Subcellular Location of Co-Expressed Plant Defensins
To determine whether the co-expressed plant defensins are either secreted extracellularly or deposited intracellularly, extracellular fluid and intracellular extract fractions were prepared from leaves of Arabidopsis plants transformed with the polyprotein construct (pFAJ3105). The cytosolic enzyme Glc-6-phosphate dehydrogenase was used as a marker to detect contamination of the extracellular fluid fraction with intracellular components. Glc-6-phosphate dehydrogenase was partitioned in a ratio of 83:17 between the intracellular extract fractions and extracellular fluid fractions. In contrast, 94% of DmAMP1-CRP content and 92% of RsAFP2-CRP content in the transgenic plants tested were found in the extracellular fluid fractions. These results indicate that both plant defensins released from the polyprotein precursors are deposited primarily in the apoplast.
Purification of Proteins Processed from Polyprotein Construct pFAJ3105
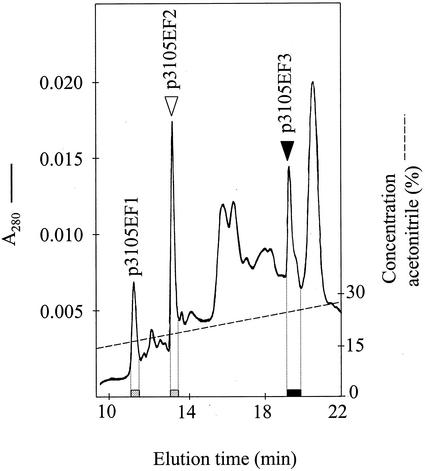
DmAMP1-CRPs and RsAFP2-CRPs were purified by reverse-phase (RP) HPLC from extracellular fluid prepared from leaves of transgenic Arabidopsis plants transformed with construct pFAJ3105 (homozygous T2 plants derived from line 12, see Table I). Extracellular fluid was obtained as described in “Materials and Methods,” using a buffer which contained a mixture of protease inhibitors (phenylmethylsulfonylfluoride, N-ethylmaleimide, ethylenediaminotetra-aceticacid, and pepstatinA). The collected extracellular fluid was analyzed by RP-HPLC on a C8-silica column and the collected fractions were tested for the presence of DmAMP1-CRPs and RsAFP2-CRPs using ELISA assays. The result of this analysis is shown in Figure 5. DmAMP1-CRPs eluted in two fractions, the latter of which eluted at a position very close to that of native DmAMP1. RsAFP2-CRPs were found in a single peak that eluted at a position very close to that of native RsAFP2 and that was well separated from the DmAMP1-CRP peaks. None of the tested fractions reacted with both the anti-DmAMP1 and anti-RsAFP2 antibodies, indicating that an uncleaved immunoreactive fusion protein was absent from the extracellular fluid. Based on comparison of integrated peak areas of the DmAMP1-CRPs and RsAFP2-CRPs with those of a series of standards consisting of native DmAMP1 and RsAFP2, respectively, it was estimated that the extract for the line transformed with the polyprotein construct pFAJ3105 contained about equal amounts of DmAMP1 and RsAFP2. This indicates that cleavage of the polyprotein precursor in this line results in about equimolar amounts of DmAMP1-CRPs and RsAFP2-CRPs. Similar chromatograms were obtained upon analysis of the extracellular fluid prepared from other transgenic lines (data not shown), indicating that the chromatographic pattern of DmAMP1-CRPs and RsAFP2-CRPs is independent from the transgenic line tested.
Figure 5.
Chromatographic purification of DmAMP1-CRPs and RsAFP2-CRPs from extracellular fluid fraction of Arabidopsis plants transformed with the double-protein construct pFAJ3105. The extracellular fluid fraction was loaded on a C8 RP-HPLC column equilibrated in 15% (v/v) acetonitrile in 0.1% (v/v) trifluoroacetic acid (TFA). The column was eluted at a flow rate of 1 mL min−1 for 15 min with 15% (v/v) acetonitrile in 0.1% (v/v) TFA followed by a linear gradient of acetonitrile in 0.1% (v/v) TFA from 15% (v/v) to 50% (v/v) acetonitrile in 35 min. The eluate was monitored by on-line measurement of the A280 (A280;—) and the acetonitrile gradient (---) was monitored with an on-line conductivity sensor. Fractions were collected and assessed for the presence of DmAMP1-CRPs and RsAFP2-CRPs by the respective ELISA assays. Dotted bars represent the presence of DmAMP1-CRPs (p3105EF1 and p3105EF2), whereas the black bar represents the presence of RsAFP2-CRPs (p3105EF3). Triangles indicate the elution position of native DmAMP1 (white triangle) and of native RsAFP2 (black triangle).
To test whether the purification procedure based on the extracellular fluid preparation reflects the true composition in DmAMP1-CRPs and RsAFP2-CRPs of the transgenic Arabidopsis leaves, an alternative purification procedure was developed starting from a crude leaf extract. The extract was fractionated by ion-exchange chromatography (IEC) and subsequently by RP-HPLC. After separation, fractions were collected and assessed for the presence of DmAMP1-CRPs and RsAFP2-CRPs using ELISA assays. IEC was performed by passing the extract over a cation exchange column at pH 6. When the column was eluted with a linear gradient of 0 to 0.5 m NaCl in 50 mm MES at pH 6, DmAMP1-CRPs were detected in fractions eluting between 0.17 and 0.33 m NaCl, whereas RsAFP2-CRPs were detected in fractions eluting between 0.24 and 0.49 m NaCl (data not shown). Fractions containing either DmAMP1-CRPs or RsAFP2-CRPs were pooled into two fractions (0.17–0.24 m NaCl and 0.33–0.49 m NaCl, respectively), which were each subjected to RP-HPLC on a C8-silica column eluted with a linear gradient of acetonitrile. DmAMP1-CRPs eluted in two peaks, the latter of which eluted at a position very close to that of native DmAMP1. RsAFP2-CRPs were found in a single peak that was well separated from the DmAMP1-CRP peaks and eluted at a position very close to that of native RsAFP2 (data not shown).
Molecular Analyses of the Purified Protein Fractions
The different DmAMP1-CRPs and RsAFP2-CRPs purified from the extracellular fluid were subjected to amino-terminal amino acid sequence analysis as well as to mass spectrometry. The carboxy-terminal amino acid was determined based on the best approximation of the predicted theoretical mass to the experimentally determined mass (Table III). The DmAMP1-CRPs, p3105EF1 and p3105EF2 (nomenclature as in Fig. 5), had masses that were consistent with the presence of a single additional Ser residue at their carboxy-terminal end as compared with native DmAMP1. However, whereas the mass of p3105EF2 corresponded exactly (within experimental error) to that calculated for a DmAMP1 derivative with an additional Ser (hereafter called DmAMP1+S) at its carboxy-terminal end, the mass of p3105EF1 was in excess by 4 D relative to the calculated mass for DmAMP1+S. Hence, this protein might be a DmAMP1+S derivative with partially reduced disulfide bridges. This hypothesis was confirmed by reduction of the disulfide bonds in p3105EF1 and p3105EF2 with excess dithiothreitol, followed by separation of the reduced forms by RP-HPLC. The reduced form of p3105EF1 and p3105EF2 eluted at exactly the same position, indicating that p3105EF1 only differs from p3105EF2 in its disulfide bond content (data not shown).
Table III.
Molecular analysis of the purified AMP fractions
| Protein Fraction | Molecular Mass Determined by:
|
Amino Acid Sequence
|
|||
|---|---|---|---|---|---|
| Matrix-assisted laser-desorption ionization time of flight MS | Electrospray ionization-MS | Theoretical prediction | Determined amino terminal | Predicted carboxy terminal | |
| D | |||||
| p3105EF1 | 5,614 ± 5 | 5,608.3 ± 1 | 5,604.25 | ELCEKAS | CYFNCS |
| p3105EF2 | 5,602 ± 5 | 5,604.9 ± 1 | 5,604.25 | ELCEKAS | CYFNCS |
| p3105EF3 | 6,223 ± 6 | nda | 6,225.15 | DVEPGQK | ICYFPC |
Molecular mass (D) of DmAMP1-CRPs and RsAFP2-CRPs purified as described in Fig. 3 was determined by matrix-assisted laser-desorption-ionization time of flight or electrospray ionization-mass spectrometry (MS) and amino-terminal sequence of the same components was determined by automated Edman degradation. Also shown are the predicted carboxy-terminal sequences that give best correspondence between experimental molecular mass and theoretical molecular mass. Amino acids of the mature DmAMP1 domain are underlined, amino acids of the linker peptide are in bold, and amino acids of the mature RsAFP2 domain are italicized.
nd, Not determined.
The RsAFP2-CRP fraction p3105EF3 represents an RsAFP2 derivative with the additional pentapeptide sequence DVEPG at its amino terminus, corresponding to the five carboxy-terminal amino acids of the linker peptide. This protein is further referred to as DVEPG+RsAFP2.
The different DmAMP1-CRPs and RsAFP2-CRPs purified from total leaf extract from plants transformed with construct pFAJ3105 were analyzed in the same way. The analyses indicated that the same three molecular species were present in the total leaf extract, namely DmAMP1+S, a reduced form of DmAMP1+S and DVEPG+RsAFP2 (data not shown).
In addition, DmAMP1-CRPs and RsAFP2-CRPs were also purified from extracellular fluid of plants transformed with construct pFAJ3340, the construct in which the order of DmAMP1 and RsAFP2 in the polyprotein was reversed. In this case, the molecular entities identified were authentic RsAFP2 including a pyro-Glu residue at its amino-terminus, RsAFP2 with an amino-terminal pyro-Glu and an additional carboxy-terminal Ser (RsAFP2+S), and finally DmAMP1 with an additional amino-terminal pentapeptide (DVEPG+DmAMP1; data not shown). In the plants transformed with polyprotein construct pFAJ3340, no reduced or partially reduced forms of either DmAMP1-CRPs or RsAFP2-CRPs could be detected (data not shown).
In Vitro Antifungal Activity of the Purified Proteins
The antifungal activity of the purified fractions from the extracellular fluid of plants transformed with construct pFAJ3105, containing the major processed products DmAMP1+S and DVEPG+RsAFP2, respectively, were assayed using Fusarium culmorum as a test fungus (Table IV). The specific antimicrobial activity, expressed as protein concentration required for 50% growth inhibition of the test organism, of purified DmAMP1+S appeared to be identical to that of native DmAMP1. The specific antimicrobial activity of purified DVEPG+RsAFP2, however, was about 2-fold lower relative to that of native RsAFP2. The slight drop in antimicrobial activity of DVEPG+RsAFP2 is most likely because of the presence of the five additional amino-terminal amino acids. Nevertheless, our data prove that processing of the polyprotein precursors in transgenic plants can result in the release of bioactive proteins.
Table IV.
Antifungal activity of the purified AMP fractions
| Protein Fraction | Antifungal Activity (IC50) |
|---|---|
| p3105EF2 | 3.2 ± 0.7 |
| DmAMP1 | 2.8 ± 0.5 |
| p3105EF3 | 3.2 ± 0.8 |
| RsAFP2 | 1.7 ± 0.4 |
Antifungal activity was determined as the concentration of protein (micrograms per milliliter) required to cause 50% growth inhibition of the fungus F. culmorum relative to a control culture. Data are given as means of triplicates with se.
DISCUSSION
In this paper, we have described the expression and processing in transgenic Arabidopsis plants of a chimeric polyprotein consisting of a leader peptide domain and two mature protein domains, corresponding respectively to the AMPs DmAMP1 and RsAFP2, separated from each other by a 16-amino acid-linker peptide corresponding to the fifth propeptide of the IbAMP precursor.
Our data indicate that the polyprotein precursor is cleaved to release bioactive derivatives of the mature AMPs DmAMP1 and RsAFP2. The processing of the polyprotein precursor encoded by construct pFAJ3105 appears to occur in at least two steps. At first, the polyprotein precursor is cleaved upon translation to release the leader peptide as occurs during the maturation of native DmAMP1. The remaining part of the polyprotein precursor is then cleaved at one or more unknown positions in the linker peptide, giving rise to a DmAMP1 derivative with an additional carboxy-terminal Ser and an RsAFP2 derivative with an additional pentapeptide (DVEPG) at its amino terminus (Fig. 6). The polyprotein precursor encoded by construct pFAJ3340, in which the order of the mature DmAMP1 and RsAFP2 domains is reversed relative to that of pFAJ3105, is cleaved at almost the same positions as for the polyprotein precursor encoded by pFAJ3105. One small difference is that cleavage of the first protein (RsAFP2 in the case of pFAJ3340) occurs both after the mature RsAFP2 domain and after the immediately adjacent Ser (Fig. 6). This indicates that cleavage within the IbAMP linker peptide is largely independent of the sequence context at both the amino- and carboxy-terminal ends.
Figure 6.
Schematic representation of the polyprotein precursor encoded by construct pFAJ3105 and pFAJ3340. The protein domain of the leader peptide of DmAMP1, DmAMP1, and RsAFP2 is represented by boxes containing the amino- and carboxy-terminal amino acids of the different subdomains. The protein domain of the linker peptide is represented by a box containing the complete amino acid sequence. Cleavage of the pFAJ3105 polyprotein precursor results in the release of a DmAMP1 derivative with an additional Ser at its carboxy terminus (DmAMP1+S) and an RsAFP2 derivative with an additional pentapeptide at its amino-terminus (RsAFP2+DVEPG). Cleavage of the pFAJ3340 polyprotein precursor results in the release of RsAFP2, an RsAFP2 derivative with an additional Ser at its carboxy terminus (DmAMP1+S) and a DmAMP1 derivative with an additional pentapeptide at its amino terminus (DVEPG+DmAMP1).
The protease (or group of proteases) responsible for these cleavages is not known. Endoproteinases or a combination of endo- and exoproteinases could be responsible for these cleavages. In the latter case, it is possible that a first cleavage occurs by the action of an endoproteinase, whereas exoproteinases would be responsible for the subsequent trimming of the cleavage products. It is possible that the carboxy terminus of the linker peptide is cleaved by an endoproteinase between two acidic amino acids, Glu and Asp. A comparison of the amino acid sequence of the five propeptides of the natural polyprotein precursor isolated from I. balsamina (Tailor et al., 1997) shows that four of five linker peptides have the doublet Glu/Asp at their carboxy-terminal end. The linker peptide used in constructs pFAJ3105, pFAJ3339, and pFAJ3340 contains this acidic doublet internally and not at the carboxy-terminal end. This doublet of acidic amino acids could form a recognition sequence for a specific endoproteinase. It is interesting that D'Hondt et al. (1993) isolated an aspartic endoproteinase from the seed of Brassica napus that cleaves between two consecutive acidic amino acids. It is possible that a first cleavage of this linker peptide in I. balsamina occurs between the acidic doublet, with subsequent action of an exocarboxypeptidase to trim the first protein up to the point of steric hindrance. The fact that cleavage of the polyprotein precursor encoded by pFAJ3340 results in the release of two forms, RsAFP2 and RsAFP2+S, points to the intervention of such an exocarboxypeptidase.
Deposition of AMPs in the apoplast of transgenic plants is preferred for the purpose of engineering disease-resistant plants because most pathogenic microorganisms occur at least during the early stages of infection in the extracellular space. The polyprotein expression system based on the IbAMP propeptide is the first of its kind to succeed in extracellular deposition of the processed mature proteins. Previous reports on polyprotein expression systems (Marcos and Beachy, 1994; Beck von Bodman et al., 1995; Urwin et al., 1998; Halpin et al., 1999) described the use of polyproteins without a leader peptide, leading to assume that in these cases processing occurred in the cytosol. It is not known whether these systems would also work in conjunction with a leader peptide because the processing environment of the cytosol is completely different from that of the secretory system. For the production of AMPs with a high content of disulfide bonding, which is the case for most AMPs of plant origin described so far (Broekaert et al., 1997; Yun et al., 1997), expression in the cytosol is not an option because it will not lead to disulfide bond formation.
In plants transformed with the polyprotein construct pFAJ3105, a minor fraction of the DmAMP1-processed product occurred in a form in which two of the four disulfide bridges are not formed, whereas this was not observed for the RsAFP2-processing product. The reason for this incorrect maturation of DmAMP1 is currently unclear. One possible explanation is that cleavage of the [DmAMP1]-[IbAMP linker]-[RsAFP2] precursor occurs in the Golgi apparatus, whereas folding and disulfide formation occurs in the endoplasmic reticulum. The nature of the precursor may hamper proper folding of the amino-terminal DmAMP1 domain in a fraction of the precursor molecules, leading to incomplete cystine formation that is known to be assisted by disulfide isomerases residing in the endoplasmic reticulum (Vitale and Denecke, 1999). In plants transformed with the polyprotein construct pFAJ3340, no such reduced forms of DmAMP1 could be detected. The overall expression levels obtained with construct pFAJ3340 were on average one order of magnitude lower than those obtained with construct pFAJ3105. The lower expression levels in plants transformed with construct pFAJ3340 may entail less overload of disulfide isomerase complexes, and hence less problems with correct disulfide bridge formation.
A striking observation from our experiments is that DmAMP1-CRP and RsAFP2-CRP levels in the lines transformed with a polyprotein construct are on average severalfold higher as compared with those in the lines transformed with the corresponding single-protein construct, respectively. The only exception was the level of DmAMP1-CRPs in plants transformed with the polyprotein construct pFAJ3340, which was not significantly different from the level of DmAMP1-CRPs in plants transformed with the single-protein construct pFAJ3433. Expression levels as high as 1.35% of total protein content, as seen in some lines transformed with pFAJ3105, have so far never been reported in the literature for a peptide expressed in leaves of transgenic plants. The fusion of a small peptide to another protein as a way to enhance its expression level has been suggested by another recent observation. Okamoto et al. (1998) describe enhanced expression of the gene coding for the antimicrobial peptide sarcotoxin IA by fusing translationally the coding sequence of this gene to that of Escherichia coli GUS. Western-blot analysis of transgenic tobacco plants demonstrated that the amounts of sarcotoxin IA present in the form of sarcotoxin IA-GUS fusion proteins were considerably higher than in tobacco plants transformed with the single sarcotoxin IA peptide construct. The levels of mRNA accumulation did not differ significantly between plants transformed with the single protein and the fusion protein constructs, and therefore it was suggested that sarcotoxin IA peptide was protected from degradation when fused to GUS. Our analysis demonstrates that the amount of DmAMP1 specific mRNA in plants transformed with the double protein construct (pFAJ3105) also does not differ significantly from the amount of DmAMP1-specific mRNA in plants transformed with the single-protein construct (pFAJ3109). Protection from degradation cannot explain the observed differences at the protein level because the eventually released mature proteins are almost identical. It is possible that, because of the increased length of the mRNA in plants transformed with the double protein construct, the mRNA becomes more efficiently translated than in the case of the smaller mRNA in plants transformed with the single-protein construct.
In our polyprotein precursor constructs in which DmAMP1 was the first protein and RsAFP2 the second (pFAJ3105 and pFAJ3339), production of DmAMP1-CRPs was consistently higher than that of RsAFP2-CRPs, as measured by ELISA assays. However, when analyzed by chromatography, the fraction of DmAMP1-CRPs and RsAFP2-CRPs appeared to be equal. We believe that the apparently lower RsAFP2-CRP content versus the DmAMP1-CRP content in those plants is because of suboptimal recognition of DVEPG+RsAFP2 by the antibodies raised against RsAFP2. ELISA assays performed with extracts containing DVEPG+RsAFP2 always showed deviating dose-response curves. Moreover, studies of the interactions of anti-RsAFP2 antibodies with a series of synthetic 15-mer peptides, representing an overlapping series of the entire RsAFP2 sequence, revealed that these antibodies react most strongly with peptides representing the amino-terminal end (A.Van Amerongen, personal communication). Hence, the DVEPG sequence in DVEPG+RsAFP2 may partially mask the most immunoresponsive epitope. In plants transformed with construct pFAJ3340, in which RsAFP2-CRPs are present with their authentic amino-terminal end, RsAFP2-CRPs and DmAMP1-CRPs were expressed at approximately equal levels.
In current experiments, we are adapting the linker peptide used in the polyprotein constructs in an attempt to ensure a more correct cleavage of the polyprotein construct with the aim to release the individual AMPs with a minimal number of additional amino acids at their amino or carboxy terminus.
MATERIALS AND METHODS
Construction of Plant Transformation Vectors
The polyprotein encoding sequences of plasmid pFAJ3105 (shown in Fig. 1) was constructed following the two-step recombinant PCR protocol of Pont-Kingdon (1994). Primers OWB175 (sense, 5′ AGGAAGTTCATTTCATTTGG) and OWB278 (antisense, 5′ GCCTTTGGCACAACTTCTGTCCTGGCTCCACGTCCTCTGGGGTAGCCACCTCGTCAGCAGCGTTGGAACAAG AAGTAACAGAAACAC) were used in a first PCR reaction with a plasmid pDMAMPD containing DmAMP1 cDNA (gift from Dr. Ian Evans, Zeneca Agrochemicals, Berkshire, UK) as a template. The second PCR reaction was done using as a template plasmid pFRG4 containing the RsAFP2 cDNA (Terras et al., 1995) and as primers a mixture of the PCR product of the first PCR reaction, primer OWB175 and primer OWB172 (antisense, 5′ TTAGAGCTCCTATTAACAAGGAAAGTAGC, SacI site underlined). The resulting PCR product was digested with XhoI and SacI and cloned into the expression cassette vector pMJB1. pMJB1 is an expression cassette vector containing in sequence a HindIII site; the enhanced CaMV35S RNA promoter (Kay et al., 1987); an XhoI site; the 5′-untranslated leader sequence of tobacco mosaic virus (Gallie and Walbot, 1992); a polylinker including NcoI, SmaI, KpnI, and SacI sites; the 3′-untranslated terminator region of the Agrobacterium tumefaciens nopaline synthase gene (Bevan et al., 1983); and an EcoRI site. The expression cassette in the resulting plasmid, called pFAJ3099, was digested with HindIII (flanking the 5′ end of the CaMV35S promoter) and EcoRI (flanking the 3′ end of the nopaline synthase terminator) and cloned in the corresponding sites of the plant transformation vector pGPTVbar (Becker et al., 1992) to yield plasmid pFAJ3105.
For the construction of plasmid pFAJ3109, the NcoI-SacI fragment of plasmid pDMAMPD containing the DmAMP1 cDNA (gift from Dr. Ian Evans, Zeneca Agrochemicals) was first cloned into the corresponding sites of the expression vector pMJB1, and the HindIII-EcoRI fragment of this plasmid was subsequently cloned in the corresponding sites of plant transformation vector pGPTVbar (see above).
For the construction of plasmid pFAJ3339, the NcoI- SacI fragment of plasmid pFAJ3099 was cloned into pMODUL3460, which is a derivative of the vector pMODUL3459 (Goderis et al., 2002) containing in sequence an I-SceI restriction site, the chimeric [uOcs]pMas promoter (De Bolle et al., 2001), a polylinker including NcoI and SacI restriction sites, the 3′-untranslated terminator region of the A. tumefaciens mannopine synthase gene (Barker et al., 1983), and an I-SceI restriction site. The expression cassette in the resulting plasmid, called pFAJ3325, was digested with I-SceI (flanking both the 5′ end of the [uOcs]pMas promoter and the 3′ end of the mannopine synthase terminator) and cloned in the corresponding site of the plant transformation vector pFAJ3337, which is a pPZP-RCS2 derivative (De Bolle et al., 2002) with a selectable marker gene cassette based on the Streptomyces hygroscopicus pat gene (White et al., 1990), to yield plasmid pFAJ3339.
For the construction of plasmid pFAJ3340, several PCR reactions were performed. At first, an inverse PCR reaction with primers OWB520 (sense, 5′ GAGGACGTGGAACCT GGTCAGAAGTTGTGCC) and OWB521 (antisense, 5′ TGGGGTAGCCACCTCGTCGGCC GCGTTGGAAC) was performed on plasmid pFAJ3099 to yield plasmid pFAJ3312 with silent mutations in the linker peptide to incorporate additional restriction sites in the coding region of the linker peptide (EagI site at the 5′ end of the linker peptide-coding region and SexAI site at the 3′ end of the linker peptide-coding region). A second PCR reaction was performed on plasmid pFAJ3099 with primers OWB452 (sense, 5′ TTACACCATGGTGAA TCGGTCGGTTGCGTTCTCCGCGTTCGTTCTGATCCTTTTCGTGCTCGCCATCTCAGATATCGCATCCGTTAGTGGACAGAAGTTGTGCCAAAGG C) and OWB519 (antisense, 5′ CGGCCGCGTTGGAACACGGGAAGTAGCAGATACAC) to attach the coding region of the leader peptide of DmAMP1 at the 5′ end of the RsAFP2 coding region and the 5′ part of the coding region of the linker peptide (together with the EagI restriction site) at the 3′ end of RsAFP2 coding region. The resulting PCR product was cloned into pGEM-T (Promega, Madison, WI) to yield plasmid pFAJ3313. A third PCR reaction was performed on plasmid pDMAMPD with primers OWB517 (sense, 5′ AACCTGGTGAACTA TGCGAGAAAGCTAGC) and OWB518 (antisense, 5′ GAGCTCTATTAGCAGTTGAAGT AACAGAAACAC) to attach the 3′ part of the linker peptide coding reigon at the 5′ end of the DmAMP1 coding region. The resulting PCR product was cloned into pGEM-T to yield pFAJ3314. The NcoI-EagI fragment of pFAJ3313 and the SexAI-SacI fragment of plasmid pFAJ3314 were cloned into the corresponding restriction sites of plasmid pFAJ3312 to yield pFAJ3324. The NcoI-SacI fragment of pFAJ3324 was cloned into the corresponding restriction sites of plasmid pMODUL3460 to yield plasmid pFAJ3328. The I-SceI fragment of plasmid pFAJ3328 was cloned into the corresponding restriction site of plasmid pFAJ3337 to yield plasmid pFAJ3340.
For the construction of plasmid pFAJ3433, the NcoI-SacI fragment of plasmid pDMAMPD containing the DmAMP1 cDNA was first cloned into the corresponding restriction sites of plasmid pMODUL3460 to yield plasmid pFAJ3432. The I-SceI fragment of plasmid pFAJ3432 was cloned into the corresponding restriction site of plasmid pFAJ3337 to yield plasmid pFAJ3433.
For the construction of plasmid pFAJ3375, a PCR reaction with primers OWB452 (sense, 5′ TTACACCATGGTGAATCGGTCGGTTGCGTTCTCCGCGTTCGTTCTGATC CTTTTCGTGCTCGCCATCTCAGATATCGCATCCGTTAGTGGACAGAAGTTGTGCCAAAGGC) and OWB172 (antisense, 5′ TTAGAGCTCCTATTAACAAGGAAAGTAGC) was performed on plasmid pFAJ3099. The resulting PCR product was digested with NcoI and SacI and cloned into the plasmid pMODUL3460 to yield plasmid pFAJ3364. The I-SceI fragment of pFAJ3364 was cloned into the corresponding restriction site of plasmid pFAJ3337 to yield plasmid pFAJ3375.
Plant Transformation
Arabidopsis ecotype Columbia-O was transformed using recombinant A. tumefaciens by the vacuum infiltration method of Bechthold et al. (1993). Transformants were selected on a sand/perlite mixture subirrigated with water containing the herbicide Basta (Agrevo NV, Machelen, Belgium) at a final concentration of 5 mg L−1 for the active ingredient phosphinothricin.
ELISA Assay and Protein Assay
Antisera were raised in rabbits injected with either DmAMP1 (purified as in Osborn et al., 1995) or RsAFP2 (purified as described in Terras et al., 1992). ELISA assays were set up as competitive type assays essentially as described by Penninckx et al. (1996). Coating of the ELISA microtiterplates was done with 50 ng mL−1 DmAMP1 or RsAFP2 in coating buffer (15 mm Na2CO3 and 35 mm NaHCO3, pH 9.6). Primary antisera were used as 1,000- and 2,000-fold diluted solutions (DmAMP1 and RsAFP2, respectively) in 3% (w/v) gelatin (Sigma-Aldrich NV, Bornem, Belgium) in phosphate-buffered saline (140 mm NaCl, 3 mm KCl, 2 mm KH2PO4, and 8 mm Na2HPO4, pH 7.4) containing 0.05% (v/v) Tween 20.
Total protein content was determined according to Bradford (1976) using bovine serum albumin as a standard.
Statistical analysis of expression levels in plant extracts (expressed as micrograms DmAMP1 or RsAFP2 equivalents per milligram of total protein) was performed. Because of the non-normal distribution of the datasets (as shown by QQplots), a natural logarithm transformation of the datasets was performed. These logarithm-transformed datasets were analyzed by using the Tukey's studentized range test with a significance level of 95%.
Northern-Blot Assay
Extraction of RNA from Arabidopsis leaves and northern-blot analysis via chemiluminiscent detection were performed as described by Eggermont et al. (1996). Quantification of band intensity was done using a NightOwl luminiscence digital camera (EG&G Berthold, Bad Wildbad, Germany).
Preparation of Extracellular Fluid, Intracellular Extract, and Crude Leaf Extract
Extracellular fluid was collected from Arabidopsis leaves by immersing the leaves in a beaker containing extraction buffer. The extraction buffer used to analyze secretion of the AMPs consisted of 100 mm KCl, 9.6 mm NaH2PO4.2H2O, 15.2 mm Na2HPO4.2H2O, and 1.5 m NaCl (pH 7). The extraction buffer used to conduct cleavage analyses consisted of 50 mm MES, 1 mm phenylmethylsulfonylfluoride, 1 mm N-ethylmaleimide, 5 mm EDTA, and 0.02 mm pepstatinA (pH 6). The beaker was placed in a vacuum chamber and subjected to six consecutive rounds of vacuum for 2 min followed by abrupt release of vacuum. The infiltrated leaves were gently placed in a centrifuge tube on a grid separated from the tube bottom. The extracellular fluid was collected from the bottom of the tube after centrifugation of the tubes for 15 min at 1,800g. The leaves were extracted in a Phastprep (BIO101/Savant, Toronto) reciprocal shaker (Eggermont et al., 1996) and the extract clarified by centrifugation (10 min at 10,000g). The resulting supernatant is referred to as the intracellular extract.
To prepare a crude leaf extract, Arabidopsis leaves were homogenized under liquid nitrogen and extracted with 50 mm MES (pH 6) containing a mixture of protease inhibitors (1 mm phenylmethylsulfonylfluoride, 1 mm N-ethylmale-imide, 5 mm EDTA, and 0.02 mm pepstatinA). The homogenate was cleared by centrifugation (10 min at 10,000g) and the supernatant is referred to as the crude leaf extract.
Glc-6-Phosphate Deydrogenase Test
Glc-6-phosphate dehydrogenase activity was measured according to a modified protocol of Simcox et al. (1977). The reaction mixture consisted of 10 mm MgCl2, 0.1% (w/v) Triton-100, 0.17 mm NADP+, and 0.33 mm Glc-6-phosphate in 20 mm N-[2-hydroxy-ethyl]piperazine-N′-[2-ethanesulfonic acid], pH 7.5. This mixture was incubated at 37°C; thereafter, the sample was added. After addition of the sample, A340 was measured 15 times with interval periods of 15 s.
Separation of Proteins Processed from Polyprotein Precursors Encoded by Constructs pFAJ3105 and pFAJ3340
Extracellular Fluid
Extracellular fluid (collected as described above) was injected on an RP-HPLC column consisting of C8 silica (0.46 × 25 cm, Varian, Palo Alto, CA) equilibrated with 15% (w/v) acetonitrile in 0.1% (v/v) TFA. The column was eluted at 1 mL min−1 in a linear gradient in 35 min from 15% to 50% (w/v) acetonitrile in 0.1% (v/v) TFA. The eluate was monitored for A280 and individual peak fractions were collected and analyzed for the presence of DmAMP1-CRPs and RsAFP2-CRPs by ELISA assays. Fractions that contained DmAMP1-CRPs and RsAFP2-CRPs were evaporated and used for amino-terminal sequence analysis, mass spectrometry, and in vitro antifungal activity assay.
Crude Leaf Extract
Crude leaf extract from plants transformed with construct pFAJ3105 was fractionated by IEC. IEC was performed by passing the extract over a cation-exchange column (Mono S, 5 × 50 mm, Pharmacia Biotech, Piscataway, NJ) equilibrated with 50 mm MES at pH 6. The column was eluted with a linear gradient of 0 to 0.5 m NaCl in 50 mm MES at pH 6 at a flow rate of 1 mL min−1 and 1-mL fractions were collected. Fractions were assessed for the presence of DmAMP1-CRPs and RsAFP2-CRPs via ELISA assay (as described above). Fractions containing either DmAMP1-CRPs or RsAFP2-CRPs were further fractionated by RP-HPLC as described above.
Assessment of Protein Levels
The concentration of purified DmAMP1 and RsAFP2 in solution was determined by weighing lyophilized proteins before dissolving. Known amounts of either purified DmAMP1 or RsAFP2 (5, 10, and 20 μg) were added to partially purified extracellular fluid from wild-type plants and the mixtures were subjected to HPLC as described above. The protein amounts of the protein standards were related to the area of the peaks corresponding to DmAMP1-CRPs or RsAFP2-CRPs as determined with an HP3396 series II integrator. This correlation of protein amount to peak area was used to determine protein amounts in HPLC chromatogram peaks, integrated using the same settings as for the protein standards, corresponding to DmAMP1-CRPs or RsAFP2-CRPs in samples consisting of partially purified extracellular fluid from transgenic plants, assuming that the peaks consisted purely of DmAMP1 or RsAFP2, respectively.
Reduction of Proteins
Reduction of proteins was performed by adding Tris-HCl (pH 8.4) and dithiotreitol to final concentrations of 100 mm, followed by incubation at 45°C for 1 h. Reagents were removed by RP-HPLC on a C8 silica column (0.46 × 25 cm, Rainin).
Antifungal Activity Assay
Antifungal activity was measured by microspectrophotometry (Broekaert et al., 1990). Spores of the test fungus Fusarium culmorum (strain MUCL30162, Mycothèque Université Catholique de Louvain, Louvain-la-Neuve, Belgium) were suspended in 12 g L−1 potato dextrose broth (Difco, Detroit) at a final concentration of 2.104 spores mL−1, supplemented with tetracycline (10 μg mL−1) and cefotaxim (100 μg mL−1). This suspension was dispensed by aliquots of 80 μL into wells of a 96-well microtiter plate containing 20 μL of the sample to be analyzed. After 24 to 48 h of incubation at 25°C, growth of the fungus was evaluated microscopically, using an inverted microscope, and by measuring the culture A595 using a microplate reader.
ACKNOWLEDGMENT
We thank Katleen Butaye for her guidance on the statistical analyses.
Footnotes
This research was partially supported by the “Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen” (grant no. SB971156), by the European Union (grant no. AIR2–CT94–1356), and by Zeneca Agrochemicals (UK). I.E.J.A.F. is the recipient of a predoctoral fellowship of the “Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen.”
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010794.
LITERATURE CITED
- Barker RF, Idler KB, Thompson DV, Kemp JD. Nucleotide sequence of the T-DNA region from Agrobacterium tumefaciens octopine Ti plasmid pTi15955. Plant Mol Biol. 1983;2:335–350. doi: 10.1007/BF01578595. [DOI] [PubMed] [Google Scholar]
- Bechthold N, Ellis J, Pelletier G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris. 1993;316:1194–1199. [Google Scholar]
- Becker D, Kemper E, Schell J, Masterson RAD. New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- Beck von Bodman S, Domier LL, Farrand SK. Expression of multiple eukaryotic genes from a single promoter in Nicotiana. Bio/Technology. 1995;13:587–591. doi: 10.1038/nbt0695-587. [DOI] [PubMed] [Google Scholar]
- Bevan M, Barnes WM, Chilton MDD. Structure and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 1983;11:369–379. doi: 10.1093/nar/11.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizily SP, Rugh CL, Meagher RB. Phytodetoxification of hazardous organomercurials by genetically engineered plants. Nat Biotechnol. 2000;18:213–217. doi: 10.1038/72678. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW. An automated quantitative assay for fungal growth inhibition. FEMS Microbiol Lett. 1990;69:55–60. [Google Scholar]
- Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW. Antimicrobial peptides from plants. Crit Rev Plant Sci. 1997;16:297–323. [Google Scholar]
- Broekaert WF, Terras FRG, Cammue BPA, Osborn RW. Plant defensins: novel antimicrobial peptides as components of the host defense system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Dougherty WG. A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc Natl Acad Sci USA. 1988;85:3391–3395. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hondt K, Bosch D, Van Damme J, Goethals M, Vandekerckhove J, Krebbers E. An aspartic endoproteinase present in seeds cleaves Arabidopsis 2S albumins in vitro. J Biol Chem. 1993;268:20884–20891. [PubMed] [Google Scholar]
- Eggermont K, Goderis IJ, Broekaert WF. High-throughput RNA extraction from plant samples based on homogenisation by reciprocal shaking in the presence of a mixture of sand and glass beads. Plant Mol Biol Rep. 1996;14:273–279. [Google Scholar]
- Gallie DR, Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis IJWM, De Bolle MFC, François IEJA, Wouters PFJ, Broekaert WF, Cammue BPA (2002) A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol Biol (in press) [DOI] [PubMed]
- Halpin C, Cooke SE, Barakate A, El Amrani A, Ryan MD. Self-processing 2A-polyproteins: a system for co-ordinate expression of multiple proteins in transgenic plants. Plant J. 1999;17:453–459. doi: 10.1046/j.1365-313x.1999.00394.x. [DOI] [PubMed] [Google Scholar]
- Kaminski A, Hunt SL, Gibbs CL, Jackson RJ. Internal initiation of mRNA translation in eukaryotes. Genet Eng. 1994;16:115–155. [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson JD. Duplication of CaMV35S promoter sequences creates a strong enhancer for plant genes. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- Mann M, Talbo G. Development in matrix-assisted laser desorption/ionization peptide mass spectrometry. Curr Opin Biotechnol. 1996;7:11–19. doi: 10.1016/s0958-1669(96)80089-9. [DOI] [PubMed] [Google Scholar]
- Marcos JF, Beachy RN. In vitro characterization of a cassette to accumulate multiple proteins through synthesis of a self-processing polypeptide. Plant Mol Biol. 1994;24:495–503. doi: 10.1007/BF00024117. [DOI] [PubMed] [Google Scholar]
- Matzke AJM, Matzke MA. Position effects and epigenetic silencing of plant genes. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Mitsuhara I, Ohshima M, Natori S, Ohashi Y. Enhanced expression of an antimicrobial peptide sarcotoxin IA by GUS fusion in transgenic tobacco plants. Plant Cell Physiol. 1998;39:57–63. doi: 10.1093/oxfordjournals.pcp.a029289. [DOI] [PubMed] [Google Scholar]
- Osborn RW, De Samblanx GW, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees SB, Broekaert WF. Isolation and characterization of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett. 1995;368:257–262. doi: 10.1016/0014-5793(95)00666-w. [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux JP, Manners JM, Broekaert WF. Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic-independent pathway. Plant Cell. 1996;8:2309–2323. doi: 10.1105/tpc.8.12.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont-Kingdon GAD. Construction of chimeric molecules by a two-step recombinant PCR method. Biotechniques. 1994;16:1010–1011. [PubMed] [Google Scholar]
- Simcox PD, Reid EE, Canvin DT, Dennis DT. Enzymes of the glycolytic and pentose phosphate pathways in proplastids from the developing endosperm of Ricinus communis L. Plant Physiol. 1977;59:1128–1132. doi: 10.1104/pp.59.6.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Mitsky TA, Houmiel KL, Hao M, Reiser SE, Taylor NB, Tran M, Valentin HE, Rodriguez DJ, Stone DA et al. Metabolic engineering of Arabidopsis and Brassica for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymer production. Nat Biotechnol. 1999;17:1011–1016. doi: 10.1038/13711. [DOI] [PubMed] [Google Scholar]
- Tailor RA, Acland DP, Attenborough S, Cammue BPA, Evans IJ, Osborn RW, Ray J, Rees SB, Broekaert WF. A novel family of small cysteine-rich antimicrobial peptides from seeds of Impatiens balsamina is derived from a single precursor protein. J Biol Chem. 1997;272:24480–24487. doi: 10.1074/jbc.272.39.24480. [DOI] [PubMed] [Google Scholar]
- Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J et al. Small cysteine-rich antifungal proteins from radisch: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terras FRG, Schoofs HME, De Bolle MFC, Van Leuven F, Rees SB, Vanderleyden J, Cammue BPA, Broekaert WF. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem. 1992;267:15301–15309. [PubMed] [Google Scholar]
- Urwin PE, McPherson MJ, Atkinson HJ. Enhanced transgenic plant resistance to nematodes by dual proteinase inhibitor constructs. Planta. 1998;204:472–479. doi: 10.1007/s004250050281. [DOI] [PubMed] [Google Scholar]
- Van den Elzen PJM, Jongedijk E, Melchers LS, Cornelissen BJC. Virus and fungal resistance: from laboratory to field. Phil Trans R Soc Lond B. 1993;342:271–278. [Google Scholar]
- Vitale A, Denecke J. The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell. 1999;11:615–628. doi: 10.1105/tpc.11.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Chang SP, Bibb MJ. A cassette containing the bar gene of Streptomyces hygroscopicus: a selectable marker for plant transformation. Nucleic Acids Res. 1990;18:1062. doi: 10.1093/nar/18.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D-J, Bressan RA, Hasegawa PM. Plant antifungal proteins. Plant Breed Rev. 1997;14:39–88. [Google Scholar]
- Zhu Q, Maher EA, Masoud S, Dixon RA, Lamb CJ. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology. 1994;12:807–812. [Google Scholar]