Abstract
Cyanobacteria and proteobacteria (purple bacteria) are the only prokaryotes known to synthesize sucrose (Suc). Suc-P synthase, Suc-phosphatase (SPP), and Suc synthase activities have previously been detected in several cyanobacteria, and genes coding for Suc-P synthase (sps) and Suc synthase (sus) have been cloned from Synechocystis sp. PCC 6803 and Anabaena (Nostoc) spp., respectively. An open reading frame in the Synechocystis genome encodes a predicted 27-kD polypeptide that shows homology to the maize (Zea mays) SPP. Heterologous expression of this putative spp gene in Escherichia coli, reported here, confirmed that this open reading frame encodes a functional SPP enzyme. The Synechocystis SPP is highly specific for Suc-6F-P (Km = 7.5 μm) and is Mg2+ dependent (Ka = 70 μm), with a specific activity of 46 μmol min−1 mg−1 protein. Like the maize SPP, the Synechocystis SPP belongs to the haloacid dehalogenase superfamily of phosphatases/hydrolases. Searches of sequenced microbial genomes revealed homologs of the Synechocystis sps gene in several other cyanobacteria (Nostoc punctiforme, Prochlorococcus marinus strains MED4 and MIT9313, and Synechococcus sp. WH8012), and in three proteobacteria (Acidithiobacillus ferrooxidans, Magnetococcus sp. MC1, and Nitrosomonas europaea). Homologs of the Synechocystis spp gene were found in Magnetococcus sp. MC1 and N. punctiforme, and of the Anabaena sus gene in N. punctiforme and N. europaea. From analysis of these sequences, it is suggested that Suc synthesis originated in the proteobacteria or a common ancestor of the proteobacteria and cyanobacteria.
Suc is found in both freshwater and marine cyanobacteria, e.g. Calothrix, Scytonema, Oscillatoria, Plectonema, Synechococcus, Anabaena, and Nostoc (Reed et al., 1984; Page-Sharp et al., 1999). It is often synthesized in these organisms in response to salt or osmotic stress and is thought to help maintain osmotic balance and stabilize protein and membrane structure and function (Reed and Stewart, 1985; Reed et al., 1986; Hagemann and Marin, 1999). Porchia and Salerno (1996) reported the first measurements of Suc-P synthase (SPS) and Suc-phosphatase (SPP) activity in cyanobacteria, in Nostoc sp. PCC 7119 (syn. Anabaena sp. PCC 7119). Both enzymes have also been found in a Scytonema sp. (Page-Sharp et al., 1999). Kaneko et al. (1996) sequenced the genome of the unicellular, freshwater cyanobacterium Synechocystis sp. PCC 6803 and found an open reading frame (ORF) that showed significant similarity to known SPS genes from higher plants. Subsequent cloning and heterologous expression proved that this ORF does encode a functional SPS enzyme, albeit one with some unusual kinetic properties (Curatti et al., 1998; Lunn et al., 1999). Mutation of the sps gene in Synechocystis sp. PCC 6803 cells abolished their ability to synthesize Suc (Hagemann and Marin, 1999). Together, these results indicate that cyanobacteria synthesize Suc by the same route as plants, via SPS and SPP.
The enzyme Suc synthase (SuSy) has also been detected in some filamentous cyanobacteria (Schilling and Ehrnsperger, 1985; Porchia et al., 1999a) and genes for the enzyme have been cloned from Anabaena (Nostoc) sp. PCC 7119 and Anabaena variabilis (Curatti et al., 2000). The reaction catalyzed by SuSy is readily reversible and, despite its name, it is thought that it usually operates in the direction of Suc breakdown. However, the relative rates of the forward and reverse reactions catalyzed by SuSy depend on the concentrations of its other reactants, so under certain conditions SuSy could catalyze the net synthesis of Suc. Schilling and Ehrnsperger (1985) found that most SuSy activity is in the vegetative cells of A. variabilis, while the N2-fixing heterocysts contain high alkaline invertase activity. Although SPS activity was not detected in either cell type of A. variabilis, Schilling and Ehrnsperger (1985) suggested that Suc is synthesized in the photosynthetic cells by SuSy and transported to the nonphotosynthetic heterocysts to support respiration.
Spatial separation of photosynthesis and N2 fixation in the different cell types of filamentous cyanobacteria is believed to protect the oxygen-sensitive nitrogenase from the oxygen generated by photosynthetic water splitting (Golden et al., 1997). If we assume that filamentous cyanobacteria evolved from unicellular ancestors, we might speculate that Suc, originally used in adaptation to osmotic stress, was later adopted as a transport compound to shuttle carbon and energy between cells in the filamentous species. This foreshadows the use of Suc as a transport carbohydrate in higher plants. However, as will be discussed later, it is most likely that plants inherited Suc metabolism from a unicellular, cyanobacterial endosymbiont. Therefore, this is most likely to be an example of parallel evolution, reflecting the suitability of Suc for a transport function. Unicellular, N2-fixing cyanobacteria overcome the incompatibility of nitrogenase with oxygenic photosynthesis by temporal separation of the two processes, governed by circadian rhythms in gene expression (Golden et al., 1997). In the unicellular cyanobacterium Cyanothece sp. strain ATCC 51142, a glycogen-like Glc polymer is synthesized during the photosynthetic phase in the light and degraded during the N2-fixing phase in the dark and is thought to act as a transient energy store (Schneegurt et al., 1994). It would not be surprising if Suc were to fulfill a similar function in some other unicellular, N2-fixing species.
There are few reports of the presence of Suc in noncyanobacterial prokaryotes (Fig. 1). Suc has been detected in two species of halotolerant methanotrophs, Methylobacter alcaliphilus 20Z and Methylobacter modestohalophilus 10S, belonging to the proteobacteria (purple bacteria) and is presumed to act as an osmoprotectant in these species (Khmelenina et al., 1999). Fru-6-P-dependent production of UDP from UTP and Glc-1-P in M. alcaliphilus 20Z cells was attributed to a combination of UDP-Glc pyrophosphorylase and SPS activities (Khmelenina et al., 2000), but otherwise little is known about the enzymology of Suc metabolism in this group of organisms. An SPS-like ORF was found in the genome of the proteobacterium Acidithiobacillus ferrooxidans (syn. Thiobacillus ferrooxidans; Mijts and Patel, 2001). An ORF found by random sequencing of the genome of Halothermothrix orenii, which belongs to the Bacillus/Clostridium group of bacteria, was also found to show some homology with the Synechocystis SPS (Mijts and Patel, 2001). However, only 56 amino acid residues were reported, and these show only 39% identity with the Synechocystis SPS (residues 24–57). This region is highly conserved in all known SPS sequences and is thought to be involved in substrate binding (Huber and Huber, 1996). Even the evolutionarily distant SPSs from Synechocystis and maize (Zea mays), which have about 43% overall identity (Lunn et al., 1999), show 59% identity in this region. A recent search indicated that the best match for the partial H. orenii sequence in the GenBank database was a chloroperoxidase from Rhodococcus sp. S9 (accession no. AF265259). Further evidence will be required before a function can be assigned to the H. orenii ORF with confidence.
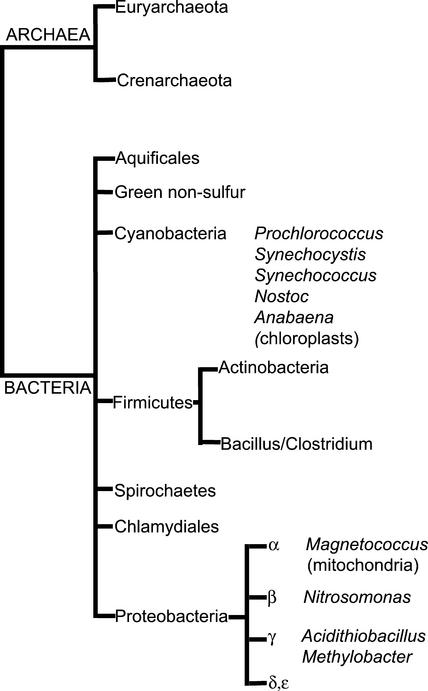
Figure 1.
Phylogenetic tree of the Archaea and Bacteria adapted from Olsen et al. (1994).
Lunn et al. (2000) reported the first cloning of an SPP gene, from maize. They also reported the presence of an ORF (GenBank accession no. AF300455) in the Synechocystis sp. PCC 6803 genome coding for a 244-amino acid polypeptide with significant similarity to the 260-amino acid N-terminal region of the maize SPP. Both the maize SPP and the putative Synechocystis SPP belong to a superfamily of phosphatases/hydrolases related to the haloacid dehalogenase (HAD) enzyme from Pseudomonas sp. YL (Aravind et al., 1998). Enzymes belonging to this superfamily are characterized by three highly conserved motifs associated with the active site. One of these motifs includes the sequence D*XDX(T/V) where D* is an Asp residue that forms an acyl-phosphate intermediate during catalysis (Collet et al., 1998). The C-terminal regions of both the maize and Synechocystis SPS are homologous to the maize SPP but lack two of the highly conserved Asp residues, including the critical active site Asp (D*). This is consistent with the observation that highly purified SPS shows no SPP activity.
In this paper, the cloning and heterologous expression of the putative Synechocystis spp gene, which show that it does encode a functional SPP enzyme, are described. Searches of microbial genome databases revealed homologs of SPS, SPP, and SuSy genes in other cyanobacteria and in several proteobacteria. Some of the putative SPS polypeptides contain all of the conserved HAD family active site residues in their SPP-like C-terminal domains, while others lack this domain altogether. The possible significance of this finding for our understanding of the origins and evolution of Suc metabolism is discussed.
RESULTS
Cloning and Expression of the Synechocystis spp Gene
The putative Synechocystis sp. PCC 6803 spp gene was isolated from genomic DNA by PCR amplification. The reaction yielded a single product with the expected size of 786 bp (data not shown), which was cloned into the bacterial expression vector pTYB2 under the control of the T7 promoter. The recombinant plasmid pTYB2/Synspp was introduced into the protease-deficient E. coli strain ER2566, which carries a chromosomal copy of the T7 RNA polymerase gene under the control of the isopropyl β-d-thiogalactopyranoside (IPTG)-inducible lacZ promoter. Protein expression was induced in early-log phase cultures by the addition of IPTG. Extracts from induced cultures showed phosphatase activity, with Suc-6F-P (Suc-6-P) as the substrate, of 3.8 μmol min−1 mg−1 protein, which was 94% inhibited by 20 mm EDTA. In contrast, extracts from cells containing the pTYB2 plasmid with no insert showed less than 0.005 μmol min−1 mg−1 protein phosphatase activity with Suc-6-P as substrate, and this activity was not inhibited by 20 mm EDTA. The extracts from cells carrying either pTYB2 or pTYB2/Synspp showed similarly low phosphatase activity (0.010 and 0.013 μmol min−1 mg−1 protein, respectively) with Fru-6-P as the substrate.
Characterization of the Synechocystis SPP
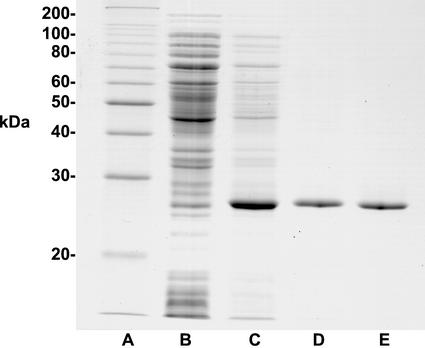
The heterologously expressed Synechocystis SPP was purified from E. coli extracts by polyethylene glycol 8000 (PEG) fractionation and acid precipitation as described in “Materials and Methods.” The purified protein had a specific activity of 46 μmol min−1 mg−1 protein and showed a single, 27-kD band on Coomassie Blue-stained SDS polyacrylamide gels (Fig. 2). Gel filtration on a Superdex 200 FPLC column (Lunn et al., 2000) did not give any greater purification (Fig. 2) but showed that the native molecular mass of the enzyme is 27 kD and, therefore, that the enzyme is monomeric. The Synechocystis SPP has a broad pH optimum around 6.8 and is highly specific for Suc-6-P with a Km of 7.5 μm (Table I). Its activity is dependent on the presence of Mg2+ with a Ka of 70 μm and, when assayed with 250 μm Suc-6-P, is inhibited 19% and 27% by 200 mm and 660 mm Suc, respectively. The purified enzyme is stable for at least 6 months when stored at −80°C.
Figure 2.
Purification of the Synechocystis SPP expressed in E. coli. Proteins were separated by SDS-PAGE in a 12% (w/v) gel and visualized by staining with Coomassie Blue R250. Lane A, 10-kD ladder protein molecular mass markers. Lane B, Total soluble extract from E. coli (pTYB2/Synspp) noninduced (20 μg). Lane C, Total soluble extract from E. coli (pTYB2/Synspp) IPTG-induced (10 μg). Lane D, 29% (w/v) PEG, pH 5 precipitate (2 μg). Lane E, Superdex 200 fraction (2 μg).
Table I.
Properties of Suc-phosphatase from Synechocystis sp. PCC 6803 and rice
| Property | Synechocystis | Ricea |
|---|---|---|
| Molecular mass (kD) | ||
| Native | 27 | 100 |
| Subunit | 27 | 50 |
| pH optimum | 6.8 | 6.8 |
| Km Suc6P (μm) | 7.5 | 65 |
| Ka Mg2+ (μm) | 70 | 8 |
| Specific activity (μmol min−1 mg−1 protein) | 46 | 1,250 |
From Lunn et al. (2000).
SPS, SPP, and SuSy Homologs in Other Cyanobacteria and Proteobacteria
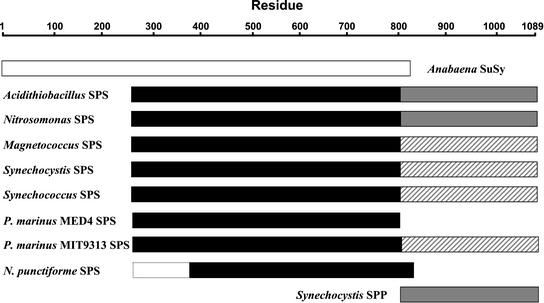
Other microbial genome sequences in public databases were searched for homologs of known SPS, SPP, and SuSy genes to provide information about the evolution of these enzymes. The Synechocystis SPS and SPP and the Anabaena SuSy are shown in alignment with their homologs from other microbial species in Appendix A. SPS-like ORFs were identified in the genomes of four cyanobacteria: Synechococcus sp. WH8102 (marine, unicellular, Joint Genome Institute [JGI]), Prochlorococcus marinus strain MED4 (marine, unicellular, high-light adapted, JGI), P. marinus strain MIT9313 (marine, unicellular, low-light adapted, JGI) and Nostoc punctiforme (filamentous, N2-fixing, JGI) (Fig. 3). SPS-like ORFs were also found in three proteobacteria, including that previously reported in A. ferrooxidans: Magnetococcus sp. MC1 (α-subdivision, magnetotactic, JGI), Nitrosomonas europaea (β-subdivision, autotrophic, chemolithotrophic [NH3-oxidizing], JGI) and A. ferrooxidans (γ-subdivision, chemolithotrophic [Fe2+/reduced sulfur-oxidizing], the Institute for Genomic Research [TIGR]) (Fig. 3). The best match for all of these SPS-like sequences in the GenBank database was the Synechocystis SPS, with expect (E) values ranging from 0 to 3 × 10−47 and sequence identities of 34% to 61% at the amino acid level (Table II). The available N. punctiforme sequence is incomplete at the 5′ end of the putative SPS coding region, which could partly account for the higher E value and lower identity. The predicted sizes of the polypeptides encoded by these SPS-like ORFs are very close to that of the Synechocystis SPS (720 amino acid residues, 81.4 kD), with the exception of those from P. marinus MIT9313 and N. punctiforme (Table II). The latter two sequences show homology only with the N-terminal, glucosyltransferase domain of the Synechocystis SPS and lack the C-terminal, SPP-like domain (Fig. 3). All of the other SPS homologs contain both the glucosyltransferase and SPP-like domains. The SPP-like domains of the putative A. ferrooxidans and N. europaea SPSs contain all of the conserved residues associated with the active site of HAD superfamily phosphatases, but all of the other sequences lack one or more of these residues (Table III).
Figure 3.
Schematic alignment of SPS and SPP from Synechocystis sp. PCC 6803 with Suc synthase from Anabaena sp. PCC 7119 and SPS-like sequences from other cyanobacterial and proteobacterial species. ░⃞, SPP and SPP-like sequences containing all of the conserved HAD phosphatase active site residues. ▨, SPP-like sequences lacking one or more of these conserved residues. The dotted lines indicate the expected size of the complete N. punctiforme SPS.
Table II.
Homologs of the Synechocystis sp. PCC 6803 sps and spp genes and Anabaena spp. sus genes in other cyanobacteria and proteobacteria
| Species | Best Database Matcha | E Value | Amino Acid Identityb | Amino Acid Residues | Molecular Wt |
|---|---|---|---|---|---|
| % | |||||
| SPS | |||||
| Synechocystis sp. PCC6803 | Synechocystis SPSc | 0 | (100) | 720 | 81,411 |
| Synechococcus sp. WH8012 | Synechocystis SPSc | 1 × 10−136 | 41 | 710 | 80,221 |
| P. marinus MIT9313 | Synechocystis SPSc | 1 × 10−143 | 43 | 710 | 80,330 |
| P. marinus MED4 | Synechocystis SPSc | 1 × 10−115 | 45 | 468 | 53,096 |
| N. punctiformed | Synechocystis SPSc | 3 × 10−47 | 34 | (419)d | |
| Magnetococcus sp. MC1 | Synechocystis SPSc | 0 | 61 | 716 | 79,637 |
| N. europaea | Synechocystis SPSc | 0 | 50 | 713 | 81,000 |
| A. ferrooxidans | Synechocystis SPSc | 1 × 10−167 | 47 | 704 | 79,655 |
| SPP | |||||
| Synechocystis sp. PCC6803 | Synechocystis SPPe | 0 | (100) | 244 | 27,761 |
| Nostoc sp. PCC7120 | Nostoc 7120 SPPf | 1 × 10−146 | 40 | 249 | 28,032 |
| N. punctiforme (1) | Synechocystis SPPe | 7 × 10−45 | 45 | 257 | 28,967 |
| N. punctiforme (2) | Nostoc 7120 SPPf | 1 × 10−115 | 39 | 252 | 28,420 |
| Magnetococcus sp. MC1 (1) | Synechocystis SPSc | 1 × 10−15g | 31 | 277 | 30,707 |
| Magnetococcus sp. MC1 (2) | Synechocystis SPPe | 1 × 10−8 | 30 | 264 | 29,469 |
| SuSy | |||||
| N. punctiforme (1) | Anabaena spp. SSh | 0 | 87 | 806 | 93,523 |
| N. punctiforme (2) | Anabaena spp. SSh | 0 | 60 | 805 | 93,062 |
| N. europaea | Rice SSi | 0 | 49 | 794 | 90,864 |
BLASTP search of GenBank nonredundant database.
Comparison with Synechocystis sp. PCC6803 SPS and SPP and Anabaena sp. PCC7119 SS, respectively.
GenBank accession no. D64006.
Partial coding sequence.
GenBank accession no. AF300455.
GenBank accession no. AJ302073.
E value for Synechocystis sp. PCC6803 SPP 3 × 10−14.
Anabaena sp. PCC7119 SS, GenBank accession no. AJ010639 and A. variabilis SS, GenBank accession no. CAC00631.
GenBank accession no. P31924.
Table III.
Conservation of HAD phosphatase superfamily active site residues in SPP and SPS from Synechocystis sp. PCC 6803 and in SPP- and SPS-like sequences from other cyanobacterial and proteobacterial species
| Enzyme | HAD-Phosphatase Active Site Residues
|
||||
|---|---|---|---|---|---|
| DXDXT9–13 | T41 | K161 | D186 | D190 | |
| Synechocystis SPP | DLDNT | T | K | D | D |
| N. punctiforme SPP1 | ELDNT | T | K | G | D |
| N. punctiforme SPP2 | DLDDT | T | K | D | D |
| Nostoc 7120 SPP | DLDHT | T | K | D | D |
| Magnetococcus SPP1 | DLDRT | T | K | D | D |
| Magnetococcus SPP2 | DMDRT | S | K | D | D |
| P. marinus MIT9313 SPS | DLDSS | T | R | S | D |
| Acidithiobacillus SPS | DIDNT | T | K | D | D |
| Synechocystis SPS | ALQGG | T | K | G | D |
| Synechococcus SPS | DLDST | T | R | S | D |
| Nitrosomonas SPS | DIDNT | T | K | D | D |
| Magnetococcus SPS | DLDQN | T | K | G | D |
Residue numbers refer to the Synechocystis sp. PCC 6803 SPP.
No SPS-like ORF was found in the sequenced genomes of species from the two major groups of Archaea (Woese et al., 1990): (a) Euryarchaeota (nine complete genomes: Archaeoglobus fulgidus, Halobacterium sp. NRC-1, Methanobacterium thermoautotrophicum delta H, Methanococcus jannaschii, Methanothermobacter wolfeii, Pyrococcus abysii, Pyrococcus horikoshii, Thermoplasma acidophilum, Thermoplasma volcanium) and (b) Crenarchaeota (two complete genomes: Aeropyrum pernix, Sulfolobus solfataricus). Likewise, no SPS-like ORFs were found in the sequenced genomes of representative species from the following major groups of Bacteria: Aquificales, green sulfur bacteria, Chlamydiales, Firmicutes (Actinobacteria: high G+C Gram-positive bacteria; and Bacillus/Clostridium group: low G+C Gram-negative bacteria), green nonsulfur bacteria, and Spirochaetes, or in the δ- and ε-subdivisions of the Proteobacteria (Olsen et al., 1994).
Two SPP-like ORFs were found in both N. punctiforme and Magnetococcus sp. MC1 (Table II). The N. punctiforme sequences matched most closely with either the Synechocystis SPP or an SPP-like sequence from Nostoc sp. PCC 7120 (GenBank accession no. AJ302073), with very low E values (Table II). One of the putative N. punctiforme SPPs (SPP1) has Glu and Gly in place of the HAD superfamily active site residues Asp9 and Asp186, respectively (Table III). The two Magnetococcus sp. MC1 SPP-like ORFs are less similar to the Synechocystis SPP, but they do contain all of the conserved, active site residues of the HAD superfamily phosphatases (Table III). No SPP-like ORFs were identified in the genomes of Synechococcus sp. WH8012, P. marinus strains MED4 or MIT9313, A. ferrooxidans, or N. europaea.
Two SuSy-like ORFs were found in the cyanobacterium N. punctiforme and one in the proteobacterium N. europaea (Table II). The former most closely matched known cyanobacterial SuSy sequences from Anabaena spp. (Curatti et al., 2000), whereas the latter matched more closely with a rice (Oryza sativa) SuSy, all with E values of zero. The putative SuSy sequences showed some homology with the N-terminal, glucosyltransferase domain of the Synechocystis SPS but not with the C-terminal, SPP-like domain or the Synechocystis SPP (Fig. 3).
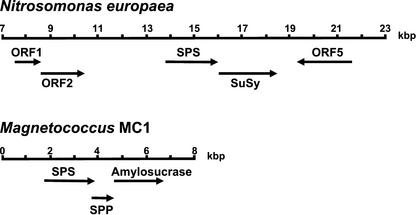
The putative N. europaea SPS and SuSy ORFs are adjacent to each other on the same strand and separated by only 70 bp (Fig. 4). The putative Magnetococcus SPS and SPP1 ORFs overlap by 4 bp on the same strand and are separated by only 130 bp from another ORF on the same strand that most closely matches amylosucrase from the proteobacterium Neisseria polysaccharea (GenBank accession no. CAA09772; Fig. 4).
Figure 4.
Putative Suc metabolism operons in the genomes of N. europaea and Magnetococcus sp. MC1. The flanking sequences in the N. europaea genome contain ORFs whose best matches in the GenBank database are as follows: ORF1, accession no. AAC73882, putative membrane protein (E. coli); ORF2, accession no. BAB34295, putative ATP-binding component of ABC transporter (E. coli); ORF5, accession no. CAC07984, CopF cation (Cu)-transporting ATPase (Ralstonia metallidurans).
Expression of a Chimeric Synechocystis SPS-SPP Protein in E. coli
As described above, the SPP-like domains of the putative SPSs in A. ferrooxidans and N. europaea contain all of the conserved residues associated with the active site of HAD superfamily phosphatases. This suggested that these enzymes might have both SPS and SPP activities. The C-terminal region of the Synechocystis SPS shows 42% identity to the Synechocystis SPP (Lunn et al., 2000) but lacks several of the conserved, active site residues and does not have SPP activity (Lunn et al., 1999). Seo et al. (2000) expressed a fusion protein of the E. coli trehalose-P-synthase (TPS) and trehalose-phosphatase (TPP) and found that the chimeric protein had both TPS and TPP activities. TPS and TPP are functionally and structurally related to SPS and SPP, leading to the question of whether a single polypeptide can have both SPS and SPP activities. A chimeric gene was constructed in which the 3′ end of the Synechocystis sps gene, coding for the SPP-like domain (Leu-474 to Val-720), was replaced with the coding region of the Synechocystis spp gene (Arg-2 to Ser-244) using a convenient SpeI site in the sps gene. This chimeric gene was expressed in E. coli, and cell extracts showed SPS and SPP activities of 0.93 and 0.33 μmol min−1 mg−1 protein, respectively. Antisera raised against either the Synechocystis SPS or SPP both recognized an 81-kD protein in the cell extracts (data not shown).
DISCUSSION
Comparison of SPP from Cyanobacteria and Plants
Heterologous expression of the putative Synechocystis spp gene (Lunn et al., 2000) in E. coli confirmed that the gene does encode a functional SPP enzyme. The enzyme is smaller than that from higher plants, showing homology only with the N-terminal region of the plant enzyme (Table I; Lunn et al., 2000). This is the region that shows homology with the HAD superfamily of phosphatases and is presumably all that is required for catalytic activity. The function of the C-terminal extension of the plant enzyme is unknown, as it does not show significant homology with any other protein of known function. The sequence of a partial cDNA clone from the bryophyte (moss) Physcomitrella patens (GenBank accession no. AW497133) encodes a protein that shows 57% identity with the maize SPP extending into this C-terminal region. This suggests that acquisition of the C-terminal extension was an early event in the evolution of SPP in plants. The native Synechocystis SPP also differs from the plant enzyme in being monomeric rather than dimeric (Table I). However, the kinetic properties of the Synechocystis SPP are similar to those of the plant enzyme; both have similar pH optima, are specific for Suc-6-P, are Mg2+ dependent (Table I), and are competitively inhibited by millimolar concentrations of Suc. The Synechocystis SPP has a 9-fold lower Km for Suc-6-P than the rice SPP, but its specific activity is only about 4% of that of the rice enzyme (Table I). The Synechocystis SPS also has a much lower specific activity than SPS from higher plants; 17 μmol min−1 mg−1 protein compared with 150 μmol min−1 mg−1 protein for the spinach SPS (Huber and Huber, 1996; Lunn et al., 1999). It is also smaller than the plant SPS, 82 kD versus 117 to 119 kD, and it is monomeric rather than di- or tetrameric (Huber and Huber, 1996; Lunn et al., 1999). The basis for the higher specific activity of the plant enzymes is unknown but is consistent with the higher Suc biosynthetic capacity expected in plants.
Origin and Evolution of Suc Metabolism
The discovery of ORFs in the genomes of the proteobacteria A. ferrooxidans, N. europaea, and Magnetococcus sp. MC1 that are homologous to known SPS genes provides further evidence that Suc metabolism is present in the α-, β-, and γ-subdivisions of this group of organisms (Khmelenina et al., 1999; Khmelenina et al., 2000; Mijts and Patel, 2001). Xiong et al. (1998) concluded, from phylogenetic analysis of genes encoding photosystem I and II reaction center proteins, that the oxygenic photosynthetic apparatus of the cyanobacteria evolved from heterologous fusion of ancestral types related to those in the heliobacteria/green sulfur bacteria (photosystem I) and proteobacteria/green nonsulfur bacteria (photosystem II). The implication of these findings, and the apparent absence of Suc-synthesizing enzymes in other groups of Bacteria or the Archaea, is that the origins of Suc metabolism probably lie in the proteobacteria or an ancestral type common to both the proteobacteria and cyanobacteria. Rickettsia spp. in the α-subdivision of the proteobacteria are thought to be closely related to the endosymbionts that evolved into the mitochondria of eukaryotic cells (Brown et al., 2001). Therefore, it is conceivable that Suc metabolism could have been acquired by eukaryotic cells during the endosymbiosis of a Suc-synthesizing mitochondrial ancestor. However, among the Eukaryota, Suc synthesis occurs only in green plants (Viridiplantae) that have oxygenic photosynthesis (Kandler and Hopf, 1980; Hawker and Smith, 1984), which points to a more likely origin in the endosymbiotic cyanobacteria that are believed to have been the ancestors of chloroplasts (Cavalier-Smith, 2000).
The C-terminal, SPP-like domains of the putative A. ferrooxidans and N. europaea SPS enzymes contain all of the conserved residues associated with the active site of HAD superfamily phosphatases (Table II). This points to the possibility that these enzymes are bifunctional with both SPS and SPP activities. Heterologous expression of an artificial, chimeric Synechocystis SPS-SPP showed that a single polypeptide can have both SPS and SPP activities. Interestingly, no SPP-like ORFs were found in the genomes of A. ferrooxidans and N. europaea, which would be consistent with the putative SPSs having SPP activity. In contrast, the putative SPS from another proteobacterium, Magnetococcus sp. MC1, does not have all the conserved residues in its SPP-like domain, but this species does have two SPP-like ORFs. While it remains to be found whether these three species do synthesize Suc, we might speculate that Suc accumulation is an adaptation to the inhospitable environments in which these organisms live. Suc could be used to maintain osmotic balance and stabilize protein and membrane structure in cells growing in high salt or dry environments. In support of this proposal, expression of the Synechocystis SPS and consequent accumulation of Suc was reported to confer desiccation tolerance in E. coli (Billi et al., 2000). Suc might also be used as a storage reserve, which could allow the cells to survive periods when environmental conditions are unfavorable and then be metabolized to allow the cell to grow and divide quickly when conditions improve. The close proximity of the putative Magnetococcus sp. MC1sps and spp1 genes with another ORF encoding an amylosucrase-like protein (Fig. 4) could indicate that all three genes form a polycistronic operon involved in synthesis of a glucan polymer via Suc. Some oral bacteria synthesize extracellular, fructan, or glucan polymer matrices from external Suc (Walker and Jacques, 1987). However, the N terminus of the putative Magnetococcus sp. MC1 amylosucrase protein does not have the characteristics of a signal peptide (Nielsen et al., 1997), suggesting that the enzyme is not secreted, so it might be involved in synthesis of intracellular polysaccharide reserves instead. Such a polymer could function as a transient storage reserve as in the cyanobacterium Cyanothece sp. ATCC 51142 (Schneegurt et al., 1994). The significance of the putative SPS(SPP)-SuSy operon in N. europaea (Fig. 4) is unclear, as it seems surprising that enzymes for Suc synthesis and breakdown should be transcribed together.
The range of SPS-like ORFs among the cyanobacteria is more complex (Fig. 3). Synechocystis sp. PCC 6803 is known to have separate SPS and SPP enzymes, and the SPP-like domain of the SPS lacks several of the conserved, HAD superfamily active site residues, including the critical Asp that is predicted to form an acyl-phosphate intermediate during the phosphatase reaction (Table II; Aravind et al., 1998; Collet et al., 1998). The putative N. punctiforme SPS lacks the C-terminal, SPP-like domain altogether and, although the coding sequence is incomplete, is likely to be smaller than the Synechocystis SPS with an estimated molecular mass of around 58 kD. A slightly smaller molecular mass, 45 to 47 kD, was reported for two forms of SPS from Nostoc (Anabaena) sp. PCC 7119 (Porchia and Salerno, 1996). N. punctiforme has at least one ORF that is likely to encode SPP (SPP2). While the other N. punctiforme SPP-like ORF (SPP1) does show similarity to the Synechocystis SPP (Table I), it has Glu and Gly residues, respectively, in the positions that are homologous to the conserved Asp9 and Asp186 of the Synechocystis SPP. Collet et al. (1998) reported that substitution of Glu or Asn for the first Asp in the conserved DXDX(T/V) motif of two HAD superfamily enzymes, phosphomannomutase and phospho-Ser phosphatase, completely abolished catalytic activity. Therefore, it seems unlikely that the N. punctiforme SPP1 ORF encodes a functional SPP.
The putative P. marinus MED4 SPS also lacks the C-terminal, SPP-like domain of other SPSs, but surprisingly no good candidate for an SPP-encoding ORF was found in the fully sequenced genome of this strain. Similarly, no SPP-like ORFs were found in the genomes of P. marinus MIT9313 or Synechococcus sp. WH8102. However, the putative SPSs from the latter two organisms do have C-terminal, SPP-like domains. Although these do not show perfect conservation of the HAD superfamily active site residues, the core active site motif DXDX(T/V) is present in the Synechococcus sp. WH8102 sequence and, with only a conservative substitution of Ser for Thr, in the P. marinus MIT9313 sequence (Table II). Therefore, it is possible that the ORFs from these two organisms could encode bifunctional enzymes with both SPS and SPP activities. The apparent lack of an SPP in P. marinus MED4 could indicate that this strain does not have the capacity to synthesize Suc. However, we cannot exclude the possibility that there is a highly divergent form of the enzyme in this organism, or that Suc-6-P could be hydrolyzed by a nonspecific phosphatase. In support of the latter possibility, it has been observed that heterologous expression of the Synechocystis SPS in E. coli led to accumulation of some free Suc in the cells, although a specific SPP was absent (Billi et al., 2000).
Two SuSy-like ORFs were found in the genome of N. punctiforme, but none were found in Synechocystis sp. PCC 6803, Synechococcus sp. WH8012, or P. marinus MED4 and MIT9313. This agrees with the report that other filamentous, heterocystic cyanobacteria, e.g. Anabaena spp., Nostoc sp. 6719, and Calothrix sp. PCC 7601, contain SuSy-like genes but unicellular cyanobacteria, e.g. Synechococcus sp. PCC 7942, do not (Curatti et al., 2000). The reason why SuSy has only been found in filamentous species of cyanobacteria is unclear. A clue might come from the apparent lack of SPS activity in A. variabilis and the suggestion that SuSy is responsible for Suc synthesis in this species (Schilling and Ehrnsperger, 1985). The equilibrium constant of the SuSy reaction is unfavorable for accumulation of high concentrations of Suc, but if the Suc were being transported out of the vegetative cells into the heterocysts then SuSy could catalyze its net synthesis. However, SPS and SPP would be required for synthesis of Suc in unicellular species or in filamentous species where high concentrations of Suc are used as an osmoprotectant. Synechocystis sp. PCC 6803 and other unicellular species contain invertase activity and so do not require SuSy to catabolize Suc. The presence of a SuSy-like ORF in the genome of N. europaea suggests that this enzyme also might have originated in the proteobacteria or a common ancestor of the proteobacteria and cyanobacteria. The function of a SuSy enzyme in this unicellular organism is unclear.
It seems likely that plants inherited the enzymes necessary for Suc synthesis from the cyanobacteria, which in turn inherited them from a proteobacteria-like ancestor. The enzymes of Suc synthesis found in the cyanobacteria show considerable diversity, with three main types: (1) an SPS containing only a glucosyltransferase domain, plus or minus a separate SPP (e.g. P. marinus MED4, N. punctiforme), (2) a bifunctional SPS-SPP enzyme (e.g. Synechococcus sp. WH8012, P. marinus MIT9313), and (3) an SPS with a noncatalytic, C-terminal, SPP-like domain plus a separate SPP (e.g. Synechocystis sp. PCC 6803).
The Prochlorophytes (e.g. Prochlorococcus spp.) are thought to be the most primitive group of cyanobacteria; therefore, it seems likely that either type 1 or 2 is closer to the ancestral situation. Between these two options, it seems more likely that separate, type 1 SPS and SPP enzymes were the first to evolve. The similarity between SuSy and the N-terminal, glucosyltransferase domain of SPS (Huber and Huber, 1996) suggests that a type 1 SPS could have evolved from SuSy or that both enzymes evolved from a common ancestor. We can speculate that a type 2 SPS could arise by fusion of a type 1 SPS and SPP, perhaps by mutation of a polycistronic SPS-SPP operon, and that type 3 SPS and SPP could arise by duplication of the active SPP domain of a type 2 SPS followed by loss of catalytic function by the SPP-like domain of the SPS. Interestingly, the gene organization in Magnetococcus sp. MC1 resembles that which might have occurred during such an evolutionary process.
At present, very little is known about Suc metabolism in eukaryotic algae and lower vascular plants. Suc does not appear to be found in the Rhodophyta (red algae; Dancer and ap Rees, 1989) but is present in the Chlorophyta (green algae). SPS enzymes have been partially purified from the green algae Chlorella vulgaris, Scenedesmus obliquus, and Dunaliella tertiolecta, and their kinetic properties have been shown to resemble those of the higher plant enzyme (Duran and Pontis, 1977; Müller and Wegmann, 1978). The algal enzyme was reported to have a native Mr of about 400,000 (Duran and Pontis, 1977), which is closer to that of SPS from higher plants rather than cyanobacteria. C. vulgaris and S. obliquus also contain SuSy activity (Duran and Pontis, 1977). SPS and SuSy activities have also been detected in permeabilized Euglena gracilis cells (Porchia et al., 1999b). Virtually nothing is known about the genes encoding the enzymes involved in Suc metabolism in green algae and primitive plants. Searches of the available databases (http://www.biology.duke.edu/chlamy_genome/crc.html and http://www.kazusa.or.jp) containing ESTs from Chlamydomonas reinhardtii (green alga) and Porphyra yezoensis (red alga) did not identify any significant matches with known SPS, SPP, or SuSy sequences. As noted previously, there is a partial SPP-like cDNA clone from the bryophyte P. patens in the EST database that shows greater similarity to SPP sequences from higher plants than from cyanobacteria. There is also an SPP-like sequence from the gymnosperm Pinus taeda (loblolly pine; GenBank accession no. BG319173).
Sequencing of microbial genomes has given us clues to how Suc metabolism might have evolved in the proteobacteria and cyanobacteria, and was then acquired by eukaryotes during the endosymbiosis of the cyanobacterial ancestor of chloroplasts. Undoubtedly, future genome sequencing efforts will reveal more about the subsequent evolution of Suc metabolism in eukaryotic species.
CONCLUSIONS
While it is clear that experimental evidence will be required to establish the true nature of the putative SPS, SPP, and SuSy genes described above, the following hypothesis for the origin and evolution of Suc synthesis is proposed. Suc synthesis probably began in a proteobacteria-like ancestor of the cyanobacteria. Mutation of some other glucosyltransferase could have given rise to either SuSy, SPS, or an enzyme that could use both Fru and Fru-6-P as substrate to produce Suc or Suc-6-P. Any Suc-6-P produced could have been hydrolyzed by a nonspecific HAD-type phosphatase, which eventually evolved into a more specific SPP. The equilibrium constant of the SuSy reaction is unfavorable for accumulation of high concentrations of Suc, so any advantage conferred by this would favor the evolution of SPS and SPP, which catalyze the irreversible synthesis of Suc (Lunn and ap Rees, 1990). Separate SPS and SPP genes were inherited and retained in some types of cyanobacteria and possibly some proteobacteria, but in others the genes became fused to form bifunctional enzymes. Subsequent duplication of the region coding for the active SPP domain and loss of SPP function by the SPS led to the separation of enzyme activities, but with an SPS that has a noncatalytic SPP-like domain. Which of the three options the endosymbiotic, cyanobacterial ancestor of chloroplasts conferred on its eukaryotic descendants is unclear, as it is possible that the same processes could have occurred during the evolution of higher plants. Characterization of SPS from green algae and lower vascular plants might resolve this question. The function of the SPP-like domain is unknown, but the presence of type 3 SPS and SPP in higher plants, in which Suc metabolism is so important, suggests that this arrangement has some advantage.
MATERIALS AND METHODS
Materials
Biochemical reagents were obtained from Roche Molecular Biochemicals (Castle Hill, NSW, Australia) and Sigma-Aldrich (Castle Hill, NSW, Australia). Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs, Inc. (Beverly, MA).
Cloning of the Synechocystis spp Gene
Standard cloning procedures were carried out as in Sambrook et al. (1989). Genomic DNA was isolated from Synechocystis sp. PCC 6803 as in Lunn et al. (1999). The slr0953 (spp) ORF was amplified from genomic DNA by PCR using forward (5′-GCATTGATCAATCATATGCGACAG-3′) and reverse (5′-GCTTTGCTTGCGAATTCGGAATTG-3′) primers designed from the available sequence (GenBank accession no. D90914). The 784-bp PCR product was ligated into the T-tailed plasmid pGEM-T Easy (Promega, Madison, WI) and sequenced on both strands by the dideoxy chain termination method (Applied Biosystems, Foster City, CA). The spp coding region was excised by incubation with NdeI and EcoRI and ligated between the NdeI and EcoRI sites of the expression vector pTYB2 (New England Biolabs). The recombinant plasmid pTYB2/SynsppA was transformed into Escherichia coli (E. coli) strain ER2566.
Purification of Synechocystis SPP Expressed in E. coli and Raising of Antiserum
A stationary phase culture of E. coli ER2566 (pTYB2/Syn spp) grown in Luria-Bertani medium was diluted 100-fold into 1 L of Luria-Bertani medium containing 100 μg ampicillin mL−1 divided equally between two 2-L flasks and incubated with shaking (200 rpm) at 37°C until the cell density reached an optical density (600 nm) of 0.5. Protein expression was induced by the addition of IPTG to a final concentration of 0.3 mm. After incubation at 37°C for 12 h, the cells were harvested by centrifugation at 5,000g for 15 min (4°C).
The pelleted cells were resuspended in 100 mL of ice-cold buffer A (25 mm HepesK+, 5 mm MgCl2, 0.5 mm EDTA, pH 7.2) containing 1 mm phenylmethylsulfonyl fluoride, and lysed by sonication for 30 s in 10-s bursts, with 30 s of cooling on ice between bursts. The crude lysate was centrifuged at 20,000g for 10 min (4°C). The supernatant was decanted, and finely powdered PEG was added slowly with constant stirring to give a final concentration of 26% (w/v; 29 g per 100 mL). After stirring for 20 min at 0°C, the precipitated protein was pelleted (20,000g, 10 min) and discarded. The clear supernatant (110 mL) was warmed to 18°C and the pH quickly adjusted to 5.05 with 1 m acetic acid-Na+, pH 4.8. The mixture was rapidly cooled to 0°C and stood for 10 min. The precipitated protein was pelleted (20,000g, 10 min) and suspended in 14 mL of buffer A and the pH adjusted to 7.2 by addition of 1 m KOH.
A rabbit was inoculated with 200 μg of the purified protein injected with Freund's complete adjuvant, and with a further three injections of 200 μg of protein in Freund's incomplete adjuvant at 3-week intervals.
Expression of a Chimeric Synechocystis SPS-SPP in E. coli
The Synechocystis sps gene was amplified by PCR from pBluescriptII/Synsps (Lunn et al., 1999) using forward (5′-CATATGAGCTATTCATCAAAATAC-3′) and reverse (5′-GTTAACGGGGTCTAACAACTC-3′) primers designed to introduce NdeI and HpaI sites at the 5′ and 3′ ends of the coding region, respectively. The 2.16-kb PCR product was ligated into the T-tailed plasmid pCR2.1 (Invitrogen Corporation, Carlsbad, CA) and sequenced as described above. The Synechocystis sps coding region was excised from pCR2.1/Synsps by incubation with NdeI and HpaI and ligated between the NdeI and SmaI sites of pTYB2.
The Synechocystis spp gene was amplified by PCR from pGEM-T Easy/SynsppA using forward (5′-TACTAGTCGACAGTTATTGCTAATTTCTG-3′) and reverse (5′-GAAAGCTTTGCTTGCGAATTCG-3′) primers designed to introduce SpeI and HindIII sites at the 5′ and 3′ ends of the coding region, respectively. The 780-bp PCR product was ligated into the T-tailed plasmid pGEM-T Easy and sequenced as described above. The Synechocystis spp coding region was excised from pGEM-T/SynsppB by incubation with SpeI and HindIII and ligated between the SpeI and HindIII sites of pTYB2/Synsps. The recombinant plasmid pTYB2/Synsps-spp was introduced into E. coli ER2566 and expressed as described above.
Gel Electrophoresis and Western Blotting
Proteins were separated by SDS-PAGE on 12% (w/v) polyacrylamide gels as described in Laemmli (1970) and either stained with Coomassie Blue R250 or transferred to a nitrocellulose membrane and probed with either anti-Synechocystis SPP or anti-Synechocystis SPS antisera (1:10,000 dilution in blocking buffer) as described in Lunn et al. (1999).
Assay of SPP and SPS Activity
SPP activity was measured as in Lunn et al. (2000). SPS activity was measured as the Fru-6-P-dependent production of UDP from UDP-Glc as described in Lunn and Hatch (1997).
Determination of Protein
Protein was measured by the dye-binding method (Bradford, 1976) with bovine γ-globulin as the standard.
Microbial Genome Databases
Completed microbial genomes in the GenBank database and contigs from unfinished genome sequences in the U.S. Department of Energy (DOE) JGI, TIGR, and the Kazusa DNA Research Institute (Cyanobase; Chiba, Japan) databases were searched for ORFs with homology to cyanobacterial SPS, SPP (Synechocystis), and SuSy (Anabaena variabilis) sequences, using the TBLASTN algorithm. The deduced amino acid sequences of hits with E values less than 1 × 10−5 were used to search the GenBank nonredundant database using the BLASTP algorithm. Preliminary sequence data were obtained from the DOE JGI (http://www. jgi.doe.gov/JGI_microbial/html), TIGR (http://www.tigr.org/), and from the Cyanobase database at the Kazusa DNA Research Institute (http://www.kazusa.or.jp/cyano/).
Supplementary Material
ACKNOWLEDGMENTS
I thank Hal Hatch (Commonwealth Scientific and Industrial Research Organization, Plant Industry) for excellent technical assistance, and Murray Badger and Dean Price (Research School of Biological Sciences, Australian National University, Canberra) for helpful discussions on the evolution and taxonomy of the cyanobacteria.
Footnotes
The online version of this article contains Web-only data. The supplemental material is available at www.plantphysiol.org.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010898.
LITERATURE CITED
- Aravind L, Galperin MY, Koonin EV. The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biochem Sci. 1998;23:127–129. doi: 10.1016/s0968-0004(98)01189-x. [DOI] [PubMed] [Google Scholar]
- Billi D, Wright DJ, Helm RF, Prickett T, Potts M, Crowe JH. Engineering desiccation tolerance in Escherichia coli. Appl Environ Microbiol. 2000;66:1680–1684. doi: 10.1128/aem.66.4.1680-1684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of dye-binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. Universal trees based on large combined protein sequence data sets. Nat Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- Collet J-F, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem. 1998;23:14107–14112. doi: 10.1074/jbc.273.23.14107. [DOI] [PubMed] [Google Scholar]
- Curatti L, Folco E, Desplats P, Abratti G, Limones V, Herrera-Estrella L, Salerno G. Sucrose-phosphate synthase from Synechocystis sp. PCC 6803: identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J Bacteriol. 1998;180:6776–6779. doi: 10.1128/jb.180.24.6776-6779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatti L, Porchia AC, Herrera-Estrella L, Salerno GL. A prokaryotic sucrose synthase gene (susA) isolated from a filamentous nitrogen-fixing cyanobacterium encodes a protein similar to those of plants. Planta. 2000;211:729–735. doi: 10.1007/s004250000343. [DOI] [PubMed] [Google Scholar]
- Dancer JE, ap Rees T. Relationship between pyrophosphate:fructose-6-phosphate 1-phosphotransferase, sucrose breakdown, and respiration. J Plant Physiol. 1989;135:197–206. [Google Scholar]
- Duran WR, Pontis HG. Sucrose metabolism in green algae I. The presence of sucrose synthetase and sucrose phosphate synthetase. Mol Cell Biochem. 1977;16:149–152. doi: 10.1007/BF01732056. [DOI] [PubMed] [Google Scholar]
- Golden SS, Ishiura M, Johnson CH, Kondo T. Cyanobacterial circadian rhythms. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- Hagemann M, Marin K. Salt-induced sucrose accumulation is mediated by sucrose-phosphate-synthase in cyanobacteria. J Plant Physiol. 1999;155:424–430. [Google Scholar]
- Hawker JS, Smith GM. Occurrence of sucrose phosphatase in vascular and non-vascular plants. Phytochemistry. 1984;23:245–249. [Google Scholar]
- Huber SC, Huber JL. Role and regulation of sucrose-phosphate synthase in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:431–444. doi: 10.1146/annurev.arplant.47.1.431. [DOI] [PubMed] [Google Scholar]
- Kandler O, Hopf H. Occurrence, metabolism, and function of oligosaccharides. In: Preiss J, editor. The Biochemistry of Plants. Vol. 3. New York: Academic Press, Inc.; 1980. pp. 221–270. [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystissp. strain PCC6803. II. Sequence determination of the entire genome and the assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Khmelenina VN, Kalyuzhnaya MG, Sakharovsky VG, Suzina NE, Trotsenko YA, Gottschalk G. Osmoadaptation in halophilic and alkaliphilic methanotrophs. Arch Microbiol. 1999;172:321–329. doi: 10.1007/s002030050786. [DOI] [PubMed] [Google Scholar]
- Khmelenina VN, Sakharovskii VG, Reshetnikov AS, Trotsenko YA. Synthesis of osmoprotectants by halophilic and alkiliphilic methanotrophs. Microbiology. 2000;69:381–386. [Google Scholar]
- Müller W, Wegmann K. Sucrose biosynthesis in DunaliellaII. Isolation and properties of sucrose phosphate synthetase. Planta. 1978;141:159–163. doi: 10.1007/BF00387883. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lunn JE, ap Rees T. Apparent equilibrium constant and mass-action ratio for sucrose-phosphate synthase in seeds of Pisum sativum. Biochem J. 1990;267:739–743. doi: 10.1042/bj2670739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Ashton AR, Hatch MD, Heldt HW. Purification, molecular cloning, and sequence analysis of sucrose-6F-phosphate phosphohydrolase from plants. Proc Natl Acad Sci USA. 2000;97:12914–12919. doi: 10.1073/pnas.230430197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Hatch MD. The role of sucrose-phosphate synthase in the control of photosynthate partitioning in Zea maysleaves. Aust J Plant Physiol. 1997;24:1–8. [Google Scholar]
- Lunn JE, Price GD, Furbank RT. Cloning and expression of a prokaryotic sucrose-phosphate synthase gene from the cyanobacterium Synechocystissp. PCC6803. Plant Mol Biol. 1999;40:297–305. doi: 10.1023/a:1006130802706. [DOI] [PubMed] [Google Scholar]
- Mijts BN, Patel BKC. Random sequence analysis of genomic DNA of an anaerobic, thermophilic, halophilic bacterium, Halothermothrix orenii. Extremophiles. 2001;5:61–69. doi: 10.1007/s007920000174. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page-Sharp M, Behm CA, Smith GD. Involvement of the compatible solutes trehalose and sucrose in the response to salt stress of a cyanobacterial Scytonemaspecies isolated from desert soils. Biochim Biophys Acta. 1999;1472:519–528. doi: 10.1016/s0304-4165(99)00155-5. [DOI] [PubMed] [Google Scholar]
- Porchia AC, Curatti L, Salerno GL. Sucrose metabolism in cyanobacteria: Sucrose synthase from Anabaenasp. strain PCC 7119 is remarkably different from the plant enzymes with respect to substrate affinity and amino-terminal sequence. Planta. 1999a;210:34–40. doi: 10.1007/s004250050651. [DOI] [PubMed] [Google Scholar]
- Porchia AC, Fiol DF, Salerno GL. Differential synthesis of sucrose and trehalose in Euglena graciliscells during growth and salt stress. Plant Sci. 1999b;149:43–49. [Google Scholar]
- Porchia AC, Salerno GL. Sucrose biosynthesis in a prokaryotic organism: presence of two sucrose-phosphate synthases in Anabaenawith remarkable differences compared with the plant enzymes. Proc Natl Acad Sci USA. 1996;93:13600–13604. doi: 10.1073/pnas.93.24.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RH, Borowitzka LJ, Mackay MA, Chudek JA, Foster R, Warr SRC, Moore DJ, Stewart WDP. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Rev. 1986;39:51–56. [Google Scholar]
- Reed RH, Richardson DL, Warr SRC, Stewart WDP. Carbohydrate accumulation and osmotic stress in cyanobacteria. J Gen Microbiol. 1984;130:1–4. [Google Scholar]
- Reed RH, Stewart WDP. Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar Biol. 1985;88:1–9. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schilling N, Ehrnsperger K. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z Naturforsch. 1985;40:776–779. [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothecesp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HK, Koo YJ, Lim JY, Song JT, Kim CH, Kim JK, Lee JS, Choi YD. Characterization of a bifunctional enzyme fusion of trehalose-6-phosphate synthetase and trehalose-6-phosphate phosphatase of Escherichia coli. App Env Microbiol. 2000;66:2484–2490. doi: 10.1128/aem.66.6.2484-2490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GJ, Jacques NA. Polysaccharides of oral streptococci. In: Reizer J, Peterkofsky A, editors. Sugar Transport and Metabolism in Gram-Positive Bacteria. Chichester, UK: Ellis Horwood Ltd.; 1987. pp. 39–68. [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Inoue K, Bauer CE. Tracking molecular evolution of photosynthesis by characterization of a major photosynthesis gene cluster from Heliobacillus mobilis. Proc Natl Acad Sci USA. 1998;95:14851–14856. doi: 10.1073/pnas.95.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.