Abstract
The secretory pathway of Pichia pastoris was genetically re-engineered to perform sequential glycosylation reactions that mimic early processing of N-glycans in humans and other higher mammals. After eliminating nonhuman glycosylation by deleting the initiating α-1,6-mannosyltransferase gene from P. pastoris, several combinatorial genetic libraries were constructed to localize active α-1,2-mannosidase and human β-1,2-N-acetylglucosaminyltransferase I (GnTI) in the secretory pathway. First, >32 N-terminal leader sequences of fungal type II membrane proteins were cloned to generate a leader library. Two additional libraries encoding catalytic domains of α-1,2-mannosidases and GnTI from mammals, insects, amphibians, worms, and fungi were cloned to generate catalytic domain libraries. In-frame fusions of the respective leader and catalytic domain libraries resulted in several hundred chimeric fusions of fungal targeting domains and catalytic domains. Although the majority of strains transformed with the mannosidase/leader library displayed only modest in vivo [i.e., low levels of mannose (Man)5-(GlcNAc)2] activity, we were able to isolate several yeast strains that produce almost homogenous N-glycans of the (Man)5-(GlcNAc)2 type. Transformation of these strains with a UDP-GlcNAc transporter and screening of a GnTI leader fusion library allowed for the isolation of strains that produce GlcNAc-(Man)5-(GlcNAc)2 in high yield. Recombinant expression of a human reporter protein in these engineered strains led to the formation of a glycoprotein with GlcNAc-(Man)5-(GlcNAc)2 as the primary N-glycan. Here we report a yeast able to synthesize hybrid glycans in high yield and open the door for engineering yeast to perform complex human-like glycosylation.
The number of protein-based therapeutics entering preclinical and clinical evaluation has shown robust growth and is expected to increase in the years to come. Fueled by advances in proteomics and genomics as well as the ability to engineer and humanize monoclonal antibodies, protein-based therapeutics constitute ≈500 candidates currently in clinical trials (1). Several therapeutic proteins can be made in a prokaryotic expression system such as Escherichia coli (e.g., insulin); however, the majority of therapeutic proteins require additional posttranslational modifications to attain full biological function. N-glycosylation in particular is essential for proper folding, pharmacokinetic stability, and efficacy for a large number of proteins (2). Most therapeutically relevant glycoproteins, including antibodies, are therefore expressed in mammalian cells. However, volumetric productivity, product heterogeneity, media cost, retroviral contamination, and the time required to generate stable cell lines are generally viewed as drawbacks of mammalian cell culture.
Fungal protein-expression systems do not suffer from the same limitations, and protein titers of 14.8 and 35 g/liter have been reported for secreted heterologous proteins in yeast and the filamentous fungus Trichoderma reesei, respectively (3, 4). However, glycoproteins derived from fungal expression systems contain nonhuman N-glycans of the high mannose (Man) type, which are immunogenic in humans and thus of limited therapeutic value (5).
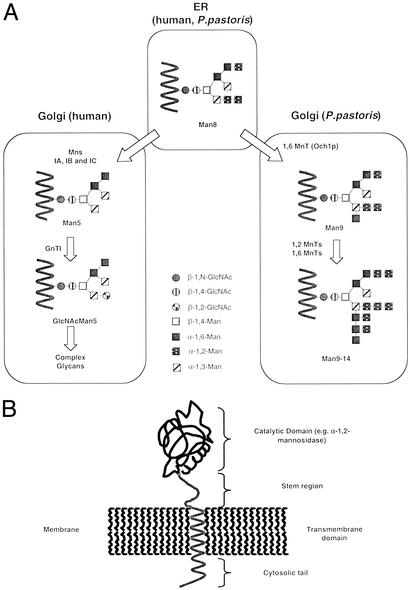
Fungi and mammals share initial steps of protein N-glycosylation, which involves the site-specific transfer of (Glc)3-(Man)9-(GlcNAc)2 from the luminal side of the endoplasmic reticulum (ER) to the de novo synthesized protein by an oligosaccharyltransferase complex. Subsequent trimming by glucosidases I and II and a specific ER-residing α-1,2-mannosidase leads to the formation of a (Man)8-(GlcNAc)2 structures (isomer Man8B) (Fig. 1), the N-glycan found on most glycoproteins leaving the ER.
Figure 1.
(A) N-linked glycosylation pathway in humans and P. pastoris. Mns, α-1,2-mannosidase; MnT, mannosyltransferase; Och1p, the initiating 1,6-mannosyltransferase. (B) Structure of typical type II membrane glycosidase or glycosyltransferase.
After the export of predominantly (Man)8-(GlcNAc)2 containing glycoproteins to the Golgi, the pathways diverge notably between mammals and yeast (6). In the human Golgi α-1,2-mannosidases (IA–IC) remove Man to yield the (Man)5-(GlcNAc)2 structure, which forms the precursor for complex N-glycans (Fig. 1A). These mannosidases are typically type II membrane proteins with an N-terminal cytosolic tail, a transmembrane domain, a stem region, and a C-terminal catalytic domain (Fig. 1B). Localization of these proteins, as with most enzymes involved in Golgi glycosylation, is mediated by the cytosolic tail, the transmembrane region, and the stem (7).
In Saccharomyces cerevisiae, N-glycosylation has been studied extensively and, unlike mammalian N-glycan processing, involves the addition of numerous Man sugars throughout the entire Golgi, often leading to hypermannosylated N-glycan structures with >100 Man residues. This process is initiated in the early Golgi by an α-1,6-mannosyltransferase (Och1p) that prefers (Man)8-(GlcNAc)2 as a substrate but is able to recognize various other Man oligomers with the notable exception of the human (Man)5-(GlcNAc)2 intermediate, which is not a substrate (8). After addition of this first α-1,6-Man by Och1p, additional α-1,6-mannosyltransferases will extend the α-1,6 chain, which then becomes the substrate for medial- and trans-Golgi-residing α-1,2- and α-1,3-mannosyltransferases as well as phosphomannosyltransferases that add yet more Man sugars to the growing N-glycan structure (9). In Pichia pastoris a very similar process occurs; however, hypermannosylation occurs less frequently and to a lower extent. In addition α-1,3-mannosyltransferase activity has not been found in this yeast, and N-glycans from P. pastoris do not have α-1,3-Man attached to the outer Man chain (10).
Humanizing the glycosylation machinery of a yeast strain will require the (i) elimination of some endogenous glycosylation reactions and (ii) the recreation of the sequential nature of human glycosylation in the ER and Golgi. Although the first step involves the generation of gene knockouts (e.g., α-1,6- and/or α-1,3-mannosyltransferases), the second step requires the proper localization of active mannosidases, glycosyltransferases, and possibly nucleotide sugar transporters to specific organelles. Moreover the formation of certain sugar–nucleotide precursor pools such as CMP-sialic acid may have to be engineered into the yeast host. Much is known about the localization of endogenous proteins in the secretory pathway of S. cerevisiae and other yeasts; however, there is no reliable method to predict whether a Golgi protein from one organism will be properly localized in the Golgi of another organism. It is also unknown whether artificial in-frame protein fusions consisting of a yeast localization sequence and a catalytic domain from a nonyeast source (e.g., glycosyltransferases or mannosidases) will (i) localize to the desired organelle and (ii) show sufficiently high activity in the targeted environment. Although proper localization is important, it is not sufficient. Because the activity of the catalytic domain has to be maintained in the environment in which it has been localized, additional aspects such as pH optima must be considered. For example, Chiba et al. (11) found that localizing a mannosidase with a pH optimum of 5 in the ER of S. cerevisiae results in only modest intracellular mannosidase activity.
To overcome these major constraints we developed two tools: a combinatorial genetic library, generating hundreds of fusion constructs at a time, and a high-throughput screen that allows us to analyze large numbers of strains in parallel for their ability to modify N-glycans of recombinant reporter proteins. The combinatorial genetic library consists of an array of different fusion-protein constructs, each of which contains a fungal cellular targeting sequence fused in frame to a catalytic domain (e.g., mannosidase). Each of the 608 generated mannosidase fusion constructs was tested individually for its ability to catalyze the trimming of higher Man structures to (Man)5-(GlcNAc)2. Those strains that were able to generate mostly (Man)5-(GlcNAc)2 on a secreted reporter protein were subjected first to a second screen to ensure that trimming occurred in vivo and then engineered further to generate GlcNAc-(Man)5-(GlcNAc)2 by screening a similar β-1,2-N-acetylglucosaminyltransferase I (GnTI)/leader library. A more comprehensive article describing the characteristics of >600 leader/mannosidase and leader/GnTI fusions is in preparation (B.-K.C., P.B., R.C.D., S.R.H., A. Stadheim, H.L., R.G.M., J.H.N., S.W., and T.U.G., unpublished data); however, some of the most important findings are reported in this article.
Here we report the re-engineering of the secretory pathway in the methylotrophic yeast P. pastoris. The engineered strain produces predominantly N-glycans that are intermediates of the human glycosylation pathway, essentially void of fungal features. Our results suggest that further implementation of the described combinatorial library approach will allow for the engineering of yeast strains with increasingly human N-glycosylation. This article reports a genetically engineered yeast capable of producing a glycoprotein with a human-like hybrid N-glycosylation structure.
Materials and Methods
Strains, Culture Conditions, and Reagents.
E. coli strains TOP10 or DH5α were used for recombinant DNA work. P. pastoris GS115 (his4, Invitrogen) or JC308 (ura3, ade1, arg4, his4, a gift from James M. Cregg, Keck Graduate Institute, Claremont, CA) were used for generation of yeast strains. Protein expression was carried out at room temperature in a 96-well-plate format with buffered glycerol-complex medium (BMGY) consisting of 1% yeast extract, 2% peptone, 100 mM potassium phosphate buffer (pH 6.0), 1.34% yeast nitrogen base, 4 × 10−5% biotin, and 1% glycerol as a growth medium. The induction medium was buffered methanol-complex medium (BMMY) consisting of 1.5% methanol instead of glycerol in BMGY. Minimal medium is 1.4% yeast nitrogen base, 2% dextrose, 1.5% agar, and 4 × 10−5% biotin and amino acids supplemented as appropriate. Restriction and modification enzymes were from New England BioLabs. Oligonucleotides were obtained from the Dartmouth College Core facility (Hanover, NH) or Integrated DNA Technologies (Coralville, IA). The enzymes, peptide N-glycosidase F, mannosidases, and oligosaccharides were obtained from Glyko (San Rafael, CA). Metal chelating HisBind resin was from Novagen. Lysate-clearing plates (96-well) were from Promega. Protein-binding 96-well plates were from Millipore. Salts and buffering agents were from Sigma. Matrix-assisted laser desorption ionization (MALDI) matrices were from Aldrich.
Cloning and Deletion of the P. pastoris OCH1 Gene.
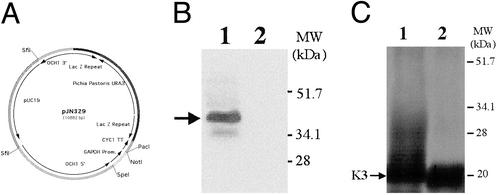
The 1,215-bp ORF of the P. pastoris OCH1 gene encoding a putative α-1,6-mannosyltransferase was amplified from P. pastoris genomic DNA (strain X-33, Invitrogen) by using the oligonucleotides 5′-ATGGCGAAGGCAGATGGCAGT-3′ and 5′-TTAGTCCTTCCAACTTCCTTC-3′, which were designed based on the P. pastoris OCHI sequence (12). Subsequently, 2,685 bp upstream and 1,175 bp downstream of the ORF of the OCHI gene were amplified from a P. pastoris genomic DNA library (gift from Judah Folkman, Harvard Medical School, Boston) by using the internal oligonucleotides 5′-ATGGCGAAGGCAGATGGCAGT-3′ and 5′-ACTGCCATCTGCCTTCGCCAT-3′ in the OCHI gene with T3 (5′-AATTAACCCTCACTAAAGGG-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′) in the backbone of the library bearing plasmid λ ZAP II (Stratagene). The resulting 5,075-bp fragment was cloned into the pCR2.1-TOPO vector (Invitrogen) and designated pBK9. To create an och1 knockout strain containing multiple auxotrophic markers, 100 μg of pJN329, a plasmid containing an och1∷URA3 mutant allele (J.F.N., S.W., and T.U.G., unpublished data; see Fig. 2A for details) was digested with SfiI and used to transform P. pastoris strain JC308 by electroporation. After incubation on defined medium lacking uracil for 10 days at room temperature, 1,000 colonies were picked and restreaked. URA+ clones that were unable to grow at 37°C but grew at room temperature were subjected to colony PCR to test for the correct integration of the och1∷URA3 mutant allele. One clone that exhibited the expected PCR pattern was designated YJN153.
Figure 2.
Knockout of OCHI in P. pastoris. (A) The OCHI knockout plasmid pJN329 contains the P. pastoris URA3 gene flanked by LacZ repeats and 2,878 bp upstream (5′) and 1,011 bp downstream (3′) of the OCHI gene of P. pastoris. (B) Immunoblot of Och1p in Pichia wild type (JC308, lane 1) and och1 mutant (BK64-1, lane 2). The same amount of cell-free extract (50 μg per lane) was used, and Och1p was detected by using an Och1p peptide antibody followed by ECL. (C) The reporter protein K3 was expressed in Pichia wild type BK64 (lane 1) and an och1 mutant, P. pastoris BK64-1 (lane 2). K3 was purified by Ni-affinity chromatography, separated by SDS/PAGE (4–20% gradient) under reducing conditions, and visualized by silver staining. The same amount of K3 (200 ng per lane) was loaded.
Reporter Protein-Expression Construct.
The Kringle 3 domain of human plasminogen (K3) was used as a model protein. A DNA fragment encoding the K3 (gift from Nick Menhart, Illinois Institute of Technology, Chicago) was amplified by using Pfu turbo polymerase (Stratagene) and cloned into EcoRI and XbaI sites of pPICZαA (Invitrogen), resulting in a C-terminal 6-His tag. To improve the N-linked glycosylation efficiency of K3 (13), Pro-46 was replaced with Ser-46 by using site-directed mutagenesis. The resulting plasmid was designated pBK64. The correct sequence of the PCR construct was confirmed by DNA sequencing.
Integration vectors and fusion constructs were generated based on the roll-in plasmids described by Lin Cereghino and coworkers (14); see details in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Generation of Yeast Strains.
To create P. pastoris strains expressing the K3, plasmid pBK64 was transformed into strains GS115 and YJN153, and colonies were selected on YPD medium containing 100 μg/ml zeocin to create strains BK64 and BK64-1, respectively. Plasmid pPB103 was linearized with EcoNI and transformed into strain BK64-1, and colonies were selected on defined medium lacking adenine. One strain containing the Kluyveromyces lactis MNN2-2 gene was designated PBP1. Plasmids pBB27 and pBC4 were linearized with SalI and transformed into strain PBP1, and colonies were selected on defined medium lacking histidine. Strains that were confirmed to contain the Caenorhabditis elegans α-1,2-mannosidase IB BB27 and BC4 fusion constructs were designated YJN188 and YJN168, respectively. Plasmid pNA15 was linearized with AatII and transformed into strain YJN168, and colonies were selected on minimal medium without amino acids. One strain that was confirmed to contain the human GnTI gene fusion was designated YJN201.
Western Blotting.
Proteins were separated by 4–20% gradient SDS/PAGE according to Laemmli (15) and then electroblotted onto nitrocellulose membrane (Schleicher & Schuell) as described (16). P. pastoris Och1p was detected by using an antibody raised against the peptide CQQLSSPKIDYDPLTL (Sigma–Genosys) with an ECL kit (Amersham Pharmacia).
Protein Purification.
K3 was purified from the medium by Ni-affinity chromatography by using a 96-well format on a Beckman BioMek 2000 laboratory robot. The robotic purification is an adaptation of the protocol provided by Novagen for their HisBind resin.
Release of N-Linked Glycans.
The glycans were released and separated from the glycoproteins by a modification of a previously reported method (17). After the proteins were reduced and carboxymethylated and the membranes were blocked, the wells were washed three time with water. The protein was deglycosylated by the addition of 30 μl of 10 mM NH4HCO3 (pH 8.3) containing 1 milliunit of N-glycanase (Glyko). After 16 h at 37°C, the solution containing the glycans was removed by centrifugation and evaporated to dryness.
MALDI/Time-of-Flight (TOF) Mass Spectrometry.
Molecular weights of the glycans were determined by using a Voyager DE PRO linear MALDI/TOF (Applied Biosciences) mass spectrometer with delayed extraction. The dried glycans from each well were dissolved in 15 μl of water, and 0.5 μl was spotted on stainless-steel sample plates and mixed with 0.5 μl of S-DHB matrix (9 mg/ml of dihydroxybenzoic acid/1 mg/ml of 5-methoxysalicylic acid in 1:1 water/acetonitrile/0.1% trifluoroacetic acid) and allowed to dry. Ions were generated by irradiation with a pulsed nitrogen laser (337 nm) with a 4-ns pulse time. The instrument was operated in the delayed extraction mode with a 125-ns delay and an accelerating voltage of 20 kV. The grid voltage was 93.00%, guide wire voltage was 0.1%, the internal pressure was <5 × 10−7 torr (1 torr = 133 Pa), and the low mass gate was 875 Da. Spectra were generated from the sum of 100–200 laser pulses and acquired with a 500-MHz digitizer. (Man)5-(GlcNAc)2 oligosaccharide was used as an external molecular weight standard. All spectra were generated with the instrument in the positive-ion mode.
Mannosidase Assays.
Fluorescence-labeled (Man)8-(GlcNAc)2 (0.5 μg) was added to 20 μl of supernatant and incubated for 30 h at room temperature. After incubation the sample was analyzed by HPLC with an Econosil NH2 4.6 × 250-mm, 5-μm bead, amino-bound silica column (Alltech, Avondale, PA). The flow rate was 1.0 ml/min for 40 min, and the column was maintained at 30°C. After eluting isocratically (68% A:32% B) for 3 min, a linear solvent gradient (68% A:32% B to 40% A:60% B) was used over 27 min to elute the glycans (18). Solvent A was acetonitrile, and solvent B was an aqueous solution of ammonium formate, 50 mM (pH 4.5). The column was equilibrated with solvent (68% A:32% B) for 20 min between runs.
Results
Generation of an α-1,6-Mannosyltransferase Deletion Mutant in P. pastoris.
In S. cerevisiae the OCH1 gene product (Och1p) is an α-1,6-mannosyltransferase that initiates the outer-chain elongation of N-linked glycans in the early Golgi. An och1 null mutant strain in S. cerevisiae shows no α-1,6-Man linkage to the core glycan structure and consequently lacks hyperglycosylation (19). To generate a deletion of the OCH1 homolog in P. pastoris an och1∷URA3 mutant allele was constructed and transformed into a ura3 strain of P. pastoris (JC308). Several och1 mutant strains were identified and confirmed by PCR and Western blotting (Fig. 2B). The P. pastoris och1 mutant strain exhibited temperature sensitivity and increased flocculation similar to that observed in S. cerevisiae (data not shown).
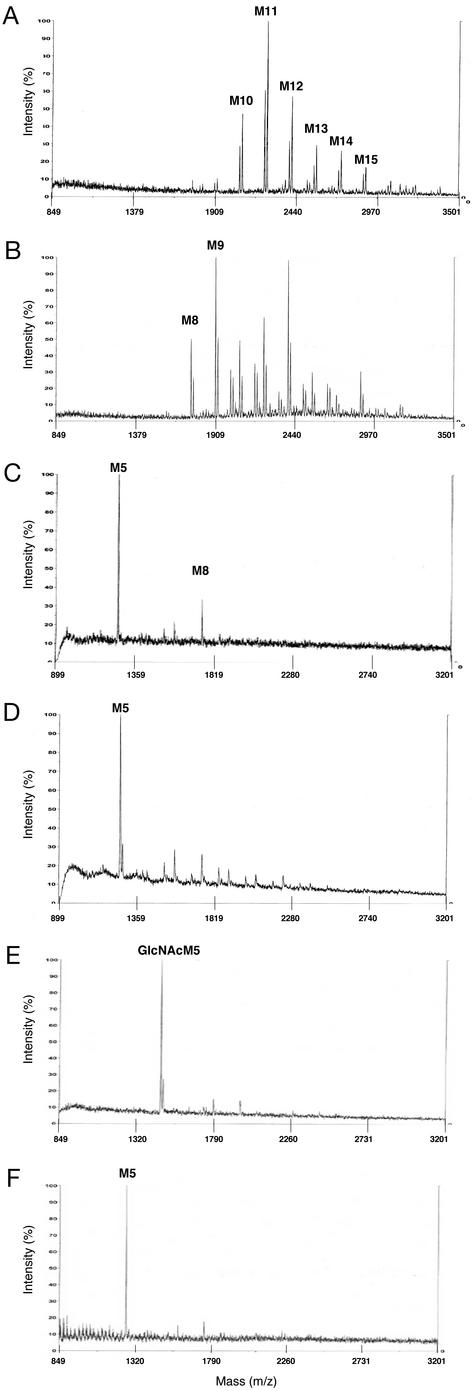
Glycan structures of heterologous proteins expressed in P. pastoris are heterogeneous, mostly consisting of (Man)10–16-(GlcNAc)2 mannans with varying degrees of charged glycans (10, 20). To monitor the effect of different engineering steps on the glycosylation of P. pastoris, a secreted form of K3 was used as a reporter protein. Hyperglycosylation observed by SDS/PAGE analysis when K3 was expressed in a wild-type P. pastoris strain was eliminated in the och1 mutant strain (Fig. 2C). N-glycans released from secreted K3 were analyzed directly by MALDI/TOF mass spectrometry. N-glycans of K3 expressed in a P. pastoris wild-type strain were found to have a molecular mass consistent with (Man)10–16-(GlcNAc)2, confirming previous findings in this yeast (Fig. 3A). By comparison, the N-glycans of K3 in an och1 mutant strain revealed predominantly (Man)8–12- (GlcNAc)2, representing a noticeable shift to smaller glycans (Fig. 3B) and a complete elimination of the smearing observed in the SDS/PAGE analysis (Fig. 2C). To rule out the possibility of protein-specific interactions a second reporter protein, full-length human IFN-β, was used and revealed identical results (data not shown).
Figure 3.
Positive-ion MALDI/TOF mass spectra of N-linked glycans released from K3. K3 was produced in P. pastoris strains BK64, BK64-1, YJN168, YJN188, and YJN201 and purified from culture supernatants by Ni-affinity chromatography. The glycans were released from K3 by peptide N-glycosidase F treatment. The released N-linked glycans were analyzed by MALDI/TOF mass spectrometry, typically appearing as the sodium or potassium adducts. (A) BK61, wild-type strain of P. pastoris expressing K3. (B) BK64-1, och1 deletion expressing K3. (C) YJN168, och1 deletion expressing K3, K. lactis UDP-GlcNAc transporter, and C. elegans α-1,2-mannosidase IB fused to MNS1. (D) YJN188, och1 deletion expressing K3, K. lactis UDP-GlcNAc transporter, and C. elegans α-1,2-mannosidase IB fused to MNN10. (E) YJN201, och1 deletion expressing K3, K. lactis UDP-GlcNAc transporter, C. elegans α-1,2-mannosidase IB fused to MNS1, and human GnTI fused to MNN9. (F) YJN201 after β-N-acetylhexosaminidase treatment. M, Man.
Construction of ER/Golgi leader, α-1,2-Mannosidase, and GnTI Libraries.
To sequentially localize different mannosidases and GnTIs along the early secretory pathway of P. pastoris three separate gene libraries were designed. The first library (the leader library) contained DNA fragments encoding N-terminal peptides of known type II membrane proteins that either localize in the ER or Golgi of S. cerevisiae and P. pastoris. They include Gls1, Mns1, Sec12, Mnn9, Van1, Anp1, Hoc1, and Mnn10, Mnn11 from S. cerevisiae and Och1 and Sec12 from P. pastoris. The generation of the respective DNA constructs is exemplified by the construction of Mns1, Mnn9, and Mnn10 leaders in Materials and Methods. A second library contained catalytic domains of α-1,2-mannosidases from Homo sapiens, Mus musculus, Aspergillus nidulans, C. elegans, Drosophila melanogaster, and Penicillium citrinium. The generation of the respective DNA constructs is exemplified by the construction of mannosidase IB from C. elegans in Materials and Methods. Finally, a third library contained catalytic domains of GnTI genes from H. sapiens, C. elegans, Xenopus laevis, and D. melanogaster. All libraries were designed in a way that any combination of a leader construct and a catalytic domain created a gene encoding a chimeric fusion protein.
Each leader fragment was represented in three lengths. The short form encoded the N-terminal cytoplasmic tail and the transmembrane domain. The long form encoded additional residues containing the complete stem region up to the respective catalytic domain, which was determined by sequence homology to known catalytically active fragments of such enzymes. The medium form was an intermediate version containing approximately half of the stem region in addition to the sequence encoded in the short form. Catalytic domains were selected to cover a wide range of pH optima as determined from literature data (e.g., P. citrinium and M. musculus) and temperature optima by selecting domains from organisms that exist at different temperatures (e.g., C. elegans and H. sapiens). Each catalytic domain was represented in several lengths and generally lacked the native N-terminal cytosolic and transmembrane domain. Some of the catalytic domains were selected solely on the basis of sequence homology to other known α-1,2-mannosidases (e.g. C. elegans) or GnTIs and had not been characterized previously.
After screening the fusion libraries chimeric constructs were identified that displayed a high degree of Man-trimming activity and UDP-GlcNAc transfer activity on the reporter protein K3. A detailed analysis of the characteristics of the fusion libraries will be published elsewhere (B.-K.C., P.B., R.C.D., S.R.H., A. Stadheim, H.L., R.G.M., J.H.N., S.W., and T.U.G., unpublished data).
Expression of α-1,2-Mannosidase Fusion Constructs in a P. pastoris och1 Mutant Strain.
After screening a library of 608 leader/α-1,2-mannosidase fusions targeted to the ER and early Golgi, several clones were identified that produced N-glycans consistent with a mass of (Man)5-(GlcNAc)2. Specifically, a putative C. elegans homolog of known α-1,2-mannosidases showed a high degree of trimming to (Man)5-(GlcNAc)2 when fused to both the S. cerevisiae MNS1 and MNN10 leader-encoding fragments. Although these constructs and several others resulted in glycans, which were 70–80% or more (Man)5-(GlcNAc)2 (Fig. 3 C and D), >56% of the fusions resulted in <10% (Man)5-(GlcNAc)2 (Table 1). These data clearly emphasize the importance of choosing the proper combination of (i) a localization sequence and (ii) an α-1,2-mannosidase catalytic domain of the proper length. Fungal α-1,2-mannosidases with acidic pH optima (e.g., P. citrinium and A. nidulans), when expressed as fusions with the leader library, generally resulted in low (Man)5-(GlcNAc)2 yields (data not shown) consistent with previous findings (11, 21).
Table 1.
Relative amount of (Man)5 on secreted K3
| Amount of (Man)5 on secreted K3, % of total glycans | Number of constructs (%) |
|---|---|
| ND* | 19 (3.1) |
| 0–10 | 341 (56.1) |
| 10–20 | 50 (8.2) |
| 20–40 | 75 (12.3) |
| 40–60 | 72 (11.8) |
| >60 | 51 (8.4)† |
| Total | 608 (100) |
Six hundred and eight different strains of P. pastoris (och1) were generated by transforming them with a single construct of a combinatorial genetic library that was generated by fusing 19 α-1,2-mannosidase catalytic domains to 32 fungal ER and cis-Golgi leaders.
Several fusion constructs were not tested because the corresponding plasmids could not be propagated in E. coli before transformation into P. pastoris.
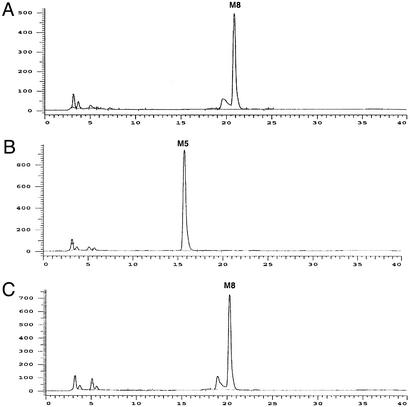
Clones with the highest degree of (Man)5 trimming (30/51) were analyzed further for mannosidase activity in the supernatant of the medium. The majority (28/30) displayed detectable mannosidase activity in the supernatant (e.g. Fig. 4B). Only two constructs displayed high (Man)5 levels while lacking mannosidase activity in the medium (e.g. Fig. 4C).
Because previous researchers have found that trimming to (Man)5-(GlcNAc)2 was often accompanied by leakage of mannosidase into the medium (22), we further investigated whether Man trimming occurred in vivo, in the Golgi, or ex vivo after secretion of the protein into the medium. To determine the extent of mannosidase activity in the medium, 2-aminobenzamide-labeled (Man)8-(GlcNAc)2 was used to assay the culture supernatant. Many of the efficient (Man)5-(GlcNAc)2-producing constructs displayed a high degree of mannosidase activity in the supernatant, suggesting that at least some of the observed (Man)5-(GlcNAc)2 structures were produced ex vivo. However, by applying a double screen we were able to identify specific chimeric fusions that were entirely retained intracellularly while at the same time displaying high in vivo α-1,2-mannosidase activity (Fig. 4C).
Figure 4.
Extracellular activity of C. elegans α-1,2-mannosidase IB. 2-Aminobenzamide-labeled (Man)8-(GlcNAc)2 was incubated with culture supernatants at 25°C for 30 h and analyzed by HPLC. (A) (Man)8-(GlcNAc)2 standard in BMMY medium (no cells). (B) Supernatant from YJN188. (C) Supernatant from YJN168. M, Man.
Expression of GnTI Fusion Constructs in a P. pastoris och1 Mutant Strain Containing an Active α-1,2-Mannosidase.
Further humanization of (Man)5-(GlcNAc)2 glycan structures involves the subsequent in vivo conversion of (Man)5-(GlcNAc)2 to GlcNAc-(Man)5-(GlcNAc)2. This step requires UDP-GlcNAc as a substrate and involves the transfer of GlcNAc to (Man)5-(GlcNAc)2 by GnTI (Fig. 1). To ensure sufficient levels of UDP-GlcNAc in the Golgi we cloned the UDP-GlcNAc transporter from K. lactis into an och1 mutant strain, which also displays high in vivo mannosidase activity.
A leader/GnTI fusion library containing 67 constructs was screened. Among several active fusion constructs, one consisting of human GnTI and a leader sequence from S. cerevisiae MNN9 was particularly active, and the corresponding strains yielded K3-containing glycans with a mass consistent with GlcNAc-(Man)5-(GlcNAc)2 almost exclusively (Fig. 3E). Moreover, these glycans were converted completely to (Man)5-(GlcNAc)2 by in vitro N-acetylhexosaminidase digestion, further indicating that this strain secretes protein almost uniformly modified with glycans of the structure GlcNAc-(Man)5-(GlcNAc)2, a well described human glycosylation intermediate (Fig. 3F). Importantly, GnTI activity was assayed in the culture medium of this strain and found to be absent (data not shown). Finally, to determine whether the UDP-GlcNAc transporter was required to provide sufficient amounts of substrate for GnTI, a strain producing (Man)5-(GlcNAc)2 was transformed with the same GnTI construct in the absence of the transporter. Although some GlcNAc-(Man)5-(GlcNAc)2 was observed from the purified K3, a large percentage of glycans consistent with (Man)5-(GlcNAc)2 remained, indicating that GlcNAc transfer was less efficient than in the strain containing the transporter (data not shown). This result confirms the purported necessity of a UDP-GlcNAc transporter for efficient GlcNAc transfer originally found in K. lactis (23).
Discussion
P. pastoris is one of several yeasts capable of high-level production of heterologous glycoproteins (24). As with any other fungal protein-expression systems, the heterologous glycoprotein is glycosylated in a fungal-like fashion generally involving the addition of α-1,6- and α-1,2-linked Man and mannosylphosphate to the (Man)8-(GlcNAc)2 core (Fig. 1; ref. 25). This yeast-type glycosylation pattern is recognized by the human immune system, which renders the underlying protein unfit for therapeutic use. In view of the current shortage of efficient glycoprotein-expression systems we have focused our efforts on engineering commercially relevant yeast and filamentous fungi to produce N-glycans with human-like glycosylation structures. In this study we report the construction of a strain that produces high levels of a reporter protein modified with a human glycosylation intermediate in the P. pastoris system.
The first step in attaining complex human-type glycans in a fungal system is to eliminate yeast-type glycosylation. To accomplish this, the P. pastoris OCH1 gene was deleted in a strain secreting the reporter protein K3. Yeast-type hyperglycosylation was abrogated in this strain as observed by silver staining of secreted K3 (Fig. 2C), and when released N-glycans were analyzed by MALDI/TOF, a general trend toward smaller structures was observed (Fig. 3B).
Once an appropriate core high-Man glycan is obtained, the next immediate step in the conversion to human-type N-glycans involves the functional expression and localization of an α-1,2-mannosidase. This enzyme will trim the (Man)8-(GlcNAc)2 core structure to (Man)5-(GlcNAc)2 and thereby generate the structure that is capable of receiving the GlcNAc that initiates the formation of hybrid N-glycans. A very similar approach was taken in a triple mutant och1 mnn1 mnn4 strain of S. cerevisiae (11). ER localization of a fungal α-1,2-mannosidase (from Aspergillus saitoi) was accomplished by adding the tetrapeptide HDEL as an ER retrieval tag to the C terminus of the gene. By using the S. cerevisiae GAPDH promoter and a multicopy number plasmid, mannosidase activity was detectable in cell-free extracts; however, only 27% of the N-glycans of an endogenous marker protein (carboxypeptidase Y) were trimmed from (Man)8-(GlcNAc)2 to (Man)5-(GlcNAc)2 in vivo. Although not entirely successful, the pioneering work of Chiba et al. (11) demonstrates that N-glycans from S. cerevisiae can be modified substantially by engineering glycosylation pathways. Here the localization of catalytic domains to the secretory pathway of the related yeast P. pastoris using yeast type II membrane protein leader domains is demonstrated. The benefit of a large library of leader domains along with an equally diverse library of catalytic domains allowed for the selection of the most active fusion constructs from a pool of >600 candidates, many of which were marginally active or not at all. Despite the lack of success of many constructs, a few particularly active enzyme fusions were able to trim the core (Man)8–12-(GlcNAc)2 glycans observed in the och1 mutant strain to (Man)5-(GlcNAc)2. Two of these constructs described here, which are fusions of a putative C. elegans α-1,2-mannosidase with different yeast type II leader domains, are able to do so at high efficiency (>75%).
The correct localization of enzymes involved in glycosylation is critical to allow for the sequential glycan modifications as glycoproteins proceed through the secretory pathway. However, one additional concern has come from another previous attempt to engineer glycans in P. pastoris, which was undertaken with an α-1,2-mannosidase from T. reesei (22). In this study immunofluorescent microscopy was used to demonstrate that a mannosidase-myc-HDEL fusion localized primarily in the ER of P. pastoris; however, leakage into the medium was also observed by Western blotting. Because the secreted glycoprotein of interest will be in the supernatant for many hours, the cosecretion of mannosidases into the medium is of concern because it can be misinterpreted as in vivo activity. It is important to generate (Man)5-(GlcNAc)2 structures in vivo, and early in the secretory pathway, if subsequent conversion to complex glycans is to be achieved. Although it is well established that the ERD2-based retrieval system is leaky and retention of the HDEL-tagged mannosidase in the secretory pathway cannot be ensured (26), the same concern is emphasized further by the striking difference demonstrated here between the two different C. elegans α-1,2-mannosidase IB fusion constructs. Thus, the use of a series of different fusion constructs with many differentially localized type II domains has allowed us to screen for chimera with activity that is completely in vivo.
After efficient trimming of the core glycan to (Man)5-(GlcNAc)2 the next step in the conversion of high Man-type glycans to hybrid- and complex-type glycans involves the expression and localization of the enzyme GnTI. Here again a library of GnTI catalytic domains was used and allowed for the screening for and selection of several particularly active GnTI fusion constructs for further study. One particularly active fusion using the human GnTI catalytic domain shown here converts the (Man)5-(GlcNAc)2 substrate to the desired GlcNAc-(Man)5-(GlcNAc)2 product almost quantitatively, an activity that was shown to be completely in vivo. Furthermore, the demonstrated activity of GnTI on the (Man)5-(GlcNAc)2 substrate is the best evidence that the α-1,2-mannosidase activity is indeed occurring in the secretory pathway. This is a demonstration of a high-level hybrid N-glycan modification of a secreted protein in yeast and represents a significant step toward the ability to express fully human glycoproteins in yeast. Although the generated structures are expected to be nonimmunogenic in humans, additional Man removal (i.e., the removal of 1,6- and 1,3-Man from the trimannose core) and further addition of β-1,2-GlcNAc will be required to generate complex N-glycans of therapeutic utility (e.g., for the production of monoclonal antibodies).
Supplementary Material
Acknowledgments
We thank Beata Bobrowicz, Teresa Mitchell, Sebastian Rausch, Andy Stadheim, Amelia Walling, and Harry Wischnewski for technical assistance. We gratefully acknowledge the gift of strains and reagents from Judah Folkman, James Cregg, Nick Menhart, Robert Barsteadt, Carlos Hirschberg, and Takashi Yoshida. We also thank Phil Robbins and Roger Bretthauer for their encouragement and continued support.
Abbreviations
- Man
mannose
- ER
endoplasmic reticulum
- GnTI
β-1,2-N-acetylglucosaminyltransferase I
- MALDI
matrix-assisted laser desorption ionization
- K3
Kringle 3 domain of human plasminogen
- TOF
time of flight
Footnotes
The member who communicated this paper is a Science Advisor for GlycoFi, Inc.
References
- 1.Walsh G. Nat Biotechnol. 2000;18:831–833. doi: 10.1038/78720. [DOI] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Werten M W T, van den Bosch T J, Wind R D, Mooibroek H, de Wolf F A. Yeast. 1999;15:1087–1096. doi: 10.1002/(SICI)1097-0061(199908)15:11<1087::AID-YEA436>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Durand H, Clanet M. Enzyme Microb Technol. 1988;10:341–346. [Google Scholar]
- 5.Ballou C E. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard S C, Ivatt R J. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- 7.Gleeson P A. Histochem Cell Biol. 1998;109:517–532. doi: 10.1007/s004180050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakayama K, Nakanishi-Shindo Y, Tanaka A, Haga-Toda Y, Jigami Y. FEBS Lett. 1997;412:547–550. doi: 10.1016/s0014-5793(97)00634-0. [DOI] [PubMed] [Google Scholar]
- 9.Dean N. Biochim Biophys Acta. 1999;1426:309–322. doi: 10.1016/s0304-4165(98)00132-9. [DOI] [PubMed] [Google Scholar]
- 10.Bretthauer R K, Castellino F J. Biotechnol Appl Biochem. 1999;30:193–200. [PubMed] [Google Scholar]
- 11.Chiba Y, Suzuki M, Yoshida S, Yoshida A, Ikenaga H, Takeuchi M, Jigami Y, Ichishima E. J Biol Chem. 1998;273:26298–26304. doi: 10.1074/jbc.273.41.26298. [DOI] [PubMed] [Google Scholar]
- 12. Koji, M., Narutoshi, S. & Tomoyasu, R. (1996) Jpn. Patent JP 8336387.
- 13.Hayes M L, Bretthauer R K, Castellino F J. Arch Biochem Biophys. 1975;171:651–655. doi: 10.1016/0003-9861(75)90076-4. [DOI] [PubMed] [Google Scholar]
- 14.Lin Cereghino G P, Lin Cereghino J, Sunga A J, Johnson M A, Lim M, Gleeson M A, Cregg J M. Gene. 2001;263:159–169. doi: 10.1016/s0378-1119(00)00576-x. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Towbin H, Gordon J. J Immunol Methods. 1984;72:313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- 17.Papac D I, Briggs J B, Chin E T, Jones A J S. Glycobiology. 1998;8:445–454. doi: 10.1093/glycob/8.5.445. [DOI] [PubMed] [Google Scholar]
- 18.Turco S J. Anal Biochem. 1981;118:278–283. doi: 10.1016/0003-2697(81)90582-0. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi-Shindo Y, Nakayama K I, Tanaka A, Toda Y, Jigami Y. J Biol Chem. 1993;268:26338–26345. [PubMed] [Google Scholar]
- 20.Trimble R B, Atkinson P H, Tschopp J F, Townsend R R, Maley F. J Biol Chem. 1991;266:22807–22817. [PubMed] [Google Scholar]
- 21.Martinet W, Maras M, Saelens X, Jou W M, Contreras R. Biotechnol Lett. 1998;20:1171–1177. [Google Scholar]
- 22.Callewaert N, Laroy W, Cadirgi H, Geysens S, Saelens X, Jou W M, Contreras R. FEBS Lett. 2001;503:173–178. doi: 10.1016/s0014-5793(01)02676-x. [DOI] [PubMed] [Google Scholar]
- 23.Guillen E, Abeijon C, Hirschiberg C B. Proc Natl Acad Sci USA. 1998;95:7888–7892. doi: 10.1073/pnas.95.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Cereghino J, Cregg J M. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 25.Miele R G, Castellino F J, Bretthauer R K. Biotechnol Appl Biochem. 1998;26:79–83. [PubMed] [Google Scholar]
- 26.Pelham H R B. Methods Enzymol. 2000;327:279–283. doi: 10.1016/s0076-6879(00)27283-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.