Abstract
Phospholamban (PLN), a regulator of sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs), interacts with both the cytosolic N domain and transmembrane helices M2, M4, M6, and M9 of SERCA. Amino acids in the transmembrane domain of PLN that are predicted to interact with SERCA1a are conserved in sarcolipin (SLN), a functional PLN homologue. Accordingly, the effects of critical mutations in SERCA1a, PLN, and NF-SLN (SLN tagged N-terminally with a FLAG epitope) on NF-SLN/SERCA1a and PLN/NF-SLN/SERCA1a interactions were compared. Critical mutations in SERCA1a and NF-SLN diminished functional interactions between SERCA1a and NF-SLN, indicating that NF-SLN and PLN interact with some of the same amino acids in SERCA1a. Mutations in PLN or NF-SLN affected the amount of SERCA1a that was coimmunoprecipitated in each complex with antibodies against either PLN or SLN, but not the pattern of coimmunoprecipitation. PLN mutations had more dramatic effects on SERCA1a coimmunoprecipitation than SLN mutations, suggesting that PLN dominates in the primary interaction with SERCA1a. Coimmunoprecipitation also confirmed that PLN and NF-SLN form a heterodimer that interacts with SERCA1a in a regulatory fashion to form a very stable PLN/NF-SLN/SERCA1a complex. Modeling showed that the SLN/SERCA1a complex closely resembles the PLN/SERCA1a complex, but with the luminal end of SLN extending to the loop connecting M1 and M2, where Tyr-29 and Tyr-31 interact with aromatic residues in SERCA1a. Modeling of the PLN/SLN/SERCA1a complex predicts that the regulator binding cavity in the E2 conformation of SERCA1a can accommodate both SLN and PLN helices, but not two PLN helices.
Sarco(endo)plasmic reticulum Ca2+-ATPases (SERCAs) are 110-kDa membrane proteins that catalyze the ATP-dependent transport of Ca2+ from the cytosol to the lumen of the sarco(endo)plasmic reticulum (1). SERCAs expressed in muscle are regulated by two members of a gene family: phospholamban (PLN) (2, 3) and sarcolipin (SLN) (4–6).
PLN is a 52-aa membrane protein that interacts with SERCA molecules to lower their apparent affinity for Ca2+ and inhibit their activity at low, but not at high, Ca2+ concentrations (2, 7). SLN is a 31-aa membrane protein that resembles PLN in these essential inhibitory features (5, 8). The two proteins have similar transmembrane sequences (4, 9) but differ at their C termini, where PLN ends with the sequence Met-Leu-Leu-52 (10), whereas SLN ends with the more hydrophilic sequence, Arg-Ser-Tyr-Gln-Tyr-31. They also differ at their N termini: phosphorylation of PLN in a well conserved 30-aa cytosolic domain disrupts inhibitory interactions, accounting, in part, for the inotropic response of the heart to adrenergic stimulation (2, 7). The poorly conserved cytosolic sequence of SLN is 7 aa long and is not phosphorylated under normal conditions. A number of physiological studies have demonstrated that PLN is a key regulator of the kinetics of cardiac muscle function (11, 12).
PLN expression is largely restricted to cardiac, slow-twitch, and smooth muscle, whereas SLN is highly expressed in fast-twitch and, to a lesser extent, in cardiac muscle (4, 13). Nevertheless, both PLN and SLN can inhibit both SERCA1a and SERCA2a with similar characteristics (14). Although PLN exists in both pentameric and monomeric forms, it is generally accepted that the monomer is the inhibitory species (15, 16). When NF-SLN and PLN are coexpressed with SERCA, superinhibition of SERCA activity is observed (5, 8).
Sites of interaction between SERCA and PLN have been identified in both cytosolic and transmembrane domains of SERCA and PLN by using cross-linking and mutagenesis (15, 17–22). Modeling from high-resolution crystal and NMR structures has identified additional amino acids that interact between PLN domain Ia and the cytosolic domains of SERCA1a and between PLN domains Ib and II and transmembrane helices M2, M4, M6, and M9 in SERCA1a (10, 23).
In this study, we investigated interaction sites between NF-SLN and SERCA1a, showing that they overlap in important ways with the transmembrane sites of PLN/SERC1a interaction. We also investigated sites involved in the superinhibition that results when PLN and SLN are coexpressed with SERCA1a or SERCA2a (8). Structural models were developed for the binary SLN/SERCA1a and ternary PLN/SLN/SERCA1a complexes.
Materials and Methods
Materials.
Enzymes for DNA manipulation were obtained from New England Biolabs and Pharmacia. G-Sepharose and a chemiluminescence kit for measurement of coimmunoprecipitation of interacting proteins were purchased from Pierce. FLAG antibody, M2, was purchased from Sigma; the anti-PLN antibody, 1D11, was a gift from Robert Johnson (Merck Research Laboratories, West Point, PA); the A52 monoclonal antibody against SERCA1a was produced in our laboratory (24).
Cell Culture and Heterologous Expression of Wild-Type (wt) and Mutant Proteins.
The culture of HEK-293 cells, their transfection with cDNAs encoding SERCA1a, SERCA2a, PLN, and NF-SLN, and the isolation of microsomal fractions from transfected cells expressing these proteins have been described in earlier publications (5, 15, 21). NF-SLN is a fusion protein of SLN with the FLAG epitope (MDYKDDDDK) at its N terminus (5). This protein has been shown to be fully functional, to be immunoprecipitated with antibody M2 against the FLAG epitope, and to be recognized in Western blots by the M2 antibody (5).
Immunoprecipitation of Proteins from Microsomal Fractions.
SERCA1a was coimmunoprecipitated with PLN and/or NF-SLN by using the 1D11 antibody against PLN or the M2 antibody against NF-SLN, as described previously (21). The relative amounts of SERCA1a, PLN, or NF-SLN in each lane were quantified in exposed films by scanning densitometry using National Institutes of Health image16.1 software. Protein expression levels in all experiments were estimated by quantitative immunoblotting using antibodies A52, 1D11, and M2.
Ca2+ Transport Activity.
Measurement of Ca2+ transport activity in microsomal fractions was carried out as described previously (15).
Modeling.
Modeling was based on our model of PLN bound to SERCA1a (10) and the solution structure of SLN derived by NMR (25). Protein Data Base ID no. 1IWO was used for the SERCA1a model. Because thapsigargin in the absence of Ca2+ does not diminish physical interactions between PLN and SERCA (26), the use of 1IWO for modeling is justified. The model was constructed manually by using turbo-frodo (http://afmb.cnrs-mrs.fr) and refined by energy minimization using presto (27).
Results
Effect of NF-SLN on the Ca2+ Affinity of wt and Mutant Forms of SERCA1a Expressed in HEK-293 Cells.
In previous papers (10, 21), we showed that the diminished Ca2+ affinity that is associated with PLN inhibition of SERCA activity was reversed partially for the SERCA1a mutants V49C, L321A, V795A, L802A, T805A, and F809A. We also showed that coimmunoprecipitation with PLN was diminished for mutants V795A and L802A. Because SLN and PLN share functional and structural properties, we used the protocols developed earlier to examine whether SLN and PLN might share binding sites in SERCA.
Ca2+ dependence of Ca2+ transport was measured in microsomal fractions from HEK-293 cells expressing wt or mutant SERCA1a, together with NF-SLN, or PLN, or both NF-SLN and PLN. The results, presented in Table 1, show that the apparent Ca2+ affinity (KCa) of wt SERCA1a was reduced by 0.22 pCa (−log10[Ca]) unit through coexpression with NF-SLN; by 0.36 pCa unit through coexpression with PLN; and by 0.93 pCa unit through coexpression with NF-SLN and PLN. The reduction in apparent Ca2+ affinity caused by interaction with NF-SLN or PLN was reversed significantly with SERCA1a mutants V89C, L321A, V795A, L802A, T805A, and F809A. These results suggest that functional interactions between NF-SLN and SERCA1a involve the same amino acids that are involved in interactions between PLN and SERCA1a.
Table 1.
Effect of M2, M4, and M6 mutants on ΔKCa
| Site | Mutation | KCa | ΔKCa
|
||
|---|---|---|---|---|---|
| +SLN | +PLN | +SLN, +PLN | |||
| wt | 6.57 ± 0.05 | −0.22 ± 0.01 | −0.36 ± 0.01 | −0.93 ± 0.04 | |
| M2 | V89C | 6.72 ± 0.04 | −0.03 ± 0.01* | −0.04 ± 0.02* | ND |
| M4 | L321A | 6.33 ± 0.05 | −0.10 ± 0.03* | −0.22 ± 0.01* | ND |
| M6 | V795A | 6.10 ± 0.01 | −0.05 ± 0.02* | −0.03 ± 0.02* | −0.74 ± 0.01** |
| L802A | 6.57 ± 0.04 | −0.03 ± 0.01* | −0.07 ± 0.02* | −0.19 ± 0.07** | |
| T805A | 6.04 ± 0.04 | −0.04 ± 0.01* | −0.03 ± 0.02* | −0.45 ± 0.05** | |
| F809A | 6.24 ± 0.05 | −0.07 ± 0.03* | −0.09 ± 0.04* | −0.61 ± 0.02** | |
The results are the mean ± SEM for three separate experiments for each mutation. ND, not determined.
, P < 0.05 against wt SERCA1a expression;
, P < 0.05 against triple expression of wt SERCA1a, NF-SLN, and PLN.
The expression of both NF-SLN and PLN with SERCA1a decreased the Ca2+ affinity of SERCA1a by nearly 1 pCa unit (5, 8) (Table 1, column 6). A significant reversal of this effect was observed after the expression of both NF-SLN and PLN with SERCA1a mutants V795A, L802A, T805A, and F809A, suggesting that these amino acids are involved functionally when both NF-SLN and PLN are bound.
Coimmunoprecipitation of SERCA1a with NF-SLN Mutants.
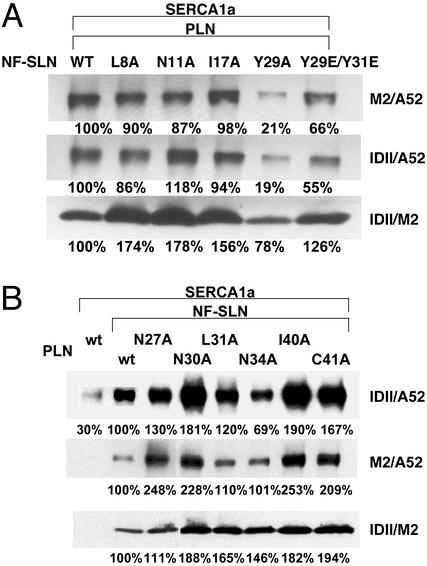
In previous studies (21), specific mutations in SERCA1a resulted in a significant reduction in the amount of SERCA1a coimmunoprecipitated with PLN: L321A, −20%; V795A, −47%; L802A, −56%; T805A, −22%; and F809A, −15%. To determine whether these and other SERCA1a mutations might decrease the ability of NF-SLN to coimmunoprecipitate SERCA1a, they were coexpressed with NF-SLN and immunoprecipitated with antibody M2 against NF-SLN. As shown in Fig. 1 A and B, coimmunoprecipitation of SERCA1a with NF-SLN was decreased with SERCA1a mutants V89C, L321A, V795A, and L802A, but increased with mutants T805A and F809A.
Figure 1.
Effects of SERCA1a and NF-SLN mutations on formation of the binary NF-SLN/SERCA1a complex. (A and B) SERCA1a mutants were expressed in HEK-293 cells in the presence or absence of NF-SLN. (C) wt SERCA1a was expressed in HEK-293 cells in the presence of wt or mutant NF-SLN. The cells were harvested after 48 h, and microsomal fractions were prepared and dissolved in Tween 20 as described (21, 26). NF-SLN was immunoprecipitated with the FLAG antibody, and the immunoprecipitate was separated by SDS/PAGE and stained with the A52 antibody against SERCA1a to determine the amount of SERCA1a coimmunoprecipitation. Numbers below each lane represent percentage of wt and are the average of three independent experiments.
In our modeling of the PLN-SERCA1a interaction (10), Val-89 in SERCA1a interacts with Val-49 in PLN; Leu-321 with Asn-27 and Asn-30; Leu-802 with Asn-34 and Phe-35; Thr-805 with Asn-34; and Phe-809 with Leu-31. Leu-795 does not interact with PLN. Thus the effects of mutation of each of these SERCA1a residues on the functional and physical interactions of SERCA1a with SLN, positive, negative, or marginal (in the case of Leu-795), are fully in line with the view that SLN and PLN fit into the same groove in the SERCA1a transmembrane domain.
Coimmunoprecipitation of SERCA1a with NF-SLN Mutants.
In previous studies (21), specific mutations in PLN decreased the amount of SERCA1a coimmunoprecipitated with PLN: L31A, −73%; and N34A, −65%. By contrast, the I40A mutation increased the amount of SERCA1a coimmunoprecipitated by +145%. Amino acids Leu-8, Asn-11, and Ile-17 in SLN are homologous to Leu-31, Asn-34, and Ile-40 in PLN. To determine whether mutation of these and other SLN amino acids might affect coimmunoprecipitation of SERCA1a, NF-SLN mutants were coexpressed with SERCA1a and immunoprecipitated with antibody M2 against NF-SLN. As shown in Fig. 1C, coimmunoprecipitation of SERCA1a with NF-SLN was unchanged with mutant L8A, but was decreased with N11A, I17A, Y29A, and Y29E/Y31E.
In our modeling of the PLN/SERCA1a interaction (10), Leu-31 in PLN (Leu-8 in SLN) interacts with Thr-805 and Phe-809 in SERCA1a; Asn-34 (Asn-11 in SLN) interacts with Thr-317, Leu-802, and Thr-805 in SERCA1a; and Ile-40 (Ile-17 in SLN) has no sites of interaction with SERCA1a. On the basis of the results presented in Fig. 1C, mutation of Leu-8 in NF-SLN provided little insight, but mutation of Asn-11 followed the same pattern as that observed with Asn-34 in PLN. The effects of the Ile-17 mutation in NF-SLN would be expected to differ from those observed with PLN mutant I40A, however, because Ile-40 is involved in PLN pentamer formation, whereas SLN has little tendency to form oligomers. The effects of Tyr-29 and Tyr-31 mutations are of great interest. Because these amino acids do not have a counterpart in PLN, their mutation provides novel insight into how the C-terminal sequence of SLN interacts with SERCA1a.
Coimmunoprecipitation of SERCA1a and PLN with NF-SLN Mutants.
To gain insight into the structural basis for the superinhibition of SERCA1a and SERCA2a that is seen when NF-SLN is coexpressed with PLN (8), we carried out triple expression of wt SERCA1a with wt PLN and the same series of NF-SLN mutants examined in Fig. 1C. In Fig. 2A, row 1, the amount of SERCA1a in the ternary complex that was coimmunoprecipitated with the M2 antibody against NF-SLN was reduced with NF-SLN mutants L8A, N11A, I17A, Y29A, and Y29E/Y31E. In row 2, the amount of SERCA1a in the ternary complex that was coimmunoprecipitated from the same samples with the 1D11 antibody against PLN was almost identical to that observed with the M2 antibody. These results are in line with the view that a binary complex of PLN and NF-SLN is interacting with SERCA1a to form a ternary complex and that the properties of the complex are altered by mutations in SLN.
Figure 2.
Effects of NF-SLN and PLN mutations on formation of the ternary PLN/NF-SLN/SERCA1a complex. (A) wt SERCA1a was expressed in HEK-293 cells in the presence of wt PLN and a series of NF-SLN mutants. Microsomal fractions were dissolved and immunoprecipitated with either antibody M2 against NF-SLN (row 1) or antibody 1D11 against PLN (rows 2 and 3). The immunoprecipitates were separated and stained with antibody A52 against SERCA1a (rows 1 and 2), or M2 against NF-SLN (row 3). (B) SERCA1a was expressed in HEK-293 cells in the absence or presence of NF-SLN and in the presence of wt PLN or a PLN mutant. Microsomal fractions were dissolved and immunoprecipitated with either antibody 1D11 against PLN (rows 1 and 3) or antibody M2 against NF-SLN (row 2). Immunoprecipitates were separated and stained with either antibody A52 against SERCA1a (rows 1 and 2) or antibody M2 against NF-SLN (row 3). Numbers below lanes represent percentage of wt and are the average of three independent experiments.
In row 3, the amount of NF-SLN coimmunoprecipitated with the 1D11 antibody against PLN increased for the NF-SLN mutants L8A, N11A, I17A, and Y29E/Y31E, but decreased for Y29A. It should be noted, however, that this protocol stains NF-SLN in a trimeric PLN/NF-SLN/SERCA1a complex in which NF-SLN and PLN do not have to interact to be precipitated together, as well as in a NF-SLN/PLN complex. Nevertheless, any apparent increase in formation of the NF-SLN/PLN complex caused by NF-SLN mutations L8A, N11A, or I17A did not appear to increase the NF-SLN/SERCA1a interaction (compare Fig. 1C with Fig. 2A, row 1). The inclusion of PLN changed the pattern of interaction of both Y29A and Y29E/Y31E mutants with SERCA1a. The Y29A mutation diminished both the apparent PLN/SLN interaction and the NF-SLN/SERCA1a interaction. Conversely, the Y29E/Y31E mutation increased both the apparent PLN/SLN interaction and the NF-SLN/SERCA1a interaction. These results are consistent with a displacement of the C terminus of NF-SLN in its interaction with SERCA1a between the binary NF-SLN/SERCA1a and ternary PLN/NF-SLN/SERCA1a complexes. The similarity of the pattern in rows 1 and 2 suggests that the proportion of the dimeric PLN/SERCA1a and NF-SLN/SERCA1a complexes is small.
Coimmunoprecipitation of SERCA1a and NF-SLN with PLN Mutants.
To gain further insight into the structural basis for the superinhibition of SERCA1a and SERCA2a that is seen when NF-SLN is coexpressed with PLN (8), we carried out triple expression of wt SERCA1a with NF-SLN and wt and mutant forms of PLN. Fig. 2B, row 1, shows that the amount of SERCA1a that was coimmunoprecipitated in the ternary complex with the 1D11 antibody against PLN was increased severalfold by comparison with the amount coimmunoprecipitated in the dimeric PLN/SERCA1a complex. Thereafter, SERCA1a coimmunoprecipitation increased or decreased, depending on the PLN mutant that was expressed. Fig. 2B, row 2, shows that the amount of SERCA1a coimmunoprecipitated in the ternary complex with the NF-SLN antibody followed the same pattern, qualitatively, if not quantitatively, as that seen with coimmunoprecipitation with the PLN antibody. These results, showing that the antibodies against either PLN or NF-SLN precipitate a very similar complex, are in line with the view that a binary complex of PLN and NF-SLN is interacting with SERCA1a. In comparison with the experiment with NF-SLN mutants, described in Fig. 2A, the amount of SERCA1a coimmunoprecipitated in the PLN/NF-SLN/SERCA1a complex was altered much more dramatically by PLN mutations than by NF-SLN mutations. Thus PLN must play a key role in binding of the PLN/NF-SLN binary complex to SERCA1a.
In Fig. 2B, row 3, the amount of SLN coimmunoprecipitated with the 1D11 antibody against PLN increased for each of the PLN mutants. Thus none of these PLN mutations appeared to diminish the PLN/NF-SLN interaction. Again, the similar pattern in rows 1 and 2 suggests that the amount of dimeric PLN-SERCA1a and NF-SLN-SERCA1a must be rather small.
Coimmunoprecipitation of SERCA1a with NF-SLN and the Superinhibitory Monomeric PLN Mutant I40A.
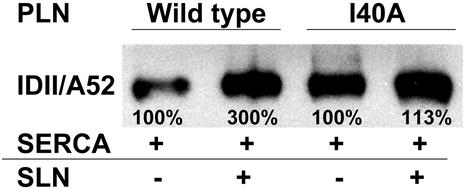
Superinhibition of SERCA can be induced by PLN mutations that diminish PLN/PLN interactions, increasing the concentration of the PLN monomer and inducing superinhibition through mass action, and by other PLN mutations that increase the binding affinity between PLN and SERCA (15). The presence of NF-SLN increases the concentration of monomeric PLN (8), but superinhibition is likely to result from an increased affinity of the PLN/NF-SLN binary complex for SERCA. It was of interest to measure the degree of superinhibition that occurs after coexpression of SERCA1a with the monomeric, superinhibitory PLN mutant, I40A, and compare it with that which occurs with the PLN I40A mutation in the presence of an approximately equal amount of NF-SLN. In Fig. 3, lanes 1 and 2 show that NF-SLN increased coimmunoprecipitation of SERCA1a by the PLN antibody by 3-fold. Lane 3 shows that PLN mutant I40A coimmunoprecipitates large amounts of SERCA1a in a binary I40A-PLN/SERCA1a complex, while lane 4 shows that the amount of SERCA1a coimmunoprecipitated in the ternary I40A-PLN/NF-SLN/SERCA1a complex is only ≈13% greater than that in the binary complex. Since we did not predict interactions between Ile-40 and amino acids in SERCA in our previous analysis (10), these data support the view that high levels of superinhibition can be achieved by two different mechanisms of superinhibition.
Figure 3.
Effects of NF-SLN and the PLN I40A mutation on formation of the ternary PLN/NF-SLN/SERCA1a complex. SERCA1a was expressed in HEK-293 cells in the presence of NF-SLN and either wt or I40A mutant PLN. Microsomal fractions were dissolved and immunoprecipitated with antibody 1D11 against PLN. Immunoprecipitates were separated and stained with antibody A52 against SERCA1a. Numbers below lanes represent percentage of wt and are the average of three independent experiments.
Modeling of the Interaction Between SLN and SERCA1a.
On the basis that mutations in SERCA1a that diminish regulatory function with PLN also diminish regulatory function with SLN and that corresponding mutations in PLN and SLN diminish functional interaction in a predictable fashion, it seemed likely that SLN would fit into the site of PLN interaction with SERCA1a (10). Indeed, it was straightforward to model the interaction of SLN with SERCA on the basis of our published model for PLN (Fig. 4).
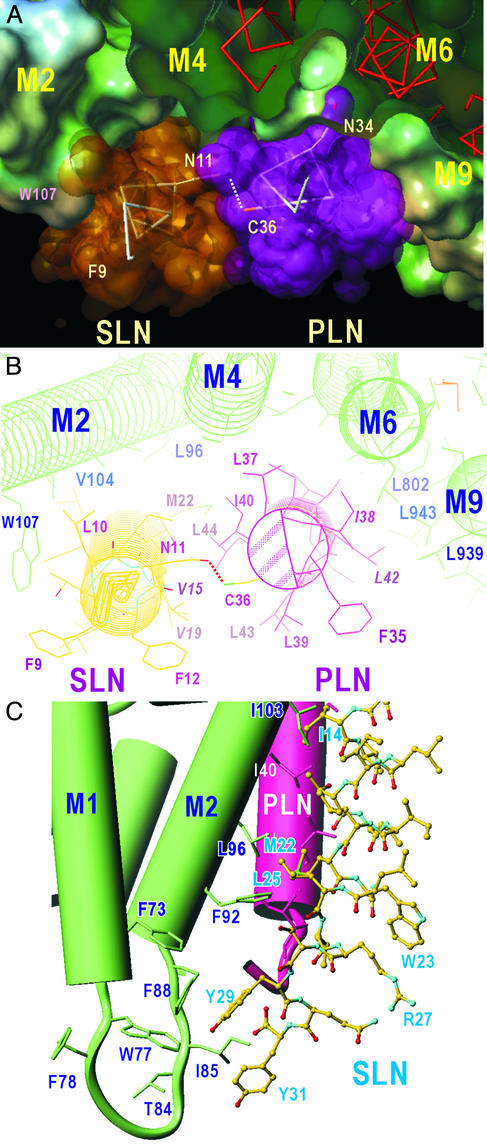
Figure 4.
A model for the binary SLN/SERCA1a complex. In this model, based on our previous model for the PLN/SERCA1a complex (10), the water-accessible surface is colored according to lipophilicity (brown, hydrophobic; sky blue, hydrophilic; green, neutral) as calculated with SYBYL6.8 (Tripos Associates, St. Louis). White dots outline SLN. The orange arrow, corresponding to 13 Å, indicates a wide space between SLN and M2 of SERCA1a. Horizontal bars show the boundaries of the hydrophobic core of the lipid bilayer.
Molecular modeling shows that the affinity of SLN for the transmembrane domain of SERCA1a might be lower than the affinity of PLN for this site, because the complementarity between the hydrophobic surfaces of SLN and SERCA1a appears less satisfactory. This is because Ile-38 and Leu-42 in PLN, which lose inhibitory activity when mutated to Ala (15), are replaced by the smaller Val (Fig. 5B, residue numbers in italics).
Figure 5.
A model for the ternary PLN/SLN/SERCA1a complex. (A) A view approximately normal to the membrane from the cytoplasmic side. The water-accessible surface of SERCA1a and van der Waals surfaces of SLN and PLN are shown. Transmembrane helices of SERCA1a (M2, M4, M6, and M9) are identified. (B) A view approximately normal to the membrane from the cytoplasmic side. Details of the interactions in the cytoplasmic half of the transmembrane region are shown. The shaded area in PLN indicates the region used for pentamer formation. Italicized amino acids (I38 and L42) show the two most important residues in hydrophobic contacts between PLN and SERCA, and the corresponding residues in SLN (V15 and V19). Dotted lines show a likely hydrogen bond between N11 (SLN) and C36 (PLN). (C) C-terminal half of SLN and the interaction with M2 of SERCA1a. Cylinders represent α-helices. The model is viewed in a shallow angle to the membrane so that the PLN transmembrane helix becomes approximately upright with the lumen of the sarcoplasmic reticulum at the bottom. M1 and M2 helices of SERCA1a are in green and the PLN transmembrane helix is in red. SLN is shown with a ball-and-stick representation. Note the cluster of aromatic residues near the luminal surface.
The C-terminal sequences of PLN and SLN diverge: Arg-Ser-Tyr-Gln-Tyr-31 in SLN replaces Met-Leu-Leu-52 in PLN. The two Tyr residues in SLN are likely to stack with other aromatic residues in SERCA1a and contribute to regulation. Obvious candidates for aromatic residues contributing to the interaction are those within the loop connecting M1 and M2 helices (Phe-73, Trp-77, Phe-88, and Phe-92 (Figs. 4 and 5). This led us to a model in which the luminal sequence of SLN interacts with the loop connecting M1 and M2, leaving Arg-27 and Gln-30 exposed to the lumen, where they may serve as a sarco(endo)plasmic reticulum retention or other docking signal, involving SLN.
Modeling of a Ternary PLN/SLN/SERCA1a Complex.
Our coimmunoprecipitation data clearly demonstrate that ternary PLN/SLN/SERCA complexes are formed when both PLN and NF-SLN are expressed together with SERCA1a or SERCA2a. It was of interest to understand how this can happen when the sites of interaction for both proteins seem to be the same. Because our data also show that a PLN/NF-SLN complex will predominate if the two proteins are expressed at similar levels, it is likely that it is this binary complex that interacts so strongly with SERCA1a.
Molecular modeling shows that a ternary PLN/NF-SLN/SERCA1a complex is not only possible (Fig. 5A) but will be more stable than either of the NF-SLN/SERCA1a or PLN/SERCA1a binary complexes. As noted previously (10) and above, the cleft formed by M2, M4, M6, and M9 of SERCA in the E2 conformation is too large for either PLN or SLN to form a very tight binding site: the hydrophobic surfaces are complementary between M9 and PLN but not between M2 and PLN. There is just enough room in this cleft to accommodate SLN between PLN and M2 (Fig. 5A).
The fact that SLN binds to the surface of PLN used for pentamer formation (shaded area in Fig. 5B) led us to propose that SLN is located on the side of PLN opposite to Ile-38 and Leu-42 (shaded area in Fig. 5B). The sequence of this face is not well conserved between PLN and SLN, consistent with the fact that SLN forms oligomers only weakly (5, 6), but is able to disrupt PLN pentamers (18). These observations suggest that the binding surface in PLN for SLN overlaps, at least partially, with the binding surface for PLN, but the affinity of SLN for this surface is higher than the affinity of the surface for other monomers of PLN. A polar residue in the transmembrane region of SLN, Asn-11, will define the orientation of SLN by interacting with Cys-36 of PLN (Fig. 5B). This will bring the face of the SLN helix used for binding to SERCA into a position where it can bind to PLN. With this orientation, Ile/Val or Leu/Leu pairs take part in hydrophobic interactions between SLN and M2. Although SLN and M2 helices cross at the middle of the transmembrane region, hydrophobic interactions are maintained along the whole transmembrane helices by using residues with long side chains (e.g., Phe-92 on M2 and Met-22 on SLN; Fig. 5C).
The luminal part of SLN can interact with the aromatic residues in the loop connecting M1 and M2. That is, Tyr-29 in SLN interacts with Phe-73, Trp-77, and Phe-88 of SERCA1a, and Tyr-31 with Phe-88 (and Ile-85), thereby providing extra interaction sites with SERCA (Fig. 5C). Also, on the cytoplasmic side, Trp-107 in SERCA and Phe-9 in SLN are likely to take part in an aromatic–aromatic interaction (Fig. 5B). Hence, the model predicts that the ternary complex will be more stable than either of the binary complexes because of the extra binding sites.
A question that arises from these predictions is whether there is room for two PLN molecules to fit into the space occupied by the PLN/SLN complex. The face of PLN that is used for PLN/PLN interaction appears to be slightly different from that used for SLN/PLN interaction and is partially blocked by Leu-37 on M4 (Fig. 5B) when PLN binds to SERCA (10), thereby preventing the binding of additional PLN monomers. Constraints arise in replacing SLN by PLN, which involve Val-15 (SLN)–Ile-38 (PLN), Val-19 (SLN)–Leu-42 (PLN), Met-22–Ile-45 (conflicts with M2), and Gly-2 (SLN)–Arg-25 (PLN). Of these, Ile-38 and Leu-42 in PLN are among the most important hydrophobic residues used for binding to SERCA (15). Changing these residues to the smaller Val will make the binding of SLN to SERCA weaker, but allow the formation of the ternary complex (Fig. 5). Moreover, the lack of C-terminal aromatic residues in PLN will make the binding of a second molecule of PLN considerably weaker than the binding of SLN. Thus, the molecular model shown in Fig. 5 is compatible with all of our current data.
Discussion
Models for the Interaction of SLN with SERCA1a in the Presence and Absence of PLN.
Several recent studies support the view that SLN and PLN are homologous members of the same gene family with similar functions, but differential expression (4–6, 8, 13). A model of the regulatory interaction between PLN and SERCA1a shows that the transmembrane sequence of PLN lies in a groove on the lipid-facing surface of SERCA that is lined by M2, M4, M6, and M9 (10). Our goals in this study were twofold: to determine whether NF-SLN and PLN fit into the same regulatory site in SERCA1a and to understand the structural basis for the superinhibition that results from coexpression of NF-SLN and PLN with SERCA1a or SERCA2a (8). This superinhibition is of potential physiological significance, because SLN and PLN are coexpressed in the heart (4, 13, 28).
Our use of mutagenesis in the studies of NF-SLN/SERCA1a and PLN/SERCA1a interactions confirmed that the two regulatory proteins were occupying the same interaction site in SERCA1a. We then proceeded to investigate the superinhibitory ternary interaction involving PLN, NF-SLN, and SERCA1a. Earlier studies (8) showed not only that PLN and NF-SLN form a heterodimer, but that the binding of SLN to monomeric PLN is tight enough to alter the PLN monomer ⇌ pentamer equilibrium in favor of PLN/SLN heterodimers and away from PLN pentamers. Because the SLN/PLN interaction has a higher affinity than the PLN/PLN interaction, and because SLN does not form oligomers itself and “disrupts” PLN pentamers, we concluded that a PLN/SLN complex will likely be the major form if the two proteins are expressed at similar levels. Under conditions where both PLN and SLN were expressed at levels that would be highly inhibitory alone, we were able to isolate substantial amounts of a ternary PLN/SLN/SERCA1a complex, which probably represents the most stable complex at equilibrium. Using mutagenesis of NF-SLN and PLN, we found that the amount of SERCA1a that was precipitated in each complex varied with each mutant, but the pattern of coimmunoprecipitation of SERCA1a was the same whether precipitation of the complex was carried out with antibodies against PLN or against NF-SLN. On this basis, we concluded that PLN and NF-SLN associate in the complex and that it is the binary PLN/NF-SLN complex that interacts so strongly with SERCA1a. In further studies, we noted that the amount of SERCA1a precipitated was very dependent on the properties of the PLN mutant, suggesting that the primary interaction of the binary PLN/NF-SLN complex with SERCA1a would be through PLN.
With these principles in mind, we modeled the NF-SLN/SERCA1a and PLN/NF-SLN/SERCA1a interactions as extensions of our earlier modeling of the PLN/SERCA1a interaction (20). Our biochemical and modeling data strongly support the view that PLN and SLN fit into the same regulatory transmembrane groove in SERCA1a. Our model of the ternary complex (Fig. 5) shows that the regulator binding site in SERCA1a will also accommodate the PLN/NF-SLN heterodimer, but not a PLN/PLN homodimer. The model accounts for an increased affinity of SERCA1a for the heterodimer, based on a tighter fit and an increased number of interactions among the three proteins. The tighter binding explains the superinhibited properties of the PLN/NF-SLN/SERCA complex (8).
Superinhibition occurs by mass action through an increase in the concentration of PLN monomers and through enhanced affinity of regulator proteins for SERCA (15). These mechanisms were compared in the experiment described in Fig. 3, which shows that superinhibition by the monomeric PLN mutant, I40A, differs from superinhibition by the I40A PLN/NF-SLN binary complex by only ≈13%. While Ile-40 does not appear to interact with other residues in SERCA1a (10), it can interact with Ile-14, Val-15, and Thr-18 in SLN. Many of these interactions are tight (<4 Å), but all are lost in the PLN I40A mutant. In a compensatory reaction, however, Ile-14 in SLN is then likely to make van der Waals contacts with Ala-100 and Val-104 in SERCA1a. Thus alterations in inhibitory properties of mutants are complex and indicative of the delicate balance that characterizes the regulation of SERCA by PLN and SLN.
Relationship of Our Models to an Earlier Model of PLN/SERCA Interaction.
Our models of PLN/SERCA and SLN/SERCA interactions do not agree with a model presented by Hutter et al. (23). Support for the Hutter et al. model was based largely on simulated annealing and energy minimization, but reliance on these techniques can be misleading if the starting model is incorrect. This is particularly important when the calculation is done in vacuo, without experimental support, because of improper handling of polar interactions. Hutter et al. (23) state that PLN does not seem to interact with the SERCA pump appropriately in the presence of thapsigargin in the E2(TG) configuration (29). While they attribute this anomaly to the presence of thapsigargin in the high-resolution crystal structure of SERCA1a in the E2(TG) configuration, the lower-resolution E2-vanadate structures on which their modeling was based (30) also contained thapsigargin to stabilize the tubular crystal and to improve resolution. Thapsigargin occupies a narrow cleft on the opposite side of the PLN/SLN binding site in the E2 conformation and its main effect is to stabilize the helices surrounding it, making them less flexible.
Acknowledgments
We are grateful to Dr. Robert Johnson for the kind gift of anti-PLN antibody 1D11, and to Dr. David B. McIntosh for helpful discussion and insights. This work was supported by Heart and Stroke Foundation of Ontario Grant T-5042 and Canadian Institutes for Health Research Grant MT-12545 (to D.H.M.), and by a grant from the Ministry of Education, Science, Sports, Culture, and Technology of Japan (to C.T.).
Abbreviations
- SERCA
sarco(endo)plasmic reticulum Ca2+-ATPase
- PLN
phospholamban
- SLN
sarcolipin
- NF-SLN
SLN tagged N-terminally with a FLAG epitope
- wt
wild type
References
- 1.MacLennan D H, Rice W J, Green N M. J Biol Chem. 1997;272:28815–28818. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- 2.Tada M, Kadoma M. BioEssays. 1989;10:157–163. doi: 10.1002/bies.950100505. [DOI] [PubMed] [Google Scholar]
- 3.Simmerman H K, Jones L R. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 4.Odermatt A, Taschner P E, Scherer S W, Beatty B, Khanna V K, Cornblath D R, Chaudhry V, Yee W C, Schrank B, Karpati G, et al. Genomics. 1997;45:541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 5.Odermatt A, Becker S, Khanna V K, Kurzydlowski K, Leisner E, Pette D, MacLennan D H. J Biol Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 6.Hellstern S, Pegoraro S, Karim C B, Lustig A, Thomas D D, Moroder L, Engel J. J Biol Chem. 2001;276:30845–30852. doi: 10.1074/jbc.M102495200. [DOI] [PubMed] [Google Scholar]
- 7.Simmerman H K, Kobayashi Y M, Autry J M, Jones L R. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 8.Asahi M, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]
- 9.Wawrzynow A, Theibert J L, Murphy C, Jona I, Martonosi A, Collins J H. Arch Biochem Biophys. 1992;298:620–623. doi: 10.1016/0003-9861(92)90457-8. [DOI] [PubMed] [Google Scholar]
- 10.Toyoshima C, Asahi M, Sugita Y, Khanna R, Tsuda T, MacLennan D H. Proc Natl Acad Sci USA. 2003;100:467–472. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo W, Grupp I L, Harrer J, Ponniah S, Grupp G, Duffy J J, Doetschman T, Kranias E G. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 12.Koss K L, Kranias E G. Circ Res. 1996;79:1059–1063. doi: 10.1161/01.res.79.6.1059. [DOI] [PubMed] [Google Scholar]
- 13.Gayan-Ramirez G, Vanzeir L, Wuytack F, Decramer M. J Physiol. 2000;524:387–397. doi: 10.1111/j.1469-7793.2000.t01-2-00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyofuku T, Kurzydlowski K, Lytton J, MacLennan D H. J Biol Chem. 1992;267:14490–14496. [PubMed] [Google Scholar]
- 15.Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 16.Autry J M, Jones L R. J Biol Chem. 1997;272:15872–15880. doi: 10.1074/jbc.272.25.15872. [DOI] [PubMed] [Google Scholar]
- 17.James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature. 1989;342:90–92. doi: 10.1038/342090a0. [DOI] [PubMed] [Google Scholar]
- 18.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:22929–22932. [PubMed] [Google Scholar]
- 19.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:3088–3094. [PubMed] [Google Scholar]
- 20.Kimura Y, Asahi M, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1998;273:14238–14241. doi: 10.1074/jbc.273.23.14238. [DOI] [PubMed] [Google Scholar]
- 21.Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- 22.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 23.Hutter M C, Krebs J, Meiler J, Griesinger C, Carafoli E, Helms V. Chembiochem. 2002;3:1200–1208. doi: 10.1002/1439-7633(20021202)3:12<1200::AID-CBIC1200>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 24.Zubrzycka-Gaarn E, MacDonald G, Phillips L, Jorgensen A O, MacLennan D H. J Bioenerg Biomembr. 1984;16:441–464. doi: 10.1007/BF00743238. [DOI] [PubMed] [Google Scholar]
- 25.Mascioni A, Karim C, Barany G, Thomas D D, Veglia G. Biochemistry. 2002;41:475–482. doi: 10.1021/bi011243m. [DOI] [PubMed] [Google Scholar]
- 26.Asahi M, McKenna E, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 2000;275:15034–15038. doi: 10.1074/jbc.275.20.15034. [DOI] [PubMed] [Google Scholar]
- 27.Morikami K, Nakai T, Kidera A, Saito M, Nakamura H. Comput Chem. 1992;16:243–248. [Google Scholar]
- 28.Minamisawa S, Wang Y, Chen J, Ishikawa Y, Chien K R, Matsuoka R. J Biol Chem. 2003;278:9570–9575. doi: 10.1074/jbc.m213132200. [DOI] [PubMed] [Google Scholar]
- 29.Toyoshima C, Nomura H. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 30.Zhang P, Toyoshima C, Yonekura K, Green N M, Stokes D L. Nature. 1998;392:835–839. doi: 10.1038/33959. [DOI] [PubMed] [Google Scholar]