Abstract
Many operons in Gram-positive bacteria that are involved in methionine (Met) and cysteine (Cys) biosynthesis possess an evolutionarily conserved regulatory leader sequence (S-box) that positively controls these genes in response to methionine starvation. Here, we demonstrate that a feed-back regulation mechanism utilizes S-adenosyl-methionine as an effector. S-adenosyl-methionine directly and specifically binds to the nascent S-box RNA, causing an intrinsic terminator to form and interrupt transcription prematurely. The S-box leader RNA thus expands the family of newly discovered riboswitches, i.e., natural regulatory RNA aptamers that seem to sense small molecules ranging from amino acid derivatives to vitamins.
Modulation of the structures of leader transcripts is a common mode of regulation of synthetic and metabolic operons in prokaryotes. Switching between two alternative RNA structures, with one causing transcription termination, is made in response to some external event, e.g., ribosome pausing (in Gram-negative bacteria), binding of a regulatory protein, or tRNA (in Gram-positive bacteria) (reviewed in ref. 1). Recently, three regulons responsible for vitamin biosynthesis have been shown to use flavinmononucleotide, thiamin pyrophosphate (TPP), and adenosyl-cobalamin to directly bind their cognate leader RNAs, changing their structures and functions (2–5). Such leader RNA aptamers were called riboswitches to reflect their unique ability to switch between two conformations in response to binding of a small molecule without the help of proteins (3, 5, 6). The evolutionarily conserved sequences encoding vitamin-sensing riboswitches have been found in the genomes of many Gram-positive and Gram-negative bacteria (7–10), and also in some archaea (10), fungi, and plant species (A.S.M. and E.N., unpublished observations). In this work, we tested the hypothesis that riboswitches are not just a peculiar attribute of vitamin biosynthetic operons but are more common in nature and are also involved in control of amino acid biosynthesis.
Grundy and Henkin (11) found that at least 11 transcription units in Bacillus subtilis that are mostly involved in Cys and Met synthesis possess a leader element that includes an intrinsic transcription terminator, competing antiterminator, and a conserved element (S-box) that functions as an anti-antiterminator. Based on their genetic and physiological studies, the authors postulated that the S-box serves as a target for repression during growth in the presence of Met (11, 12). Northern blot experiments substantiated a model of S-box gene regulation by transcription antitermination in response to Met (13). It has also been shown that overexpression of S-adenosyl-methionine (SAM) synthase leads to Met auxotrophy in B. subtilis, suggesting that SAM is an effector of Met biosynthesis in this bacteria (14). This notion was corroborated recently. The transcription profile of S-box genes in several Met auxotroph strains of B. subtilis was analyzed by using DNA arrays. The results argued that S-box regulon was repressed in the presence of Met most likely in response to SAM (15).
The mechanism of Met or SAM sensing, however, is unknown. The striking similarity in overall organization of the S-box leader to the recently characterized thiamin (thi-box) and riboflavin (rfn-box) riboswitches (4, 5) and the apparent absence of a protein candidate for its regulation prompted us to test whether the S-box leader RNA also functions as a riboswitch. Using a minimal reconstituted transcription system lacking any additional factors, we demonstrate that SAM, but not other metabolites, directly binds S-box RNA to stabilize its anti-antitermination structure, thus causing termination of the leader transcript.
Experimental Procedures
Templates and Proteins.
The metE transcription template (Fig. 1A) was generated by PCR from genomic DNA of the B. subtilis strain RK99 (lysA42) by using the “forward” 5′-attaagcttatataatatacaccacatcatg and “reverse” 5′-tttgatggatccattgtttctcctccc oligos. For the truncated template used in the experiment of Fig. 3, the reverse oligo was 5′-tatatagaattccccatgagagaaag. The thi template was generated by PCR with the following primers: 5′-aggctgaaacttggagaccg (forward) and 5′-ttctgaaggatccacaaatcattccc (reverse). Mutations in the S-box leader were introduced by PCR by using the following oligos: 5′-ggtcaaaaggccaatgtgatgcccaactctcgata for the “56–60” template and 5′-gtcaaacactgcctcatagatctctcagacac for the “188–191” template. The amplified DNA products were resolved in a 1.2% (wt/vol) NuSieve GTG agarose gel (BMA Biomedicals) followed by electroelution, extraction with phenol/chloroform, and precipitation with ethanol. The DNA pellet was diluted in 50 μl of TE buffer (10 mM Tris⋅HCl, pH 7.9/1 mM EDTA). The final concentration of DNA was ≈2 pmol/μl as determined by spectrometry. The B. subtilis His-6-β′-tagged RNAPσA was purified according to Anthony et al. (16).
Figure 1.
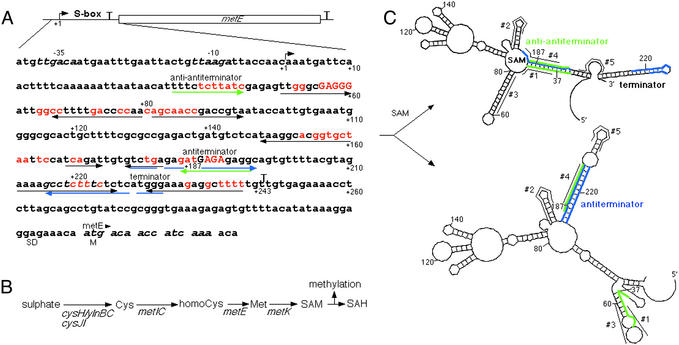
Model for the SAM-mediated attenuation control of the S-box regulon. (A) S-box leader region of the metE gene. Numbers indicate positions beginning at the +1 start of transcription. The S-box is defined as a region that includes the evolutionarily conserved bases shown in red (11). Mutations that were analyzed in detail in this work are capitalized. Blue arrows show the antiterminator and green arrows the anti-antiterminator. Black arrows show potential stem-loops including that of the terminator. The termination points are indicated by “T.” SD, Shine-Dalgarno sequence; M, the first Met of metE gene. (B) The pathway of sulfate assimilation and Met and SAM biosynthesis. Only S-box family genes that are repressed by SAM are indicated (11, 15). The function of these genes is as follows: ylnB, ATP sulfurylase; ylnC, adenosine phosphosulfate kinase; cysH, 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase; cysJI, NADH-sulfite reductase; metI, cystathionine γ-synthase/O-acetylhomoserine sulfhydrylase; metC, cystathionine β-lyase; metK, SAM synthetase. Various methyl transferases convert SAM to SAH during methylation. (C) SAM-directed terminator/antiterminator riboswitch. Two alternative structures of S-box, the anti-antiterminator (Upper) and antiterminator (Lower), were calculated by using the Zuker–Turner algorithm of free energy minimization (18). The antiterminator sequence is in blue and anti-antiterminator in green. The program predicts the anti-antiterminator structure by default and the antitermination structure by forcing the complementary “blue” bases to be paired. According to the model, the anti-antiterminator structure folds only in the presence of SAM. Numbers indicate the positions in the RNA in relation to the 5′ terminus (+1). These reference numbers are also shown in (A). #1–#5, the antisense DNA oligos that were used to probe the structure of S-box in the experiment of Fig. 3. The oligos are shown next to their annealing targets.
Figure 3.
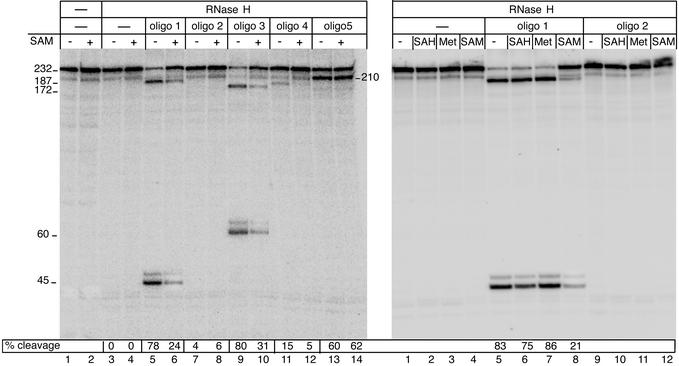
The effect of SAM on the structure of the pure S-box RNA. RNA structural transition is monitored by using the antisense DNA oligonucleotides and RNase H (5). The truncated S-box-leader template lacking the terminator was used to obtain the 232-nt transcript. Its 5′ proximal region was 32P-labeled (see Experimental Procedures). The transcript was extracted from the transcription reaction with phenol/chloroform, followed by ethanol precipitation. The pure RNA was dissolved in TB with or without SAM. The mixture was then heated to 80°C for 5 min and slowly cooled to room temperature. The changes in RNA structure during refolding in the presence of SAM (5 μM) were monitored by annealing oligos 1–5, followed by RNase H cleavage. As the radiogram shows, the sensitivity of refolded RNA to RNase H was greatly suppressed by SAM (Left, lanes 6 and 8) but not by Met (100 μM) or SAH (100 μM; Right, lanes 6 and 7). This experiment indicates that the formation of a specific binary complex between the S-box RNA and SAM that triggers the conformational change in the leader RNA occurs during S-box RNA folding, independently of other components of the elongation complex. Numbers on the right and left indicate the size of the RNase H cleavage products. The annealing sites for oligos are shown in Fig. 1C.
In Vitro Transcription.
The start-up elongation complex on the S-box leader templates was prepared as follows: 2 pmol DNA (1 μl) and 1 pmol RNA polymerase (RNAP; 1 μl) were mixed in 10 μl of transcription buffer (TB; 150 mM KCl/3 mM MgCl2/10 mM Tris⋅HCl, pH 7.8) and incubated 5 min at 37°C. Next, 1 μl of the starting mixture was added to give the final concentration of CpApA primer (Oligos Etc., Guilford, CT) of 20 μM, ATP and GTP of 30 μM, and [α-32P]UTP [3,000 Ci/mmol (1 Ci = 37 GBq)] of 0.3 μM. After 5 min at 37°C, 5 μl of TB-preequilibrated Ni-agarose beads (Qiagen, Valencia, CA) were added and incubated at 25°C for 5 min, followed by multiple washings of the beads with high salt TB (1 mM KCl) and equilibration with TB. Aliquots were then taken to tubes containing all four NTPs (100 μM final concentration). The chase reactions were stopped after ≈15 min at 37°C by adding an equal volume of sequencing gel loading buffer containing EDTA (200 mM) and formamide (95%). Transcription on the thi template was performed essentially as described above, except that the primer for the start-up complex was GpGpU and [α-32P]CTP was used instead of UTP. Positions of the terminated products were determined by the transcription sequencing reaction with 3′-dNTPs. Synthesis of truncated S-box leader RNA for the RNase H experiment of Fig. 3 was performed in solution without Ni-agarose beads. Two picomoles of DNA (1 μl) and 1 pmol of RNAP (1 μl) were mixed in 10 μl of TB and incubated 5′ at 37°C. Transcription was initiated with 20 μM CpApA primer; 30 μM ATP, GTP, and UTP; and 0.3 μM [α-32P]CTP (3,000 Ci/mmol) for 1 min at 37°C, followed by the chase reaction with all NTPs (300 μM) at room temperature for 5 min. During the pulse reaction, the 5′ portion (≈150 nt) of the transcript was uniformly radiolabeled. The transcript was then purified as described in the legend of Fig. 3. The antisense oligos used in this experiment were as follows: 1, 5′-tctcgataag; 2, 5′-cacaatctga; 3, 5′-gttggggtca; 4, 5′-ctctctcatc; 5, 5′-ttctacgtaa. Relative amounts of [32P]RNA were determined by using PhosphorImager and software from Molecular Dynamics. Exposed x-ray films (Kodak) were processed by using an Epson Expression 636 scanner and multianalyst software from Bio-Rad. The efficiency of termination (%T) was calculated by dividing the amount of radioactivity in a particular terminated band by the total radioactivity present in that and all read-through bands. Met, SAM, Cys, S-adenosyl-homocysteine (SAH), and TPP were purchased from Sigma/Aldrich. RNase H (Roche Applied Sciences) experiment was performed as in ref. 17.
Results
SAM-Dependent Transcription Termination.
We developed a minimal in vitro system to study transcription regulation of S-box genes. The effect of Met and other related small molecules on termination was monitored by a single-round run-off assay with a highly pure 6His-tagged RNAP from B. subtilis and a PCR-generated template containing the metE (methionine synthase gene; Fig. 1B) promoter followed by a complete S-box leader sequence (Fig. 1A). This leader sequence has a classical intrinsic transcription terminator (a stable hairpin followed by a stretch of U residues) and an upstream region, the antiterminator (shown by the blue arrow), that is complementary to the ascending portion of the termination hairpin. Annealing of the antiterminator to the left shoulder of the terminator would prevent hairpin folding and thus inhibit termination. In addition, the potential anti-antiterminator sequence (shown by the green arrow), which is fully complementary to the antiterminator, is located 5′ proximal to the antiterminator. We used a Zucker–Turner algorithm (18) to calculate two stable mutually exclusive structures of S-box leader RNA with similar thermodynamic characteristics (Fig. 1C). In one case, the antiterminator sequesters the left shoulder of the terminator; alternatively, the anti-antiterminator prevents the antiterminator from interfering with the terminator.
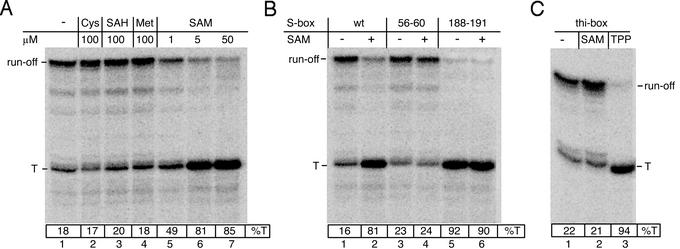
The RNA transcript was 32P-labeled near its 5′ end during formation of the initial elongation complex, which was stalled at position +10. The complex was immobilized on Ni2+-agarose beads, washed with transcription buffer (TB; 150 mM KCl/3 mM MgCl2/10 mM Tris⋅HCl, pH 7.8), and then chased to the S-box leader terminator by adding all four substrate NTPs to 100 μM each. Under these conditions, transcription was partially terminated at positions +243–245, corresponding to the downstream portion of the T-stretch of the terminator. The %T was only ≈18%. The presence of 100 μM Met, Cys, or SAH had no effect on termination. However, addition of 5 μM SAM to the chase reaction increased %T to >80% (Fig. 2A). SAM, even at much higher concentrations, failed to potentiate termination of the thi-box leader transcript (Fig. 2C). Furthermore, point substitutions of the conserved bases of the S-box element abolished the stimulating effect of SAM on termination (Fig. 2B, lanes 3 and 4). Substitutions in the potential antiterminator induced termination to the level that occurred on the original template in the presence of SAM (Fig. 2B, lane 5). SAM was unable to increase termination further (Fig. 2B, lane 6). Taken together, these results indicate that SAM acts as a highly specific, S-box-dependant termination factor; i.e., it prevents antitermination by stabilizing the anti-antitermination configuration of S-box (Fig. 1C).
Figure 2.
SAM-mediated transcription termination in vitro. (A) Effect of small molecules on termination by B. subtilis RNAP. The autoradiogram of the 6% sequencing PAGE shows 32P-labeled full size (run-off) and terminated (T) RNA products from a reconstituted single round transcription reaction (see Experimental Procedures). T, terminated transcript; SAM, Met, Cys, and SAH were added to the final concentrations indicated at the top. (B) Effect of S-box mutations on termination. Mutations are as follows: “56–60,” G56A, A57T, G58C, G59A, G60C; and “188–191” (the antiterminator), G188C, A189T, G190A, and A191T. The concentration of SAM was 5 μM. (C) Effect of SAM and TPP on termination of the thi-box leader transcript. The autoradiogram of the 6% sequencing PAGE shows 32P-labeled full size (run-off) and terminated (T) RNA products from the reconstituted single round transcription reaction (see Experimental Procedures). TPP and SAM were added to 50 μM.
Direct Sensing of SAM by S-Box RNA.
To confirm that the S-box functions as a SAM-dependent riboswitch, i.e., it changes its structure in response to SAM binding, as shown in the model of Fig. 1C, we purified the S-box RNA by phenol/chloroform extraction, denatured, and then renatured it in the presence or absence of SAM. The pure RNA contained the entire S-box leader sequence except the terminator. The RNA structure was hybridized to short (10 nt) DNA oligos complementary to different parts of the leader sequence (Fig. 1C), followed by treatment with RNase H, which specifically cleaves RNA:DNA heteroduplexes. Oligo 1 is complementary to the anti-antiterminator (positions +39–48); oligos 2 and 3, to conserved regions of S-box, +167–176 and +70–79, respectively; oligo 4, to the antiterminator (positions +185–194); and oligo 5, to the nonconserved region +203–212. As Fig. 3 shows, the leader RNA became less available for oligos 1 and 3 to anneal if SAM was present during the RNA refolding process (lanes 5, 6 and 9, 10). Met and SAH had no effect on RNase H sensitivity (Fig. 3 Right). These results provide direct evidence that the specific binary complex between the S-box RNA and SAM forms during RNA folding, independently of other components of the transcription reaction (RNAP or DNA), and that such a complex changes the folding pathway of the RNA. The results with oligos also support the structural model of Fig. 1C. Positions complementary to oligos 1 and 3 are mostly base-paired in the anti-antitermination structure and unavailable for annealing, whereas the alternative antitermination structure maintains many of these bases unpaired, making them easily accessible for the oligos to anneal. Alternatively, positions complementary to oligo 2 are involved in the same stable hairpin in both variants and should not be accessible to this oligo. This result is confirmed by the absence of RNase H cleavage products with or without SAM (Fig. 3, lanes 7 and 8). According to the structural model of Fig. 1C, oligo 4 should also be poorly accessible for base pairing. Indeed, in both cases, the antiterminator must be fully sequestered by either the anti-antiterminator or terminator. Again, this prediction was consistent with the RNase H results (lanes 11 and 12). Finally, oligo 5 rendered the transcript sensitive to RNase H to the same extent with or without SAM. This result also corroborates the Fig. 1C model, which predicts that oligo 5 is complementary to the weakly structured segments in both variants. Although these experiments prove the existence of a binary complex between SAM and the S-box RNA, which stabilizes its anti-antitermination configuration, it remains to be determined how exactly SAM binds to the RNA and what features of SAM and the RNA are essential for binding.
Discussion
At least 60 transcription units from a variety of bacteria species have been found to be members of the S-box regulon (19). Most of these genes are directly involved in sulfur metabolism. This family was characterized by the presence of the evolutionarily conserved regulatory leader sequence (S-box). An unknown factor was postulated to acquire an ability to interact with the S-box RNA on binding to Met and attenuate transcription of downstream genes (12). The present study demonstrates that the S-box RNA from B. subtilis functions as a SAM-dependent riboswitch; i.e., it directly senses the level of SAM, a metabolite of Met. The attenuation mechanism relies on binding of SAM to the nascent S-box transcript leading to stabilization of its anti-antitermination structure, which in turn allows the formation of the stem-loop of the attenuator and early transcription termination (Fig. 1C). Previous mutational analysis in vivo (11) and the present in vitro studies (Fig. 2B) confirm the anti-antitermination/antitermination model. Mutations that disrupted the potential terminator or anti-antiterminator resulted in constitutive high-level expression of the S-box genes. Alternatively, mutations that interfered with the potential antiterminator structure resulted in uninducible, low-level expression (11). In addition, mutations of conserved bases of the S-box that are not directly involved in sequestration of the antiterminator also resulted in high level constitutive expression (11) and weak termination in the presence of SAM (Fig. 2B), which is consistent with the role of those bases in direct binding of SAM. Finally, recent in vivo analysis of the metIC operon revealed that its induction is independent of the promoter located upstream of the S-box motif (13). The Northern blot analysis showed two transcripts: a small one corresponding to early termination at the end of the S-box leader sequence and a large one corresponding to the transcript of the entire metIC operon. The ratio between the two transcripts depended on Met availability (13).
Interestingly, in Escherichia coli, the regulation of Met and Cys biosynthesis also depends on SAM concentration although the mechanism of regulation is different. Here, SAM controls transcription initiation rather than elongation. MetJ repressor binds SAM to acquire its ability to bind Met box sequence present in the promoter regions of the met genes (20, 21).
Our reconstituted S-box transcription system contains only one protein, the RNAP, which does not bind SAM. Although, SAM potentiates termination almost 6-fold, making it >80% (Fig. 2), it is possible that additional factors (e.g., NusA) may stabilize the SAM-RNA complex and thus further improve the efficiency of S-box-mediated transcription attenuation in vivo. The RNA refolding experiment of Fig. 3 provides direct evidence for the formation of the SAM–S-box RNA binary complex. It also supports the computer generated structural model of S-box leader RNA of Fig. 1C. It is likely that the SAM–S-box RNA complex assembles contranscriptionally during the early phase of the S-box structure formation and exists for a brief period. As transcription proceeds to the terminator, this time window should be sufficient for selection of the appropriate folding pathway, either anti-antitermination or antitermination, that the RNA would follow. The specificity of the SAM–S-box complex is exceptional. Met or SAH have no effect on S-box structure and function (Fig. 2A). Furthermore, TPP, but not SAM, could stimulate thi-box-dependant termination (Fig. 2C; ref. 5), and single substitutions in S-box-conserved regions completely abolished the effect of SAM on termination (Fig. 2B).
SAM is an essential coenzyme in all organisms. It is synthesized directly from Met by SAM synthetase (Fig. 1B) and serves as a source of methyl groups for protein and nucleic acid modification. The basic Met and Cys synthetic pathways (Fig. 1B) are preserved in evolution, suggesting that analogous riboswitches may control sulfur metabolism in many bacteria species and possibly in higher organisms as well.
The S-box RNA examined seems to be capable of specific binding of SAM to adopt the structure of the intrinsic terminator. It is thus the fourth riboswitch described to date (3–5). Each riboswitch represents a distinct RNA sequence that can adopt two structural arrangements, one of which has a transcription terminator or inhibitor of translation initiation. They all seem to recognize a unique small molecule (TPP, flavinmononucleotide, B12, and SAM), suggesting that the realm of natural riboswitches and their roles in gene regulation could be vast. The presence of potential riboswitch sequences in eukaryotic genomes (A.S.M. and E.N., unpublished observation) is consistent with this notion. Considering the numerous metabolites and regulatory small molecules that are present in any type of living cell, the exciting possibility arises that the known riboswitches are just a few examples in the yet unexplored large array of regulatory RNA sensors.
Acknowledgments
This work was supported by National Institutes of Health Grant GM58750 and Fogarty International Collaboration Research Award TW06122.
Abbreviations
- TPP
thiamin pyrophosphate
- SAM
S-adenosyl-methionine
- RNAP
RNA polymerase
- %T
efficiency of termination
- SAH
S-adenosyl-homocysteine
References
- 1.Henkin T M, Yanofsky C. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 2.Nou X, Kadner R J. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nahvi A, Sudarsan N, Ebert M S, Zou X, Brown K L, Breaker R R. Chem Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 4.Winkler W, Nahvi A, Breaker R R. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 5.Mironov A S, Gusarov I, Rafikov R, Lopez L E, Shatalin K, Kreneva R A, Perumov D A, Nudler E. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 6.Weisberg R A, Storz G. Mol Cell. 2002;10:1266–1268. doi: 10.1016/s1097-2765(02)00788-8. [DOI] [PubMed] [Google Scholar]
- 7.Gelfand M S, Mironov A A, Jomantas J, Kozlov Y I, Perumov D A. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 8.Miranda-Rios J, Navarro M, Soberon M. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitreschak A G, Rodionov D A, Mironov A A, Gelfand M S. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodionov D A, Vitreschak A G, Mironov A A, Gelfand M S. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 11.Grundy F J, Henkin T M. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 12.Grundy F J, Henkin T M. Front Biosci. 2003;8:D20–D31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- 13.Auger S, Yuen W H, Danchin A, Martin-Verstraete I. Microbiology. 2002;148:507–518. doi: 10.1099/00221287-148-2-507. [DOI] [PubMed] [Google Scholar]
- 14.Yocum R R, Perkins J B, Howitt C L, Pero J. J Bacteriol. 1996;178:4604–4610. doi: 10.1128/jb.178.15.4604-4610.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auger S, Danchin A, Martin-Verstraete I. J Bacteriol. 2002;184:5179–5186. doi: 10.1128/JB.184.18.5179-5186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anthony L C, Artsimovitch I, Svetlov V, Landick R, Burgess R R. Protein Expression Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- 17.Gusarov I, Nudler E. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M, Mathews D H, Turner D H NATO ASI Series, editors. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark B F C, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]
- 19.Murphy B A, Grundy F J, Henkin T M. J Bacteriol. 2002;184:2314–2318. doi: 10.1128/JB.184.8.2314-2318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saint-Girons I, Belfaiza J, Guillou Y, Perrin D, Guiso N, Barzu O, Cohen G N. J Biol Chem. 1986;261:10936–10940. [PubMed] [Google Scholar]
- 21.Green R C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 542–560. [Google Scholar]