Abstract
The membrane-bound glycoprotein dipeptidyl peptidase IV (DP IV, CD26) is a unique multifunctional protein, acting as receptor, binding and proteolytic molecule. We have determined the sequence and 1.8 Å crystal structure of native DP IV prepared from porcine kidney. The crystal structure reveals a 2-2-2 symmetric tetrameric assembly which depends on the natively glycosylated β-propeller blade IV. The crystal structure indicates that tetramerization of DP IV is a key mechanism to regulate its interaction with other components. Each subunit comprises two structural domains, the N-terminal eight-bladed β-propeller with open Velcro topology and the C-terminal α/β-hydrolase domain. Analogy with the structurally related POP and tricorn protease suggests that substrates access the buried active site through the β-propeller tunnel while products leave the active site through a separate side exit. A dipeptide mimicking inhibitor complexed to the active site discloses key determinants for substrate recognition, including a Glu–Glu motif that distinguishes DP IV as an aminopeptidase and an oxyanion trap that binds and activates the P2-carbonyl oxygen necessary for efficient postproline cleavage. We discuss active and nonactive site-directed inhibition strategies of this pharmaceutical target protein.
Keywords: serine protease‖oxyanion hole‖substrate channeling‖drug design‖diabetes mellitus
Dipeptidyl peptidase IV (DP IV) is a multifunctional type II transmembrane glycoprotein with a broad tissue distribution (1). It is identical to the T cell activation antigen CD26 (2) and to the adenosine deaminase (ADA) binding protein (3). The porcine and human proteins have an identical length of 766 aa. Soluble DP IV migrates as a homodimer with a molecular weight range of 210–290 kDa (4), but can form higher molecular weight assemblies migrating as 900-kDa complexes (5). DP IV is anchored to the membrane by a single hydrophobic helix with a short N-terminal cytoplasmic extension. A flexible stalk links the membrane anchor with a highly glycosylated and cysteine-rich domain. The C-terminal serine protease domain is homologous to α/β-hydrolases. Dimerization was found to be a prerequisite for enzyme activity (6), albeit enzymatically active heterodimers with the fibroblast activation protein α (FAP α) are observed (7).
DP IV is a serine aminopeptidase that cleaves preferentially Xaa–Pro dipeptides from oligopeptides with typical length of <30 aa, but tolerates other small residues at P1-position (8). DP IV specifically processes regulatory peptides, leading to their biological activation or inactivation (9). This mechanism relates to its role in diseases like diabetes mellitus (10–14), obesity (15), tumor growth (16–18), and HIV infection (19).
Despite the enormous data store available on DP IV, important questions remain open including the structural basis of substrate selectivity and specificity; substrate access to and product egress from the active site; the role of the dimerization and higher molecular weight assemblies; and the interaction with additional components. To address these and other questions, we set out to determine the crystal structure of natively prepared DP IV which we present in this work.
Materials and Methods
Preparation of DP IV.
DP IV was purified from 2-kg cortex of pig kidney. The procedures included tissue homogenization, membrane autolysis, differential centrifugation, ammonium sulfate precipitation, size exclusion chromatography, and ion exchange chromatography similar to methods referred to in ref. 8. Finally, naturally glycosylated protein purified by preparative isoelectric focusing using the Rotofor system (Bio-Rad). One run yields ≈60 mg of ≈280-fold purified protein with a specific activity of >42 units/mg.
Sequencing of Porcine DP IV cDNA.
To obtain the cDNA sequence of porcine DP IV, total RNA was extracted from porcine kidney, and RT-PCR was performed as described elsewhere (J. Bär, T.H., L.W., and H.-U.D., unpublished data). The sequence was submitted to GenBank (accession no. AY198323).
Inhibitor Synthesis.
A tight-binding DP IV-inhibitor, p-Iodo-Phe-Pyr-CN *TFA, was synthesized according to known chemical protocols (20). Synthesis was carried out starting from Boc-p-Iodo-Phe-OH and H-Pro-NH2*HCl. The final 1-(2-(S)-cyanopyrrolidin-1-yl)-3-(4-iodophenyl)-1-oxopropan-2-aminium trifluoroacetate was obtained as a white solid. The structure was confirmed by 1H-NMR, 13C-NMR, and electrospray ionization (ESI)-MS. The chemical purity was verified by using HPLC on RP-18 material using a water/ACN gradient. Mp: 138–140°C (uncorrected); ESI-MS: cal 369.0, found (M+H)+ = 370.0; 1H-NMR: (D2O), d (ppm): 1.55–1.61 (m, 1H), 1.7–1.82 (m, 1H), 1.91–2.19 (m, 2H), 2.49–2.62 (m, 1H), 2.89–3.09 (m, 1H), 3.19–3.21 (m, 1H), 3.21–3.34 (m, 1H), 4.31–4.39 (m, 1H), 4.61–4.69 (m, 4H), 6.91–7.00 (m, 2H), 7.60–7.71 (m, 2H); 13C-NMR: (D2O), d (ppm); 167.832, 131.656, 118.055, 93.173, 65.934, 52.250, 47.061, 46.428, 36.322, 29154, 24.063.
DP IV-Activity Determination and Inhibition.
DP IV-activity was determined as in ref. 8. Inhibition constants of the tight-binding inhibitor p-Iodo-Phe-Pyr-CN were determined according to ref. 21 and the activity assay as described above. The overall Ki value of the inhibitor was 25.1 ± 0.9 nM.
Crystallization and Crystal Transformation.
Triclinic crystals were obtained at room temperature within several days by mixing equal volumes of protein at a concentration of 20 mg/ml with the reservoir solution (20–22% PEG 2000/0.1 M ammonium sulfate/0.1 M Tris⋅HCl, pH 8.0) by using the sitting drop vapor diffusion method. We covered the crystallization drop with perfluoropolyether (PFPE) oil and harvested the crystals by using a loop with humidity control (22). The relative humidity was ramped down from 96.5% to 86.5% by using a gradient of 0.5% (150 s−1), leading to a dramatically improved diffraction pattern, typically from below 10 Å to 3 Å. At an optimal relative humidity crystals were frozen for data collection. For ligand complex studies, DP IV-crystals were soaked with the inhibitor before the crystal transformation procedure. Data were processed and scaled by using denzo and scalepack (23).
Structure Determination.
The structure was determined by multiple wavelength anomalous dispersion using a mercury derivative and subsequent noncrystallographic symmetry (NCS) averaging (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Details of the method will be described elsewhere (H.B., unpublished data). Briefly, a local Harker section perpendicular to the molecular dimer axis (program glrf; ref. 24) was cut out of the three-dimensional anomalous Patterson map (program main; ref. 25), averaged along the orthogonal local two-fold axes, and subsequently input to rsps (26) for automatic local doublet sites detection. This enhancedthe signal to noise ratio about 50- to 100-fold. The relative position of the two symmetry-related Hg-doublets was determined by translational search (26). By construction, the resulting sites follow the local symmetry and determine the translational NCS parameters. After heavy atom refinement and phasing (program mlphare) and solvent flipping (solomon) (27, 28), phases were extended to 2.0-Å resolution by NCS averaging (main; ref. 25), which rendered the electron density readily interpretable (Table 1 and Fig. 6, which are published as supporting information on the PNAS web site).
Model Building and Refinement.
We placed the catalytic domain of POP in the electron density (main; ref. 25) which served to jump start model building and sequence assignment. The model was refined by using the program cns (29) with current R values of 21.7% (working set) and 24.9% (test set) and deviations from ideality of 0.008 Å (bond length) and 1.4° (angle deviation).
Results
Sequence of the Porcine DP IV.
Sequence comparison of the porcine DP IV with the human DP IV-sequences reveals a very high degree of sequence conservation with no sequence insertion or deletion (Fig. 1). The overall sequence identity between these two species is 88%. Significant particularities of the porcine sequence are detailed further in the text.
Figure 1.
Alignment of the newly determined porcine DP IV sequence with those of human and mouse DP IV as well as with the related FAP-α and prolyl oligopeptidase. The comparison rationalizes the lack of dimerization of POP and correlates the lack of ADA binding in rodents with the glycosylation site Asn-279 (281).
Overall Structure and Subunit Assembly.
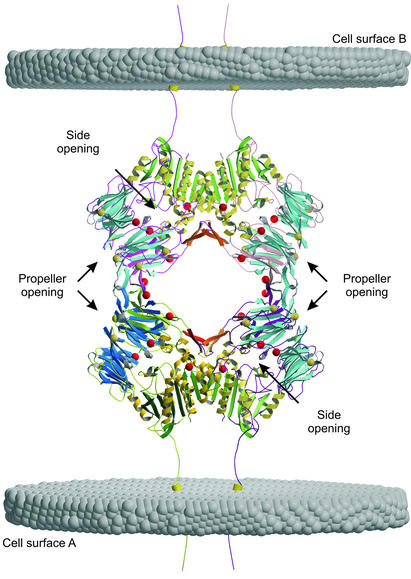
The monomer comprises an N-terminal β-propeller domain (Arg-54–Asn-497) followed by the catalytic domain Gln-508–Pro-766. Notably, the crystal structure reveals a dimer of dimers in the crystallographic unit cell obeying 222 symmetry with all axes intersecting (Fig. 2). Propeller blade IV is involved in both the dimerization and tetramerization interaction (Fig. 3). The more extensive dimer contact is dominated by hydrophobic residues of the catalytic domain, including Trp-734 and Phe-713, with a contact area of 2,270 Å2 versus 2 × 570 Å2 = 1,140 Å2 of the dimer-to-tetramer interface.
Figure 2.
Soluble DP IV forms a 222 symmetric assembly as a dimer of dimers. The view is along one two-fold axis. Potential glycosylation sites are indicated as gray spheres, and red spheres are the sites modified in our crystal structure. The transmembrane helices and their orientation to the membrane were modeled to illustrate how tetramerization of DP IV can mediate cell–cell contacts. The figures were prepared by using the programs MAIN (25), MOLSCRIPT (55), and RASTER3D (56).
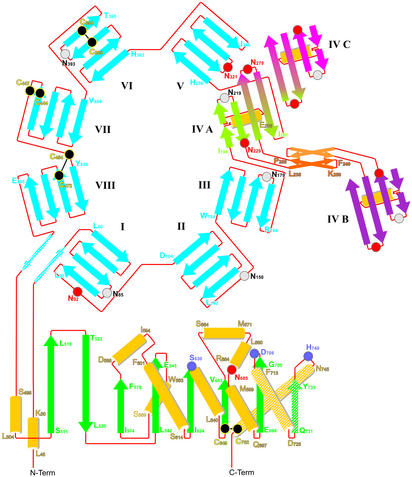
Figure 3.
Topology diagram illustrating the domain structure of DP IV. Blade IV of the propeller is involved in both the dimer contact (IV A–IV B: L235-P255, together with the highlighted C-terminal three secondary structure elements F713-C762) and the tetramerization of DP IV (IV A–IV C and IV B–IV C, not shown).
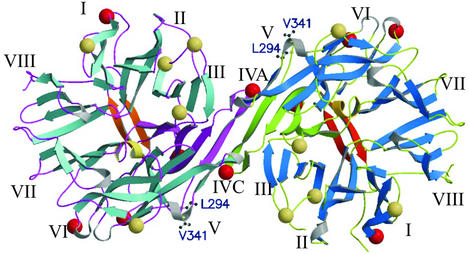
The tetramer interface is more hydrophilic. In its center, the strands Asn-279–Gln-286 of each DP IV-dimer form an antiparallel β-sheet, thus extending the propeller blade IV to an eight-stranded antiparallel sheet (Fig. 4). The outer strands of blade V additionally contribute to the tetrameric assembly Significantly, Asn-279 is located at the tetramerization interface and is glycosylated (Figs. 2 and 4).
Figure 4.
View along the two-fold axis on the tetramerization interface. Blades IV of each subunit align to form an eight-bladed antiparallel β-sheet. The highlighted Leu-294 and Val-341 are involved in ADA binding.
Subdomain Structure.
The β-propeller.
The N-terminal β-propeller domain contains eight blades with four antiparallel strands each. The first and the last blade of a regular β-propeller are clamped together either covalently by disulfide bond formation (four-bladed β-propellers) or by strand exchange between the first and last blade (five- to eight-bladed propellers). So far there are three exceptions to this closed propeller topology rule, namely the seven-bladed β-propeller of POP (30), the seven- and six-bladed propellers of the tricorn protease (31), and the five-bladed propeller of α-l-arabinase 43A (32). The β8-propeller of DP IV is also irregular with an open Velcro-type topology, because no segment C-terminal to blade VIII interacts with the first propeller blade. Interestingly, however, the N-terminal extension of blade I (Phe-53–Tyr-58) tightens up the propeller structure by interacting with the immediate C-terminal extension to blade VIII (Glu-499–Met-503). A similar, yet shorter, external clamp has been described for the β7-propeller of the tricorn protease (31).
With the exception of Cys-649–Cys-762 all disulfide bonds are located in the β-propeller domain where they form intrablade stabilizing crosslinks (Fig. 3). All glycosylation sites but Asn-685 are located on the β-propeller. Five of 10 total potential glycosylation sites cluster at the surface oriented away from the catalytic domain, as detailed in Figs. 2 and 3. Intriguingly, of these five potential glycosylation sites only Asn-279, which is involved in tetramerization, is posttranslationally modified. Further glycosylated residues are found on blade I (Asn-92 at the end of strand 4), blade IV (Asn-229 close to the tetramerization motif), and blade V (Asn-321 on the loop connecting strand 2 and 3) (Fig. 2 and 3). Asn-150 on the exit strand of blade II is not modified in our crystals.
The shape of the DP IV β8-propeller is asymmetric where blades VI, VII, VIII, and I, and blades II, III, IV, and V form more compact subdomains, respectively (Figs. 2 and 3), consistent with a three domain organization of DP IV (5). The ellipsoidal tunnel through the propeller is continuously open. At the solvent-exposed opening, its diameter measures 9 and 15 Å from blade IV to VIII and from blade II to VI, respectively. The tunnel widens toward the catalytic domain with diameters of 15 and 25 Å. By its dimensions, the tunnel allows for direct passage of an extended peptide or a hairpin loop, but not for a folded α-helix. Remarkably, a sulfate is bound to an oxyanion pocket formed by the amide nitrogens of Glu-361 and Ile-407 and Nɛ2 of His-363.
Catalytic domain.
The catalytic domain spans residues Gln-509 to Pro-766. It adopts a typical α/β-hydrolase fold with a central eight-stranded β-sheet sandwiched by several α-helices, Figs. 2 and 3. The β-sheet exerts a significant twist of more than 90°, in line with observations on related α/β-hydrolases (33–35).
Within the catalytic domain a single disulfide bond Cys-649–Cys-762 crosslinks the C-terminal helix Met-746–Ser-764 with the β-sheet, thus stabilizing its tertiary arrangement. Helix Met-746–Ser-764, together with helix Gln-714–Asp-725 and strand Asp-729–Thr-736 from the C-terminal region, constitute the central dimerization motif, which is further stabilized by blade IV of the β-propeller.
The catalytic Ser-630 is embedded in the surrounding secondary structure framework where it participates both in the preceding strand 4 (Arg-623–Trp-629) as well as in the following helix Tyr-631–Ala-642. This causes a strained backbone conformation of the active site Ser-630 with dihedral angles (φ, ψ) = (61.4, −115.7), presumably providing an energy reservoir for catalysis (35).
Active Site and Substrate Recognition.
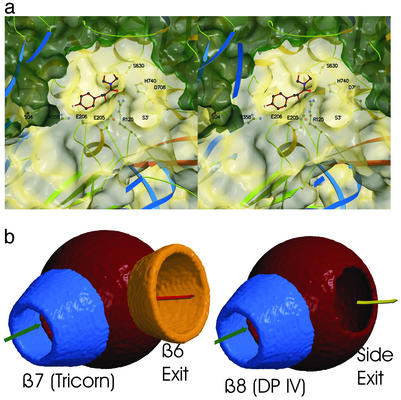
The sequential and three-dimensional arrangement of the catalytic residues Ser-630, His-740, Asp-708 corresponds to that of related α/β-hydrolases (33, 35, 36). The oxyanion hole is formed by the amide Tyr-631 and the hydroxyl Oη of Tyr-547 is occupied by a water molecule in the uninhibited structure. To detail the exact mechanism of substrate recognition, we determined the structure of a dipeptide mimetic, the iodinated Phe-cyanopyrrolidide inhibitor in complex with DP IV (Fig. 5a). The active site nucleophile, the hydoxyl residue of Ser-630, forms a covalent bond with the scissile carbonyl carbon of the inhibitor resulting in a stable carbaminic acid adduct. The pyrrolidine ring is accommodated by a hydrophobic pocket formed by side chains of Tyr-666, Tyr-662, Val-711, Val-656, and Trp-659. Although this environment is almost perfectly suited for the imino acid proline as P1-residue, the hydroxyl Oη of Tyr-662 is correctly positioned to interact with the normal amide nitrogen atom of an amino acid in P1. The inhibitor also unambiguously maps the S2-site. The P2-carbonyl oxygen atom gets trapped in an electrostatic sink formed by the side chains of Arg-125 and Asn-710. Glu-205 and Glu-206, and to a lesser extent the carbonyl oxygen atom of Glu-205, interact with the free amino terminus of the P2-residue, thus determining the dipeptidyl “amino”-peptidase activity of the enzyme. It is, therefore, the β-propeller that provides essential determinants for P2-recognition, namely Arg-125, which is positioned on the hairpin loop between strands 2 and 3 of blade II and Glu-205-Glu-206, positioned on a short helical insertion within strand 1 of the β-propeller blade IV. Ample space is available to accommodate voluminous side chains such as Tyr or Trp in P2 (Fig. 5a). In our inhibitor the phenyl ring of the P2-residue is iodated and forms an ionic interaction with Arg-358 (Fig. 5a).
Figure 5.
Substrate recognition by DP IV. (a) The peptidomimetic inhibitor p-Iodo-Phe-Pyr-CN is covalently bound to active site Ser-630. The accessible surface is indicated and cut open (dark green) for better visibility. (b) Schematic representation of the active site access in tricorn and DP IV.
Substrate Access to and Product Egress from the Active Site.
The β-propeller domain covers the active site and thereby restricts substrate access. There are two possible routes to the active site, namely through the tunnel of the β-propeller and through a side opening. Similar as the propeller tunnel, the shape of the side entrance is oval with dimensions of 15 and 22 Å. The side opening to the active site is generated by the kinked arrangement of blade I and II (Fig. 2). The distance from the protein surface to the active site measures 20 and 37 Å through the side opening and the propeller tunnel, respectively. From its dimensions, the side opening might give active site access to helical substrates. After substrate cleavage, two products leave the active site chamber. Clearly, exit and entrance routes may differ.
Discussion
Oligomerization of Membrane-Bound and Soluble DP IV.
Tetramerization on the cell surface involves, for geometric reasons, a membrane-bound and a soluble DP IV dimer pair or dimers located on the surface of two different cells, as illustrated in Fig. 2. DP IV functions as a cell–cell communication molecule. Thus, DP IV may be involved in mediating such cell–cell contacts by tetramerization of two DP IV dimers present on the surfaces of interacting cells. If so, addition of soluble DP IV should prevent such cell–cell interactions. This is indeed observed when applying soluble DP IV in a cell adhesion model (37, 38), consistent with the proposed cell–cell contact model (Fig. 2). Alternatively, soluble dimers can assemble to form a homotetramer, as observed in our crystal structure. The tetramer encloses a large cavity and may explain why the high molecular weight form of DP IV behaves as a hexamer on gel filtration chromatography (5). Glycosylation of Asn-279 might provide the missing regulatory link that was proposed to control its assembly (5).
Dimerization is mediated by the three C-terminal secondary structure elements positioned on the catalytic domain, and a finger-like insertion motif within strands 2 and 3 of propeller blade IV. An N-terminal extension of 60 aa present in POP covers the dimer contact area of DP IV, explaining why POP does not dimerize. Consequently, N-terminally truncated isoforms of POP should be able to dimerize. Likewise, it should be possible to construct a monomeric DP IV-variant by adding a dimerization mimetic motif to DP IV. Furthermore, DP IV is known to form heterodimers with FAPα, seprase (7). FAPα shares the essential elements of the DP IV-dimerization motif, including the extension of strands 2 and 3 of propeller blade IV (Fig. 1). Therefore, we expect the FAPα/FAPβ-heterodimer and FAPα-homodimers to resemble the FAPβ(DP IV)-homodimer.
Functional Role of Oligomerization.
The crystal structure shows that dimerization is not required to complete the active site architecture of DP IV, as for example in the case of tricorn (31). Instead, dimerization and tetramerization will affect interaction with other components, including proteolytic substrates and ADA, and possibly mediate cell–cell contacts. Moreover, dimerization of DP IV is likely to enhance the receptor–ligand affinity by bivalent interaction which may be critical for signal transduction into the cell.
Substrate Preference and Catalytic Mechanism.
The hydrophobic S1-pocket fits proline as well as other small uncharged residues such as alanine or serine as a P1-residue. The hydroxyl Oη atom of Tyr-662 is able to form a hydrogen bond with the P1-amide nitrogen atom and thus optimally presents the substrate for catalysis. Proper orientation of proline in P1-position is achieved by its side-chain interaction in addition to the binding to the oxyanion pocket.
The main chain of the P2-residue interacts with two prominent anchor sites, namely Glu-205–Glu-206, which form a twin-single salt bridge with the free amino terminus of the P2-residue; and Arg-125 together with Nδ2 of Asn-710, which stabilize and activate the P2-carbonyl oxygen atom. In POP, the latter two residues are structurally and functionally substituted for by Arg-643, whereas it lacks an N terminus binding site equivalent to Glu-205–Glu-206. The Glu-205–Glu-206 motif has been shown by site directed mutagenesis to be essential for enzymatic activity (39). This dual Glu–Glu recognition motif remarkably resembles the functionally equivalent sites of tricorn and its associated factor F1. In F1, the Glu-213–Glu-245 charge array serves as the docking site for the free N terminus (35), whereas in tricorn, a carboxy-dipeptidase, an Arg-131–Arg-132 motif provides the anchor site for the substrate C terminus (31).
As a postproline processing enzyme DP IV has to meet an additional requirement for efficient catalysis. Proline-containing peptides can adopt cis- and trans-peptide bond conformations. As highlighted by our inhibitor structure, only a peptide in the trans conformation is able to productively bind to the active site, in agreement with earlier observations (40). Expectedly, locking the trans conformation of an N-alanyl-prolyl, O-acyl hydroxamine by the substitution of the P2–P1 peptide bond with a fluoroolefin group results in a superior irreversible DP IV-inhibitor (41–43). Contrasting their fluoroolefin mimetics, the “normal” N-aminoacyl-prolyl, O-acyl hydoxylamines are processed by DP IV to >99.9% as substrates, whereas they inhibit other proteases very potently (44, 45).
These unexpected results suggest a conformational two step mechanism for the substrate hydrolysis where a trans substrate binds to the active site, followed by a trans–cis isomerization to enable proteolytic cleavage of the substrate (8). Such conformational rearrangement can occur without steric conflicts, given the voluminous active site geometry of DP IV (Fig. 5a). By stabilizing the negative charge on the carbonyl oxygen of the P2 residue, the P2-oxyanion trap (Arg-125) reduces the valence of the partial C⩵N double bond and thus destabilizes the trans conformation.
The active site can accommodate bulky side chains such as tryptophan or tyrosine in P2 position. In fact, substrates or inhibitors with modified long or bulky side-chains of the P2 amino acid showed an even enhanced binding (8). Substrates possessing acidic residues in P2 position are poorly turned over, possibly caused by the attractive interaction with Arg-125, thus disrupting the P2 oxyanion trap, which is needed for efficient catalysis or by repulsion with Glu-131–Glu-132. Accordingly, phosphorylation of P2 amino acid side chains as in Ser–Pro- and Thr–Pro-p-nitroanilides reduces the catalytic efficiency of the enzyme by up to 99.5% (8, 46).
Comparison of β-Propeller Architectures.
The tunnel through the eight-bladed β-propeller widens from the surface toward the active site of DP IV. A similar conical widening of the propeller tunnel toward the catalytic domain was also observed for the β7-propellers of tricorn and POP (30, 31). Tricorn's β6-propeller tunnel has also a conical shape but widens toward the solvent. Because tricorn's β6 serves as an exhaust system feeding its products into downstream protease factors (47, 48), this seeming exception actually confirms the rule of tunnel widening toward the processing active site.
Substrate Access to and Product Egress from the Active Site.
DP IV is expressed as a fully active enzyme. The lack of a zymogen–enzyme activation step and of endogenous DP IV-inhibitors shifts the problem of activity regulation toward the level of gene expression, protein synthesis and substrate selection. Similar to proteases with caged active sites, such as POP, the proteasome, or tricorn, substrate access to the active site serves as a major regulatory element of the enzymatic activity. Two openings give access to the active site. The shorter route through the side opening appears to provide the easier and kinetically favored access to and egress from the active site. Functionally, however, the “side opening-only” model fails to explain DP IV's high substrate selectivity, as even secondary structured peptides could access the active site through the side opening. Moreover, this model is ignorant of the strong preference given by POP (β7) and tricorn (β7 and β6), where all three topologically open β-propellers channel substrates to and products away from the active site. The situation in DP IV is most closely resembled by the tricorn protease where a seven-bladed and six-bladed β-propeller provide a separate entrance to and exit from the active site, respectively. Tricorn protease is a serine protease with low but significant structural homology to the family of α/β-hydrolases. We superimposed the catalytic core elements, including the active site serine and histidine, the strictly conserved helix following the active site serine (Ser-630–Ala-642 and Ser-965–Leu-977, respectively), and tricorn's five-stranded parallel β-sheet onto the equivalent strands of the eight-stranded DP IV-sheet. Both sheets have identical polarity. Significantly, both tricorn propellers superimpose onto the two DP IV openings, the tricorn β7-propeller onto the DP IV β8-propeller, and the tricorn β6-propeller onto the side exit (Fig. 5b). This similarity suggests that the β8-propeller provides substrate access to and the side opening product release from the DP IV active site. The tricorn-derived model explains the high substrate selectivity critical for DP IV-function to activate or inactivate regulatory peptides. Passage through the β-propeller tunnel requires the substrates to unfold thereby providing a chemical “fingerprint” to DP IV. Once the amino terminus of the peptide approaches the active site, it is still held in place by its C terminus interacting with the β-propeller which may contribute to bend the substrate for cleavage. After the nucleophilic attack, the acyl enzyme intermediate forms, whereas the primed product is directly released through the side exit. This explains why degradation of glucagon by DP IV is not processive, but occurs sequentially in two independent steps (glucagon 3–29, glucagon 5–29) (49). Clearly, the final determination of the functional roles of the DP IV openings awaits further experiments.
Interaction with Other Components.
DP IV binds ADA to the T cell surface, thereby preventing the cell from adenosine mediated inhibition of proliferation (50). Interestingly, murine and rat DP IV do not bind ADA (51, 52). By using site-directed mutagenesis, Leu-294 and Val-341 were identified as two ADA-binding sites (53). Leu-294 and Val-341 are positioned at the outer strand of the tetramerization blade IV and blade V, respectively. Therefore, ADA binding will interfere with tetramerization (Fig. 4). Similarly, the glycosylation of Asn-279 (Asn-281 in the human sequence) is likely to influence ADA binding. This suggests that tetramerization of DP IV and proper glycosylation of Asn-279 serve as major control mechanism for ADA binding. Interestingly, murine and rat DP IV lack this glycosylation site, consistent with their failure to bind ADA (53).
DP IV as a Target for Drug Design.
DP IV-activity strongly correlates with many diseases, such as diabetes, obesity, and tumor progression (13, 14, 16–18) making it a top target of pharmaceutical research. Our inhibitor structure identified important recognition elements at DP IV's active site and represents an excellent starting point for rational design of active site-directed inhibitors. Compounds such as the cyanopyrrolidides block the enzyme covalently. However, DP IV's involvement in a great variety of physiological processes poses a high challenge to avoid unwanted side effects for any DP IV-drug development program. Ideally, one would like to target a particular DP IV-substrate rather than the complete DP IV-activity. Nonactive site-directed inhibition strategies depict a solution to this problem. The sulfate bound to the oxyanion pocket within the β-propeller tunnel, as indicated in Fig. 5a, identified an excellent target point for the development of inhibitors that block substrate passage through the β-propeller tunnel.
Supplementary Material
Acknowledgments
We thank Marianne Braun for expert help in crystallization, Stefan Strobl for help in the early stages of the project, and Gleb Bourenkov for help with data collection at beamline BW6 (Deutsches Elektronen Synchrotron, Hamburg). This work was supported by the German Department of Science and Technology, Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie Grant 0312302.
Abbreviations
- ADA
adenosine deaminase
- FAP
fibroblast activation protein
- DP IV
dipeptidyl peptidase IV
Note
While submitting the manuscript, we became aware of the crystal structure of recombinantly produced human DP IV, which was determined in the dimeric state at 2.5-Å resolution (54).
Footnotes
Data deposition: The structure factors and coordinates reported in this paper have been deposited in the Protein Data Base, www.rcsb.org (PDB ID codes 1ORV and 1ORW).
References
- 1.Yaron A, Naider F. Crit Rev Biochem Mol Biol. 1993;28:31–81. doi: 10.3109/10409239309082572. [DOI] [PubMed] [Google Scholar]
- 2.Fleischer B. Immunol Today. 1994;15:180–184. doi: 10.1016/0167-5699(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 3.Kameoka J, Tanaka T, Nojima Y, Schlossman S F, Morimoto C. Science. 1993;261:466–469. doi: 10.1126/science.8101391. [DOI] [PubMed] [Google Scholar]
- 4.Dukecohan J S, Morimoto C, Rocker J A, Schlossman S F. J Immunol. 1996;165:1714–1721. [PubMed] [Google Scholar]
- 5.Lambeir A M, Pereira J F D, Chacon P, Vermeulen G, Heremans K, Devreese B, VanBeeumen J, Demeester I, Scharpe S. Biochim Biophys Acta. 1997;1340:215–226. doi: 10.1016/s0167-4838(97)00045-9. [DOI] [PubMed] [Google Scholar]
- 6.Püschel G, Mentlein R, Heymann E. Eur J Biochem. 1982;126:359–365. doi: 10.1111/j.1432-1033.1982.tb06788.x. [DOI] [PubMed] [Google Scholar]
- 7.Scanlan M J, Raj B K, Calvo B, Garin-Chesa P, Sanz-Moncasi M P, Healey J H, Old L J, Rettig W J. Proc Natl Acad Sci USA. 1994;91:5657–5661. doi: 10.1073/pnas.91.12.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demuth H-U, Heins J. In: Dipeptidyl Peptidase IV (CD26) in Metabolism and the Immune Response. Fleischer B, editor. Austin, TX: R. G. Landes; 1995. pp. 1–35. [Google Scholar]
- 9.Mentlein R. Regul Pept. 1999;85:9–24. doi: 10.1016/s0167-0115(99)00089-0. [DOI] [PubMed] [Google Scholar]
- 10.Drucker D J. Exp Opin Invest Drugs. 2003;12:87–100. doi: 10.1517/13543784.12.1.87. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann T, Glund K, McIntosh C H S, Pederson R A, Hanefeld M, Rosenkranz B, Demuth H-U. In: Cell-Surface Aminopeptidases: Basic and Clinical Aspects. Mizutani M, Turner A J, Nomura S, Ino K, Matsuo M, editors. Amsterdam: Elsevier; 2001. pp. 381–393. [Google Scholar]
- 12.Hoffmann T, Demuth H-U. In: Ectopeptidases: CD13/Aminopeptidase N and CD26/Dipeptidylaminopeptidase IV in Medicine and Biology. Langner J, Ansorge S, editors. New York: Kluwer Academic/Plenum; 2002. pp. 259–278. [Google Scholar]
- 13.Pederson R A, White H A, Schlenzig D, Pauly R P, McIntosh C H, Demuth H U. Diabetes. 1998;47:1253–1258. doi: 10.2337/diab.47.8.1253. [DOI] [PubMed] [Google Scholar]
- 14.Pospisilik J A, Stafford S G, Demuth H U, McIntosh C H, Pederson R A. Diabetes. 2002;51:2677–2683. doi: 10.2337/diabetes.51.9.2677. [DOI] [PubMed] [Google Scholar]
- 15.Medeiros-Mdos S, Turner A J. Neurochem Res. 1996;21:1125–1132. doi: 10.1007/BF02532423. [DOI] [PubMed] [Google Scholar]
- 16.Cheng J D, Dunbrack R L, Valianou M, Rogatko A, Alpaugh R K, Weiner L M. Cancer Res. 2002;62:4767–4772. [PubMed] [Google Scholar]
- 17.Kajiyama H, Kikkawa F, Suzuki T, Shibata K, Ino K, Mizutani S. Cancer Res. 2002;62:2753–2757. [PubMed] [Google Scholar]
- 18.Ho L, Aytac U, Stephens L C, Ohnuma K, Mills G B, McKee K S, Neumann C, LaPushin R, Cabanillas F, Abbruzzese J L, Morimoto C, Dang N H. Clin Cancer Res. 2001;7:2031–2040. [PubMed] [Google Scholar]
- 19.Schols D, Proost P, Struyf S, Wuyts A, DeMeester I, Scharpe S, VanDamme J, DeClercq E. Antiviral Res. 1998;39:175–187. doi: 10.1016/s0166-3542(98)00039-4. [DOI] [PubMed] [Google Scholar]
- 20.Ashworth D M, Atrash B, Baker G R, Baxter A J, Jenkins P D, Jones D M, Szelke M. Bioorg Med Chem Lett. 1996;6:1163–1166. [Google Scholar]
- 21.Steinmetzer T, Silberring J, Mrestani-Klaus C, Fittkau S, A, B, Demuth H-U. J Enzyme Inhibition. 1993;7:77–85. doi: 10.3109/14756369309040750. [DOI] [PubMed] [Google Scholar]
- 22.Kiefersauer R, Than M E, Dobbek H, Gremer L, Melero M, Strobl S, Dias J M, Soulimane T, Huber R. J Appl Crystallogr. 2000;33:1223–1230. [Google Scholar]
- 23.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Tong L, Rossmann M G. Acta Crystallogr A. 1990;46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 25.Turk D. Ph.D. thesis. München, Germany: Technische Universität München; 1992. [Google Scholar]
- 26.Knight S D. Acta Crystallogr D. 2000;52:42–47. doi: 10.1107/s0907444999012913. [DOI] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 28.Abrahams J P, Leslie A G W. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 29.Brünger A T, Adams P D, Clore G M, Delano W L, Gros P, Grossekunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Fülöp V, Böcskei Z, Polgár L. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 31.Brandstetter H, Kim J-S, Groll M, Huber R. Nature. 2001;414:466–469. doi: 10.1038/35106609. [DOI] [PubMed] [Google Scholar]
- 32.Nurizzo D, Turkenburg J P, Charnock S J, Roberts S M, Dodson E J, McKie V A, Taylor E J, Gilbert H J, Davies G J. Nat Struct Biol. 2002;9:665–668. doi: 10.1038/nsb835. [DOI] [PubMed] [Google Scholar]
- 33.Medrano F J, Alonso J, Garcia J L, Romero A, Bode W, Gomis-Rüth F X. EMBO J. 1998;17:1–9. doi: 10.1093/emboj/17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito K, Inoue T, Kabashima T, Kanada N, Huang H-S, Ma X, Azmi N, Azab E, Yoshimoto T. J Biochem. 2000;128:673–678. doi: 10.1093/oxfordjournals.jbchem.a022800. [DOI] [PubMed] [Google Scholar]
- 35.Goettig P, Groll M, Kim J-S, Huber R, Brandstetter H. EMBO J. 2002;21:5343–5352. doi: 10.1093/emboj/cdf552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fülöp V, Jones D. Curr Opin Struct Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 37.Cheng H C, Abdel-Ghany M, Elble R C, Pauli B U. J Biol Chem. 1998;273:24207–24215. doi: 10.1074/jbc.273.37.24207. [DOI] [PubMed] [Google Scholar]
- 38.AbdelGhany M, Cheng H C, Levine R A, Pauli B U. Invasion Metastasis. 1998;18:35–43. doi: 10.1159/000024497. [DOI] [PubMed] [Google Scholar]
- 39.Abbott C A, McCaughan G W, Gorrell M D. FEBS Lett. 1999;458:278–284. doi: 10.1016/s0014-5793(99)01166-7. [DOI] [PubMed] [Google Scholar]
- 40. Fischer, G., Heins, J. & Barth, A. (1983) Biochim. Biophys. Acta742. [DOI] [PubMed]
- 41.Lin J, Toscano P J, Welch J T. Proc Natl Acad Sci USA. 1998;95:14020–14024. doi: 10.1073/pnas.95.24.14020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao K, Lim D S, Funaki T, Welch J T. Bioorg Med Chem. 2003;11:207–215. doi: 10.1016/s0968-0896(02)00384-x. [DOI] [PubMed] [Google Scholar]
- 43.Demuth H-U, Neumann U, Barth A. J Enzyme Inhibition. 1989;2:239–248. doi: 10.3109/14756368909088477. [DOI] [PubMed] [Google Scholar]
- 44.Brömme D, Demuth H-U. Methods Enzymol. 1994;244:671–685. doi: 10.1016/0076-6879(94)44050-6. [DOI] [PubMed] [Google Scholar]
- 45.Demuth H-U, Baumgrass R, Schaper C, Fischer G, Barth A. J Enzyme Inhibition. 1988;2:129–142. doi: 10.3109/14756368809040718. [DOI] [PubMed] [Google Scholar]
- 46.Kaspari A, Diefenthal T, Grosche G, Schierhorn A, Demuth H-U. Biochim Biophys Acta. 1996;1293:147–153. doi: 10.1016/0167-4838(95)00238-3. [DOI] [PubMed] [Google Scholar]
- 47.Brandstetter H, Kim J-S, Goettig P, Groll M, Huber R. Biol Chem. 2002;383:1157–1165. doi: 10.1515/BC.2002.127. [DOI] [PubMed] [Google Scholar]
- 48.Kim J-S, Groll M, Musiol H-J, Behrendt R, Kaiser M, Moroder L, Huber R, Brandstetter H. J Mol Biol. 2002;324:1041–1050. doi: 10.1016/s0022-2836(02)01153-1. [DOI] [PubMed] [Google Scholar]
- 49.Pospisilik J A, Hinke S A, Pederson R A, Hoffmann T, Rosche F, Schlenzig D, Glund K, Heiser U, McIntosh C H S, Demuth H-U. Regul Pept. 2001;96:133–141. doi: 10.1016/s0167-0115(00)00170-1. [DOI] [PubMed] [Google Scholar]
- 50.DeMeester I, VanHoof G, Lambeir A M, Scharpe S. J Immunol Methods. 1996;189:99–105. doi: 10.1016/0022-1759(95)00239-1. [DOI] [PubMed] [Google Scholar]
- 51.Dinjens W N, ten-Kate J, Wijnen J T, van-der-Linden E P, Beek C J, Lenders M H, Khan P M, Bosman F T. J Biol Chem. 1989;264:19215–19220. [PubMed] [Google Scholar]
- 52.Iwaki-Egawa S, Watanabe Y, Fujimoto Y. Cell Immunol. 1997;178:180–186. doi: 10.1006/cimm.1997.1123. [DOI] [PubMed] [Google Scholar]
- 53.Abbott C A, McCaughan G W, Levy M T, Church W B, Gorrell M D. Eur J Biochem. 1999;266:798–810. doi: 10.1046/j.1432-1327.1999.00902.x. [DOI] [PubMed] [Google Scholar]
- 54.Rasmussen H B, Branner S, Wiberg F C, Wagtmann N. Nat Struct Biol. 2003;10:19–25. doi: 10.1038/nsb882. [DOI] [PubMed] [Google Scholar]
- 55.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 56.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 57.Esnouf R M. Acta Crystallogr D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 58.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.