Abstract
Heterotrimeric G proteins transduce signals from activated transmembrane G protein-coupled receptors to appropriate downstream effectors within cells. Signaling specificity is achieved in part by the specific α, β, and γ subunits that compose a given heterotrimer. Additional structural and functional diversity in these subunits is generated at the level of posttranslational modification, offering alternate regulatory mechanisms for G protein signaling. Presented here is the identification of a variant of the γ2 subunit of G protein heterotrimer purified from bovine brain and the demonstration that this RDTASIA γ2 variant, containing unique amino acid sequence at its N terminus, is a substrate for ubiquitylation and degradation via the N-end rule pathway. Although N-end-dependent degradation has been shown to have important functions in peptide import, chromosome segregation, angiogenesis, and cardiovascular development, the identification of cellular substrates in mammalian systems has remained elusive. The isolation of RDTASIA γ2 from a native tissue represents identification of a mammalian N-end rule substrate from a physiological source, and elucidates a mechanism for the targeting of G protein γ subunits for ubiquitylation and degradation.
The G protein heterotrimer, composed of an α, β, and γ subunit, is complexed with the nucleotide GDP in its inactive state. Upon activation, GDP is exchanged for GTP, resulting in subunit dissociation, and the GTP-bound α or the βγ dimer can both subsequently modulate the activity of a variety of downstream effectors (1, 2). The γ subunit, although generally associated in the cell with a β subunit, is the smallest and most variable heterotrimeric G protein subunit (3, 4) and has been the least well understood with regard to its specific biological function. Several well established lipid modifications of γ subunits include farnesylation or geranylgeranylation of the C terminus (5, 6), and these modifications play an important role in membrane association, receptor coupling, and effector regulation (7–9). Recent work suggests a role for the γ subunit of transducin in ubiquitylation and destabilization of Gtβγ dimers (10). Although it is unclear whether such observations can be extended to other distinct γ isoforms, the crucial question of how a γ subunit could be targeted to the ubiquitylation machinery in a regulated fashion also remains unanswered. The amino terminal region of γ subunits is the region most divergent between the 12 known γ isoforms (11), an observation which suggests that the first 20 amino acids may be crucial for a specific function or subcellular localization. Indeed, presented here is the identification of an N-terminal variant of the Gγ2 subunit isolated from bovine brain. Our discovery that this RDTASIA γ2 variant undergoes N-end rule degradation highlights regulated degradation as an important function for the γ subunit of the G protein heterotrimer, offering a mechanism by which G protein subunits can be targeted to enzymatic components of the ubiquitylation pathway.
The N-end rule predicts the stability of a protein, based on the identity of its N-terminal amino acid (12). Determinants for ubiquitylation and degradation of an N-end substrate include not only a destabilizing N-terminal residue but also an internal lysine to which ubiquitin (or multiubiquitin chains) is covalently attached (13). Destabilizing N-terminal amino acids may be exposed by a proteolytic cleavage event or can be generated by posttranslational modification of asparagine and glutamine (via deamidation), or aspartic acid, glutamic acid, and cysteine residues (via arginylation) at the N terminus of a polypeptide (14). Multiubiquitylation of an N-end-targeted protein frequently results in degradation via a proteasome-dependent mechanism. The first bona fide N-end substrate to be identified was the proteolytic C-terminal fragment of the cohesin subunit, SCC1, which bears an N-terminal arginine and is a target for N-end ubiquitylation and degradation in Saccharomyces cerevisiae (15). Although it has been well established that mammalian systems contain the enzymatic components of the N-end pathway (16, 17), there previously have been no substrates isolated from a physiological source that offer proof that the hierarchy of N-end modifications (including deamidation and arginylation) do indeed occur, resulting in the production of a native substrate. In addition to elucidating a role for the ubiquitin modification of the Gγ2 heterotrimer subunit, our identification of RDTASIA γ2, which bears unique N-terminal amino acid sequence consistent with the proposed modifications of the N-end rule pathway, offers important physiological evidence for the existence of native N-end rule mammalian substrates.
Experimental Methods
Matrix-Assisted Laser Desorption Ionization (MALDI)–MS and Electrospray MS of γ subunits.
Gγ subunits from 2.5 mg of purified bovine brain G protein heterotrimer were resolved by using reverse-phase chromatography as described, and the elution flow was split to simultaneously allow fraction collection and tandem MS analysis (18). Aliquots from fractions were lyophilized, resuspended in 1–2 μl of sample solvent [47.6% (vol/vol) n-propanol/47.6% (vol/vol) water/4.76% (vol/vol) acetonitrile/0.095% (vol/vol) trifluoroacetic acid (TFA)], and then mixed 1:1 or 1:3 with α-cyano-4-hydroxycinnamic acid (50 mM in 70% acetonitrile/30% water/0.1% TFA) before MALDI-MS analysis by using an Applied Biosystems Voyager-DE instrument. Collected fractions were dried and resuspended in 50% methanol, 26% water, 20% n-propanol, and 4% acetic acid and loaded into a pulled glass capillary for nanospray ionization. A custom nanospray source was used on a ThermoFinnigan (San Jose, CA) LCQ Classic ion-trap mass spectrometer. The protein of interest was isolated according to its m/z ratio and fragmented in the ion trap. Fragment ions were recorded with an average of 15–30 spectra accumulated. Predicted ions were determined by using the SHERPA 3.3.1 program (19). For Edman sequencing, ≈250 μl of the HPLC fraction was prepared by using a Prosorb (PE Applied Biosystems, Foster City, CA) filtration device and analyzed on an Applied Biosystems Procise 494 automated protein sequencer.
Ubiquitin-Fusion Plasmid Design.
Construction of ubiquitin-fusion expression plasmids allowed the recombinant production of γ2 proteins with different residues at the N terminus (20). Gγ2 fusions were created by insertion of five copies of a myc epitope tag (5xmyc) and ubiquitin (K48R) cDNA into a pcDNA3-γ2 vector at the N terminus of the γ2 coding region. QuikChange site-directed mutagenesis (Stratagene) was used to delete the intervening sequence, resulting in an in-frame fusion between the last glycine of the ubiquitin sequence and the first amino acid of the γ2 sequence. Various mutagenic primers were used to create the γ2 variant constructs (pc5mUb48R-RD and pc5mUb48R-MD). In the case of RDTASIA noK (lysine-less construct), the γ2 cDNA sequence was synthesized by Applied Bio Products (Bionexus, Oakland, CA) and encoded an arginine substitution at each lysine residue (K14R, K20R, K29R, K39R, K46R, K64R, and K65R); the 5xmycUb-fusion construct was subsequently generated using standard subcloning and mutagenesis techniques as described above. The constructs depicted in Fig. 4, which contain one or two lysine residues in the background of the RDnoK construct, were similarly created by using mutagenic primers and QuikChange site-directed mutagenesis.
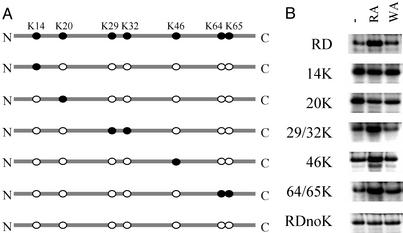
Figure 4.
Lysine residues required for the ubiquitin-dependent degradation of RDTASIA γ2. (A) RDTASIA γ2 (containing seven lysine residues), RDnoK (arginine substitution at all residues indicated), and various constructs containing lysine residues that were reintroducted by mutagenesis techniques. (B) [35S]Methionine-labeled protein produced from RDTASIA γ2 constructs depicted after translation in reticulocyte lysate, in the absence or presence of dipeptide inhibitors.
In Vitro Translation/Stability Assays.
One microgram of each pc5mUb48R-γ2 fusion (Fig. 2B) was translated used the Quick Coupled Transcription/Translation system (Promega) in the presence of 35S-labeled methionine. All translation reactions contained 0.15 mM bestatin and either buffer control or 3 mM of the appropriate dipeptide inhibitor [Arg-β-Ala (RA) or Trp-Ala (WA)], previously prepared as a 50-mM stock solution. Translations were incubated at 30°C for 20, 60, or 120 min, and at each time point an aliquot of reaction was removed and stopped in SDS sample buffer. Reaction products were resolved by 12% denaturing polyacrylamide electrophoresis, and dried gels were analyzed by fluorography.
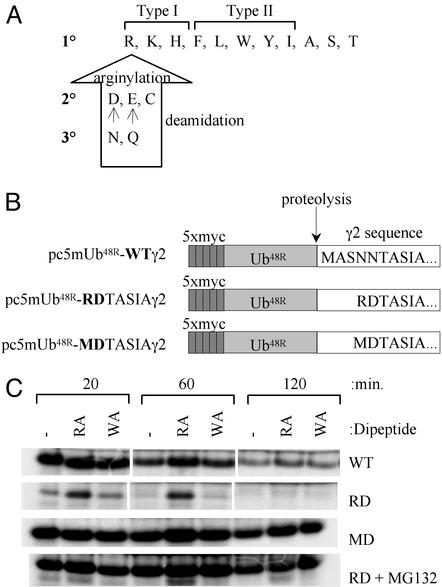
Figure 2.
N-terminal variant of γ2 undergoes proteasome-dependent degradation. (A) Destabilizing N-terminal amino acids of the N-end rule pathway. Primary destabilizing amino acids can be subdivided into the type I (basic) residues or type II (bulky hydrophobic) residues. Secondary destabilizing amino acids require N-terminal arginylation, whereas tertiary amino acids are modified by deamidation and arginylation before N-end degradation (14). (B) The ubiquitin-fusion constructs used to generate γ2 in reticulocyte coupled transcription/translation system with varying N termini. Ub48R, ubiquitin containing K48R substitution, which does not support formation of multiubiquitin chains. (C) Accumulation of γ2 or its N-terminal variants after translation in rabbit reticulocyte lysate. Dipeptide inhibitors of the N-end ubiquitin ligase (type 1 inhibitor, RA; type 2 inhibitor, WA) are included in the indicated reactions.
Results
Identification and Characterization of N-Terminal Variant of Gγ2.
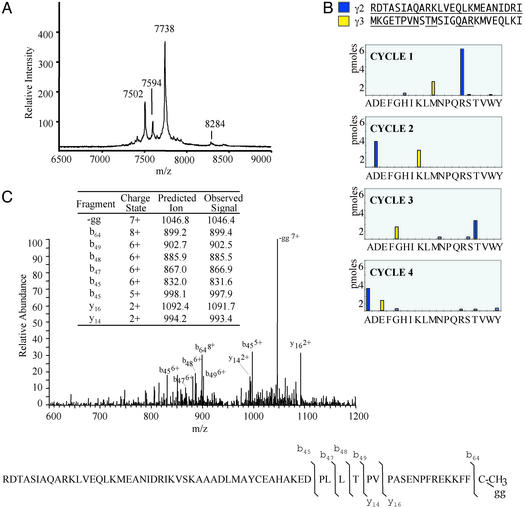
In the characterization of the structural diversity of G proteins, the individual α-, β-, and γ subunits were isolated from purified Go/Gi heterotrimers by using HPLC, and fractions that contained γ isoforms were subsequently analyzed by using MS and other protein characterization techniques (18). Gγ2, a primary component of the brain G protein Go, is one of the most abundant Gγ subunits in brain (21). The majority of γ2 protein isolated was physiologically processed as predicted at the N terminus by removal of the N-terminal methionine and subsequent acetylation (22); however, a small population of protein (molecular mass of 7,592.7 ± 1.0 Da; mean ± SEM, n = 4) copurified with γ isoforms but had a mass that did not correspond to known γ isoforms (Fig. 1A). Edman sequencing of HPLC fractions containing this variant confirmed its γ2 similarity (Fig. 1B), although the N-terminal amino acids, arginine and aspartic acid, were not those predicted by γ2 cDNA sequence. A γ2 variant beginning with the sequence RDTASIA, instead of the predicted sequence MASNNTASIA, has a predicted mass of 7,593 Da, which is in good agreement with the observed matrix-assisted laser desorption ionization–MS mass of 7,592.7 Da. MS fragmentation of the C terminus (Fig. 1C) and γ2 immunoreactivity (data not shown) confirmed the identification of an N-terminal variant of γ2, containing two amino acids on a modified N terminus, likely a result of posttranslational modification. Given that this newly described N-terminally processed protein was isolated from a biological source (bovine brain), its function and mechanism of generation are likely related to the fundamental role of the G proteins of which it is a part.
Figure 1.
Identification of the RDTASIA variant of γ2 subunit isolated from bovine brain heterotrimer. (A) Matrix-assisted laser desorption ionization–MS of an HPLC fraction containing γ variant (m/z 7,594), unmethylated γ2 (m/z 7,738), γ5 C-terminal variant (m/z 7,502; ref. 35), and γ3 (m/z 8,284). (B) Edman sequencing analysis of an HPLC fraction containing truncated/modified γ isoforms for the first four cycles. The sequenced fraction contained two independent sequences, γ3 (minor) and RDTASIA γ2 (major). (C) Electrospray ionization tandem MS analysis of RDTASIA variant of γ2. A tandem MS spectrum of RDTASIA γ2 isoform is shown. [M + 8H]8+ ion, m/z 950.3 selected, average of 18 scans. (Inset) Table of predicted and observed ions.
RDTASIA γ2 Is a Type I N-End Substrate.
Although nascent polypeptides are translated starting with a stabilizing methionine residue, posttranslational processing at the N terminus or internal cleavage of a protein may expose an N-terminal amino acid predicted to be destabilizing by the N-end rule pathway (Fig. 2A). An exposed secondary destabilizing residue (Asp, Glu, Cys) may be modified by the conjugation of Arg, or a tertiary destabilizing residue (Asn, Gln) may undergo deamidation followed by arginylation, culminating in the ubiquitin-dependent degradation of the polypeptide substrate (23). Given the observation that the RDTASIA γ2 variant bears an N-terminal destabilizing amino acid (Fig. 2A), wild-type γ2 or its RDTASIA variant were produced in a reticulocyte lysate translation system by using a ubiquitin-fusion strategy (Fig. 2B; ref. 20), and accumulation of each was evaluated in the presence of dipeptide inhibitors known to target the type I (RA) or II (WA) binding sites of the N-end ubiquitin ligase, Ubr1/E3α (24). If RDTASIA γ2 is a specific Ubr1/E3α substrate, its accumulation would be predicted to be altered only by type I inhibitors. Although near uniform levels of wild-type γ2 protein are observed up to 2 h after the start of translation, the RDTASIA γ2 protein (RD) accumulates only in the presence of the type I dipeptide inhibitor, RA (Fig. 2C). A γ2 construct identical to RDTASIA γ2, with a substitution of Met for the N-terminal Arg (MD), restores the wild-type phenotype, confirming that RDTASIA is degraded as a consequence of its N-terminal Arg, not due to the absence of amino acids 2–5 from the wild-type γ2 sequence. It is important to note that the accumulation of translated protein is in fact correlated to stability; translation of RDTASIA γ2 in the presence of 50 μM MG132, an inhibitor of proteasome activity, results in uniform accumulation of RDTASIA γ2 (Fig. 2C), indicating that the absence of protein in select treatments reflects its susceptibility to proteasome-dependent degradation.
Degradation in Vitro Correlates with High-Molecular-Weight Multiubiquitin Conjugates.
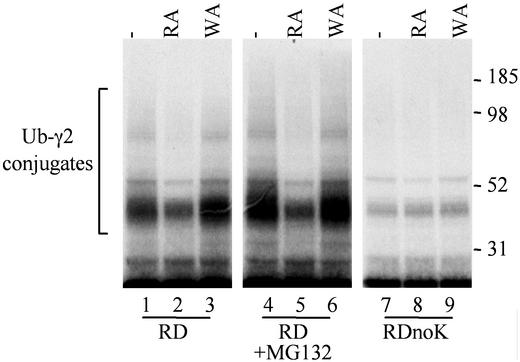
If the differences in stability/accumulation between γ2 and its RDTASIA variant are indeed a result of ubiquitin-dependent degradation, a pattern of high-molecular-weight multiubiquitylated conjugates, which are specifically altered by dipeptide inhibitors, should be visible in the upper portion of the gels. The dipeptide inhibitor RA, which targets the type I site on the N-end ubiquitin ligase, clearly reduces the formation of multiubiquitylated RDTASIA γ2 as seen in Fig. 3. This attenuation of ubiquitylation is not observed with an inhibitor for a distinct substrate binding site (WA; Fig. 3, lane 3). A similar (and perhaps more marked) pattern of multiubiquitylation is observed for RDTASIA γ2 translated in the presence of a proteasome inhibitor (Fig. 3, lanes 4–6), consistent with the MG132 block of degradation, a step downstream of ubiquitin modification. Finally, N-end-dependent degradation includes a requirement for a lysine residue within the substrate molecule, as a site of ubiquitylation (13). An RDnoK γ2 variant (containing substitution of all seven lysine residues with arginine) lacks the selective accumulation of higher-molecular-weight ubiquitin conjugates (Fig. 3, lanes 7–9), because it lacks the amino acid to which ubiquitin is covalently attached. Consistently, the accumulation of this RDnoK γ2 protein was unaltered by either type I or II dipeptide inhibitors (Fig. 4), confirming that a lysine residue (and likely ubiquitin modification) is indeed required for the degradation of RDTASIA γ2 in reticulocyte extract.
Figure 3.
High-molecular-weight ubiquitin conjugates formed in reticulocyte lysate after translation of RDTASIA γ2, RDTASIA γ2 + 50 μM MG132, or RDTASIA γ2 noK (all lysine residues substituted with arginine). Gels shown are from [35S]methionine-labeled reaction aliquots terminated 20 min after the start of translation.
Lysine Residues in the C Terminus of Gγ2 Mediate Ubiquitin-Dependent Degradation.
Although these data establish the γ2 subunit as a substrate for ubiquitylation, we sought to further these observations by identifying the specific lysine residues required for ubiquitin-dependent degradation. Our strategy consisted of reincorporating lysine residues in the context of the RDnoK γ2 construct (see Fig. 4A) and assaying the stability of these translated protein products in the presence of dipeptide inhibitors, with the goal of identifying those lysine residues sufficient to recapitulate the RDTASIA γ2 phenotype. As seen in Fig. 4B, the protein levels of RDTASIA γ2 containing a lysine residue at either position 14 or 20 were unaltered by dipeptide inhibitors, suggesting that these sites are not important for modulation of degradation. However, lysine residue(s) incorporated at positions 29/32, 46, or 64/65 (Fig. 4, rows 4–6) were each sufficient to effect degradation of RDTASIA γ2, as shown by its selective accumulation in the treatment (RA) group that blocks access to the type I site of the ubiquitin ligase. These functional residues reside in the C-terminal half of the γ subunit (Fig. 5), which, in the context of the G protein heterotrimer structure, has an orientation in close proximity to the plasma membrane (25, 26). In addition to a predicted role in degradation, we suggest that ubiquitin modification of the C-terminal residues of γ2 may also modulate association of the γ (and associated β) subunits with the plasma membrane.
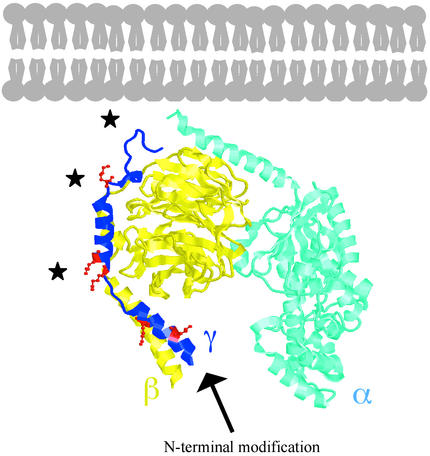
Figure 5.
N-end modification and ubiquitylation of the Gγ2 subunit. Depicted above is the crystal structure (IGP2) for the Gαi1, β1, γ2 heterotrimer, with the N terminus of α and C terminus of γ oriented toward the plasma membrane (25). Structural figure includes amino acids 8–61 of the γ2 subunit (amino acids 1–71) with the remainder of residues absent because of disordered structure. Lysine residues within γ2 are represented in red ball and stick format (with the exception of K64 and K65). The N terminus of γ2, the site of posttranslational modification, extends toward the interior of the cell, whereas the sites of ubiquitylation (indicated by black stars) are more closely oriented toward the plasma membrane.
Discussion
We have presented the identification of an N-terminal variant of the heterotrimeric Gγ2 protein, present within a physiological tissue source, bovine brain. Our demonstration that the RDTASIA γ2 variant undergoes ubiquitin-dependent degradation by the N-end rule pathway highlights a novel function for the γ2 subunit and offers important insight into the mechanism of targeting of heterotrimeric G protein subunits for ubiquitin-dependent degradation. Our work supports a model in which the N terminus of the γ subunit undergoes posttranslational modification, resulting in the production of a destabilizing N terminus capable of targeting the γ2 subunit for ubiquitylation (on a series of C-terminal lysine residues) and degradation by the N-end rule pathway (Fig. 5). Given the short predicted half-life of an N-terminal Arg-containing protein [t1/2 = 2 min (14)], our isolation and detection of the RDTASIA γ2 variant from brain extracts, although perhaps surprising, is clearly important, and it suggests that a large fraction of bovine γ2 protein could be processed through this pathway in the brain.
The proposed role for ubiquitin-dependent regulation of the γ subunit of the heterotrimer is particularly significant, given accumulating evidence for a role for ubiquitin in the regulation of distinct components of the G protein signaling pathway. Nearly two decades ago, it was first observed that a heterotrimer α subunit, yeast Gpa1, was a substrate for N-end rule degradation. This degradation, although dependent on the N-end enzyme Ubr1, was not targeted by the N-terminal amino acid of Gpa1 (27), a phenomenon believed to be the result of targeting by an internal degron. Recently, the site of ubiquitin modification for Gpa1 was elegantly mapped to residue 165, substitution of which results in moderate pheromone resistance, an observation consistent with its reduced level of ubiquitylation (28). However, given that the ubiquitylation site in Gpa1 is present within a loop absent in mammalian α subunits, should N-end ubiquitin-dependent degradation of mammalian heterotrimeric G protein subunits occur, it is likely to use a different ubiquitylation site and distinct targeting mechanism than that identified thus far for Gpa1 (α subunit) in S. cerevisiae. In fact, recent evidence suggests that modification of a distinct G protein subunit may occur in the mammalian retina. During the preparation of this article, transducin Gtγ was demonstrated to be an in vitro substrate for ubiquitylation, suggesting a role for ubiquitin-dependent degradation of βγ subunits in the rod inner segments after illumination (10). Although the targeting signal, or degron, for transducin subunits is proposed to reside within the β subunit, our data demonstrate the N terminus of a distinct isoform, Gγ2, is posttranslationally modified, resulting in the production of a substrate that can be recognized by an N-end ubiquitin ligase such as Ubr1/E3α. These observations cumulatively highlight an important role for ubiquitin modification in the regulation of heterotrimeric G protein subunits, although the mechanism of targeting and resulting cellular effects are likely to vary significantly.
The regulated degradation of one or more subunits of a G protein heterotrimer is likely to have important functional implications, suggested by the precise regulation of α and βγ levels, which has been observed during early mouse development or pertussis toxin treatment of cells (29, 30). There is clear precedence that distinct components of the G protein-coupled receptor (GPCR) signaling pathway, including GPCRs, β-arrestin, and RGS7 (regulator of G protein signaling), are regulated by ubiquitylation (31, 32), thus potentially altering the duration of the GPCR-dependent response. One hypothesis generated from the present study is that regulated βγ degradation (in response to agonist stimulation) may contribute to the attenuation of cellular signaling events, at the level of the G protein, by removal of an activated βγ dimer. In addition to contributing to the cessation of the primary signal, βγ removal may alter subsequent reformation of the heterotrimer either temporally (delay during the translation of new protein subunits) or spatially (allowing distinct subunits already present at a location in the cell to preferentially associate). Although our data suggest that N-end targeting of Gγ2 is likely to result in degradation, ubiquitylation may result in internalization within a cell rather than degradation; thus, there exists the possibility that ubiquitylation of Gγ2 at C-terminal lysine residues may have an important function in altering the cellular localization of particular modified or associated subunits, events which also have the potential to alter βγ function.
In addition to elucidating a targeting mechanism for ubiquitylation of the G protein γ2 subunit, this study is significant in that it identifies a Gγ2 variant found in bovine brain as a mammalian N-end rule substrate isolated from a physiological tissue source. RGS4, a negative regulator of Gα subunit activity, has been recently identified (by using a cDNA-based expression cloning strategy) as an N-end substrate in rabbit reticulocyte lysate and is N-terminally arginylated when expressed exogenously in mouse cells (33, 34). However, it remains unclear whether native RSG4 protein is processed in such a way for N-end rule recognition/degradation within a cell. Our identification of a Gγ2 variant in bovine brain, which contains a unique amino acid sequence at its N terminus, offers proof in principle for the proposed hierarchy of posttranslational modifications that converts a tertiary destabilizing residue to a type I N-end substrate (23), providing physiological evidence that such a series of N-end modifications does indeed occur in vivo. We acknowledge that elucidation of the upstream events that generate this variant of γ2 are of extreme importance. Because mammalian enzymes that could supply the N-terminal amidase and arginyltransferase activities have already been described (16, 17), we postulate that a currently unidentified protease may be the initiating step that activates the cascade of modifications that results in ubiquitylation and degradation. Such an activity is likely to be a significant part of the complexity of G protein subunit regulation and therefore signaling specificity within a cell.
Acknowledgments
We thank R. Ettling for Edman sequencing and the Biomolecular MS and Protein Facilities at the Medical University of South Carolina. This work was supported in part by a research grant from the National Institute of Diabetes and Digestive and Kidney Diseases, training grants from the National Institute of General Medical Sciences and the National Heart, Lung, and Blood Institute, and research resources support by the Medical University of South Carolina.
Abbreviations
- RD
RDTASIA γ2 protein
- RA
Arg-β-Ala
- WA
Trp-Ala
- noK
lysine-less construct
References
- 1.Helper J R, Gilman A G. Trends Biochem Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 2.Birnbaumer L. Cell. 1992;71:1069–1072. doi: 10.1016/s0092-8674(05)80056-x. [DOI] [PubMed] [Google Scholar]
- 3.Simon M I, Strathmann M P, Gautam N. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 4.Hurowitz E H, Melnyk J M, Chen Y-J, Kouros-Meir H, Simon M I, Shizuya H. DNA Res. 2000;7:111–120. doi: 10.1093/dnares/7.2.111. [DOI] [PubMed] [Google Scholar]
- 5.Yamane H K, Farnsworth C C, Xie H Y, Howald W, Fung B K, Clarke S, Gelb M H, Glomset J A. Proc Natl Acad Sci USA. 1990;87:5868–5872. doi: 10.1073/pnas.87.15.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mumby S M, Casey P J, Gilman A G, Gutowski S, Sternweis P C. Proc Natl Acad Sci USA. 1990;87:5873–5877. doi: 10.1073/pnas.87.15.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iniguez-Lluhi J A, Simon M I, Robishaw J D, Gilman A G. J Biol Chem. 1992;267:23409–23417. [PubMed] [Google Scholar]
- 8.Kisselev O G, Ermolaeva M V, Gautam N. J Biol Chem. 1994;269:21399–21402. [PubMed] [Google Scholar]
- 9.Dietrich A, Brazil D, Jensen O N, Meister M, Schrader M, Moomaw J F, Mann M, Illenberger D, Gierschick P. Biochemistry. 1996;35:15174–15182. doi: 10.1021/bi960305j. [DOI] [PubMed] [Google Scholar]
- 10.Obin M, Lee B Y, Meinke G, Bohm A, Lee R H, Gaudet R, Hopp J A, Arshavsky V Y, Willardson B M, Taylor A. J Biol Chem. 2002;277:44566–44575. doi: 10.1074/jbc.M205308200. [DOI] [PubMed] [Google Scholar]
- 11.Cook L A, Schey K L, Cleator J H, Wilcox M D, Dingus J, Hildebrandt J D. Protein Sci. 2001;10:2548–2555. doi: 10.1110/ps.ps.26401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 13.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 14.Varshavsky A. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 16.Grigoryev S, Stewart A E, Kwon Y T, Arfin S M, Bradshaw R A, Jenkins N A, Copeland N G, Varshavsky A. J Biol Chem. 1996;271:28521–28532. doi: 10.1074/jbc.271.45.28521. [DOI] [PubMed] [Google Scholar]
- 17.Kwon Y T, Kashina A S, Varshavsky A. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cook, L. A., Wilcox, M. D., Dingus, J., Schey, K. L. & Hildebrandt, J. D. Methods Enzymol. 344, 209–233. [DOI] [PubMed]
- 19.Taylor J A, Walsh K A, Johnson R S. Rapid Commun Mass Spectrom. 1996;10:679–687. doi: 10.1002/(SICI)1097-0231(199604)10:6<679::AID-RCM528>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 20.Varshavsky A. Methods Enzymol. 2000;327:578–593. doi: 10.1016/s0076-6879(00)27303-5. [DOI] [PubMed] [Google Scholar]
- 21.Asano T, Morishita R, Ohashi K, Nagahama M, Miyake T, Kato K. J Neurochem. 1995;64:1267–1273. doi: 10.1046/j.1471-4159.1995.64031267.x. [DOI] [PubMed] [Google Scholar]
- 22.Wilcox M D, Schey K L, Busman M, Hildebrandt J D. Biochem Biophys Res Commun. 1995;212:367–374. doi: 10.1006/bbrc.1995.1979. [DOI] [PubMed] [Google Scholar]
- 23.Gonda D K, Bachmair A, Wunning I, Tobias J W, Lane W S, Varshavsky A. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- 24.Baker R T, Varshavsky A. Proc Natl Acad Sci USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall M A, Coleman D E, Lee E, Iniguez-Lluhi J A, Posner B A, Gilman A G, Sprang S R. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 26.Lambright D G, Sondek J, Bohm A, Skiba N P, Hamm H E, Sigler P B. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 27.Madura K, Varshavsky A. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 28.Marotti L A, Newitt R, Wang Y, Aebersold R, Dohlman H G. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]
- 29.Allworth A E, Hildebrandt J D, Ziomek C A. Dev Biol. 1990;142:129–137. doi: 10.1016/0012-1606(90)90156-d. [DOI] [PubMed] [Google Scholar]
- 30.Watkins D C, Northup J K, Malbon C C. J Biol Chem. 1989;264:4186–4194. [PubMed] [Google Scholar]
- 31.Shenoy S K, McDonald P H, Kohout T A, Lefkowitz R J. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 32.Kim E, Arnould T, Sellin L, Benzing T, Comella N, Kocher O, Tsiokas L, Sukhatme V P, Walz G. Proc Natl Acad Sci USA. 1999;96:6371–6376. doi: 10.1073/pnas.96.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon Y T, Kashina A S, Davydov I V, Hu R-G, An J Y, Seo J W, Du F, Varshavsky A. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 34.Davydov I V, Varshavsky A. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 35.Cook L A, Schey K L, Wilcox M D, Dingus J, Hildebrandt J D. Biochemistry. 1998;37:12280–12286. doi: 10.1021/bi980230e. [DOI] [PubMed] [Google Scholar]