Abstract
The hyperthermophilic archaeon Methanococcus jannaschii encodes two putative transcription regulators, Ptr1 and Ptr2, that are members of the Lrp/AsnC family of bacterial transcription regulators. In contrast, this archaeon's RNA polymerase and core transcription factors are of eukaryotic type. Using the M. jannaschii high-temperature in vitro transcription system, we show that Ptr2 is a potent transcriptional activator, and that it conveys its stimulatory effects on its cognate eukaryal-type transcription machinery from an upstream activating region composed of two Ptr2-binding sites. Transcriptional activation is generated, at least in part, by Ptr2-mediated recruitment of the TATA-binding protein to the promoter.
The core components of archaeal transcription closely resemble those of eukaryotic RNA polymerase II (1). Archaeal promoters consist of an A+T-rich TATA-like element recognized by archaeal TATA-binding protein (TBP); the TFIIB-related transcription factor B (TFB) binds to the TBP–DNA complex and directs a eukaryotic-type RNA polymerase (RNAP) to specifically initiate transcription at an initiator sequence located some 25 bp downstream of the TATA element. Efficient preinitiation complex (PIC) assembly is ensured by the adjacent purine-rich BRE element, which mediates sequence-specific interactions with TFB upstream of the TATA box (2) and dictates the directionality of transcription complex assembly and initiation (3). TBP, TFB, and RNAP are necessary and sufficient to direct transcription at many archaeal promoters in vitro; a modest stimulatory effect of TFE, the archaeal homologue of the α subunit of the RNA polymerase II transcription factor TFIIE, is discerned under conditions of suboptimal TBP–TATA box interaction (4, 5).
On the other hand, all archaeal genomes sequenced to date encode potential transcription regulators of bacterial type, underscoring the chimeric nature of the archaeal transcription apparatus (6, 7). Particular interest is attached to the question of how these bacterial-type effectors, especially activators, generate regulation of a eukaryote-like transcription system. All of the putative regulators of transcription that have been characterized in vitro, the metal-dependent repressor 1 (MDR1) from Archaeoglobus fulgidus (8), as well as the homologues LrpA from Pyrococcus furiosus (9, 10), Lrs-14 from Sulfolobus solfataricus (11), and Ptr1 from Methanococcus jannaschii (unpublished results), have only been shown to repress transcription by their cognate RNA polymerases.
Here we show that Ptr2, a site-specific helix–turn–helix DNA-binding protein from the hyperthermophilic archaeon M. jannaschii and homologue of the bacterial leucine-responsive regulatory protein (Lrp) family of transcription factors, is a potent transcriptional activator in vitro. We also show that Ptr2 conveys its stimulatory effects on its cognate transcription machinery through direct recruitment of TBP.
Materials and Methods
Protein Purification.
The RNA polymerase from Methanococcus/Methanocaldococcus jannaschii was purified from 10 g of cells (wet weight) under exclusion of oxygen in an anaerobic chamber. A cellular extract (S-100) was prepared in TMK buffer [50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/50 mM KCl/20% (wt/vol) glycerol] (12), and loaded onto DEAE-cellulose. Bound proteins were eluted with a linear gradient of KCl (0.05–1 M). Active fractions were pooled, diluted with 3 vol of TMK buffer, and applied to a heparin-Sepharose column. Bound proteins were again eluted with a linear KCl gradient, and RNA polymerase-containing fractions were concentrated on Mono Q. Final purification was achieved by gel filtration on Superdex 200, preequilibrated with TMK buffer containing 300 mM KCl. This purification yielded the highly purified RNA polymerase (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org) used for promoter-specific in vitro transcription. Ptr2 overproduction and purification have been described (13). M. jannaschii transcription factors TBP and TFB were overproduced in Escherichia coli BL21(DE3)Gold(pLysS) harboring plasmids pLJ-MJ-TBP and pLJ-MJ-iTFB, respectively. These plasmids express fusion genes encoding an N-terminal His6 tag, followed by the products of M. jannaschii MJ0507 ORF and an inteinless derivative of MJ0782, respectively. The His-tagged factors were affinity purified on Ni-nitrilotriacetate-agarose, and were eluted with 250 mM imidazole. These proteins were at least 95% pure, as judged by SDS/PAGE analysis (see Fig. 5). Protein concentrations were measured by using the Micro BCA assay (Pierce), with BSA as the standard.
DNA Templates.
DNA fragments encompassing the entire fdxA–ptr2 intergenic region were amplified by PCR using M. jannaschii chromosomal DNA and oligonucleotides ON54 (5′-CTCAACCGCCATTTTCTTCCTCC-3′) and ON60 (5′-TCTCTCATAAGAATTTCGAT-3′). DNA containing only the fdxA promoter region was generated by using oligonucleotides ON114 (5′-CTAAATACATATAGTTCATTGCAAAATG-3′) and ON60. DNA fragments encompassing the rb2 promoter region were generated by using oligonucleotides ON116 (5′-GGGAATACGAAAAAGAGATTCTGC-3′) and ON117 (5′-CGGCTCACCTTTGTCTTCATCATAC-3′). All DNA templates were purified by native PAGE.
Hydroxyl Radical (•OH) Footprinting.
Protein–DNA complexes were assembled in 50 μl of binding buffer (20 mM K-Hepes, pH 7.8, 10 mM MgCl2/300 mM NaCl) with 200 fmol of 5′-end-32P-labeled DNA (4 nM) and specified quantities of Ptr2. Reaction mixtures were incubated at 65°C for the indicated periods of time and were subjected to hydroxyl radical (•OH) cleavage for 30 sec at the same temperature (13).
In Vitro Transcription.
Reaction mixtures were assembled in 50 μl (final volume) of transcription buffer [20 mM K-Hepes, pH 7.8/10 mM MgCl2/500 mM NaCl/1 mM DTT/2.5% (wt/vol) glycerol and thermoprotectants as noted below]. Preinitiation complexes were assembled by using 4 nM linear template DNA (200 fmol), 20 nM TBP, 20 nM TFB, and ≈30 nM RNA polymerase, for 20 min at 65°C. Single rounds of transcription were then initiated by the addition of nucleoside triphosphates to 0.4 mM each of ATP, CTP, and GTP; 32 μM [α-32P]UTP [3,000 Ci (1 Ci = 37 GBq)/mmol], poly(dI-dC)⋅poly(dI-dC) to 80 μg/ml; and 18 units of RNase inhibitor, for 15 min. The addition of 80 μg/ml poly(dI-dC)⋅poly(dI-dC) prevents reinitiation, thus limiting transcription to a single round (results not shown). Sample preparation (extraction with phenol/chloroform/isoamyl alcohol, and precipitation with ethanol), resolution by electrophoresis on denaturing 5% polyacrylamide gel, visualization by phosphor imaging, and quantification followed standard procedures. Although they are of hyperthermophilic origin, we noted that M. jannaschii RNA polymerase, TBP, and TFB were relatively rapidly inactivated in dilute solutions at 65°C. Exploration of thermoprotectants led to a mixture combining 600 mM trehalose, 500 mM betaine, and 5% (wt/vol) polyethylene glycol (PEG 3300), which conferred complete stability on the time scale of these transcription experiments. Reanalysis of salt-concentration dependence of transcription and activation in the presence of these thermoprotectants and macromolecular crowding agents led to selection of 350 mM NaCl as optimal for activation of fdxA and rb2UAS/tRNAval-44 transcription, and 500 mM NaCl for transcriptional activation at Prb2. However, transcriptional activation is not restricted to these conditions and is readily observed at NaCl concentrations as low as 200 mM (data not shown).
For transcription of M. jannaschii genomic DNA, PICs were assembled in 50 μl (final volume) of transcription buffer containing 350 mM NaCl, using ≈1.7 μg of EcoRI-digested chromosomal DNA (≈1.5 fmol), 33 nM TBP, 33 nM TFB, and ≈90 nM RNA polymerase, for 20 min at 65°C. Transcription was then initiated by the addition of nucleoside triphosphates to 0.4 mM each, for 20 min. Prb2 transcripts were detected by primer extension using 5′-end-32P-labeled oligonucleotide ON117 and avian myeloblastosis virus reverse transcriptase.
Results
The DNA-binding properties of Ptr2 have been characterized (13). A high-temperature (65°C) SELEX (systematic evolution of ligands by exponential enrichment) (14) has identified a palindrome, 5′-GGACGATTTTCGTCC-3′, as the consensus Ptr2-binding site. Ptr2 binds this symmetrical site from one side of the DNA helix, protecting ≈25 bp of DNA from DNase I cleavage, with evidence for DNA distortion at the edges of the protected segment (13).
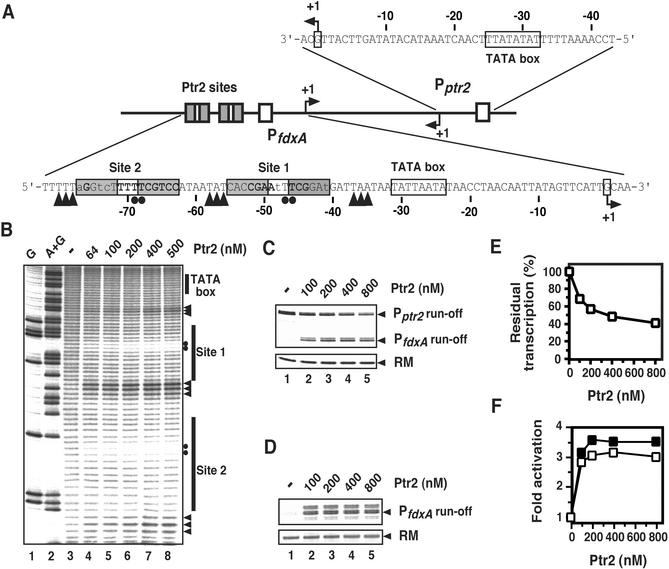
Ptr2 is encoded by MJ0723, an ORF that is adjacent to the ferredoxin-encoding ORF MJ0722 (fdxA), and is divergent from it (ref. 15; Fig. 1A). Ptr2 binding to DNA fragments encompassing the intergenic region between fdxA and ptr2 has been shown in electrophoretic mobility-shift assays (EMSAs) (13), suggesting a mode of Ptr2 autoregulation similar to what has been reported for its bacterial (16) and archaeal homologues (9–11). We used •OH footprinting to examine Ptr2 binding to the fdxA–ptr2 promoter region in more detail. As shown in Fig. 1B, two adjacent Ptr2-binding sites were mapped to base pairs −48 (site 1) and −70 (site 2), relative to the fdxA start site of transcription (base pairs +135 and +157 relative to the ptr2 start, respectively; Fig. 1A). Additional weaker Ptr2 binding to the DNA segment separating the fdxA and ptr2 transcriptional start sites has been detected in EMSAs (results not shown).
Figure 1.
Effects of Ptr2 on initiation of transcription at the divergent promoters PfdxA and Pptr2. (A) The fdxA–ptr2 intergenic region and its regulatory elements. Ptr2-binding sites (centered at base pairs −48 and −70 relative to the PfdxA start site of transcription) are represented with their hexamer motifs boxed and shaded. Nucleotides matching and differing from the SELEX-derived consensus for Ptr2 binding are shown in bold and lowercase, respectively. The PfdxA and Pptr2 sequences shown correspond to their respective nontranscribed (nontemplate) DNA strands, with TATA elements boxed, and mapped start sites of in vitro transcription (see Fig. 6, which is published as supporting information on the PNAS web site) indicated by bent arrows. The 87-bp segment between the ptr2 and fdxA start sites is transcribed off both strands. (B) High-temperature •OH footprinting of the Ptr2-binding sites upstream of PfdxA. A DNA probe, 5′-end-labeled on the nontranscribed strand, was incubated at 65°C for 15 min in the absence (lane 3) or presence of increasing concentrations of Ptr2 (specified for the monomer; lanes 4–8) and subjected to •OH cleavage for 30 sec at 65°C. On the right (and also below the PfdxA sequence in A), filled circles indicate positions of strongest protection by Ptr2, and arrowheads indicate increased susceptibility to cleavage. Lanes 1 and 2, G and A+G chemical sequencing ladders, respectively. (C) Effects of Ptr2 on single rounds of transcription at PfdxA and Pptr2. A DNA template encompassing the fdxA and ptr2 promoter regions (base pair −104 of PfdxA to base pair −60 of Pptr2) was incubated in the absence (lane 1) or presence (lanes 2–5) of increasing concentrations of Ptr2 before addition of TBP, TFB, and purified RNA polymerase. The 191-nt Pptr2 and the 150-nt PfdxA runoff transcripts and the recovery marker DNA (RM) are indicated on the right. The multiple PfdxA and Pptr2 runoff transcripts are generated through utilization of alternative start sites of transcription (Fig. 6) and 3′-end heterogeneity that results from utilization of PCR-derived linear DNA templates. (D) Effect of Ptr2 on fdxA transcription from a DNA template encompassing PfdxA only (bp −104 of PfdxA to bp −20 of Pptr2). The 107-nt runoff transcript and the RM are indicated on the right. (E) Quantitative analysis of Ptr2 repression of transcription of its gene (data are from C). (F) Quantitative analysis of Ptr2-activated relative to basal fdxA transcription. Open squares correspond to levels of activation elicited by Ptr2 on DNA encompassing both the fdxA and ptr2 promoters (data are from C), and filled squares correspond to those elicited on a template containing PfdxA only (data are from D).
To investigate the transcriptional effects of Ptr2 binding to these sites, we developed a reconstituted M. jannaschii in vitro transcription system composed of highly purified RNA polymerase and recombinant transcription factors TBP and TFB. Surprisingly, the addition of Ptr2 to linear DNA templates encompassing the entire fdxA–ptr2 intergenic region (Fig. 1A) not only inhibited transcription initiating at the ptr2 promoter (by ≈60%; Fig. 1 C and E) but also stimulated initiation at the oppositely oriented, weak fdxA promoter (≈3-fold; Fig. 1 C and F). To eliminate the possibility that increased fdxA transcription is merely the passive consequence of a shift in the competition between the strong ptr2 and weak fdxA promoters resulting from repression at Pptr2, we examined transcription of a DNA segment (base pairs −104 of PfdxA to base pairs −20 of Pptr2) containing only PfdxA and its upstream Ptr2-binding sites. Ptr2 elicited similar levels of activation with this DNA (Fig. 1 D and F). This finding represents direct evidence of an archaeal transcription regulator exerting dual transcriptional effects on divergent genes through binding to their intergenic region. Ptr2 is also, to our knowledge, the first bacterial-type regulator of archaeal origin shown to activate its cognate eukaryote-like transcription machinery in vitro.
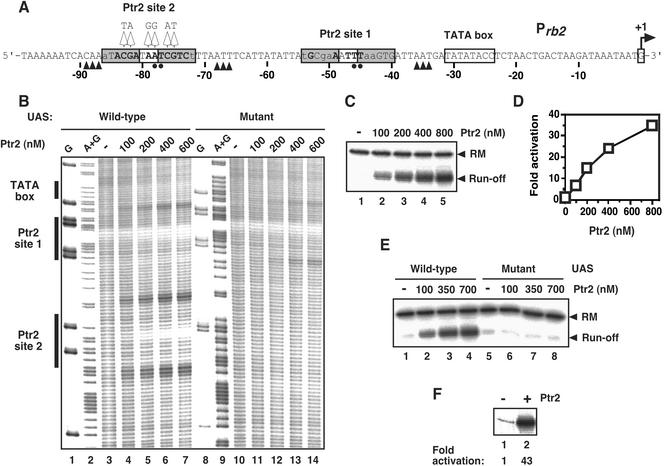
Ferredoxins and rubredoxins are small, acidic, soluble iron–sulfur proteins that serve as electron carriers in diverse redox processes, and they are important components of anabolic and catabolic electron transfer reactions (17). A putative Ptr2-binding site upstream of rb2, the rubredoxin 2-encoding gene (MJ0740), had been identified on the basis of its high degree of similarity to the SELEX-derived consensus sequence for Ptr2 binding (13). We turned to the rb2 promoter as a potential second site for Ptr2-mediated transcriptional activation. By using •OH footprinting, two adjacent Ptr2-binding sites were mapped upstream of the rb2 promoter, centered at base pairs −47 (site 1) and −79 (site 2) relative to the start site of transcription, respectively (Fig. 2 A and B, lanes 4–7). Site 2, which conforms more closely to the consensus, is the previously identified “putative” Ptr2-binding site. Initiation of transcription at the very weak Prb2 was stimulated almost 40-fold at the highest concentrations of Ptr2 (Fig. 2 C and D). Mutations driving Ptr2 site 2 farther from the SELEX-derived consensus (Fig. 2A) abolished Ptr2 binding to site 2 (Fig. 2B, lanes 11–14) and abolished transcriptional activation by Ptr2 (Fig. 2E, lanes 6–8). (Quantitative changes of the Ptr2 site 1 footprint, and the appearance of a new site of increased susceptibility to •OH cleavage when Ptr2-binding site 2 is eliminated suggest an interaction between Ptr2 binding at sites 1 and 2.) Taken together, these data specify that Ptr2 is a potent positive regulator of rb2 gene transcription, and that it conveys its stimulatory effect on the transcription machinery through a cis-acting site that includes Ptr2-binding site 2. The requirement for a bipirtate binding locus has been recently shown for the transcriptional activation of the ilvIH operon by E. coli Lrp (18).
Figure 2.
Ptr2-dependent activation of transcription at the rb2 promoter. (A) The nucleotide sequence of the nontranscribed DNA strand is shown, with the TATA element boxed, and the mapped start site of transcription indicated by the bent arrow. Ptr2-binding sites (centered at base pairs −47 and −79 relative to the rb2 start site of transcription, respectively) are represented as in Fig. 1. Changes at Ptr2-binding site 2 to generate the mutant upstream activation sequence (UAS) are indicated above the sequence. Filled circles below the sequence indicate positions of strongest protection from •OH cleavage by Ptr2, and arrowheads indicate increased susceptibility. (B) •OH footprinting of the Ptr2-binding sites upstream of Prb2. DNA probes containing the wild-type (lanes 3–7) or mutant Prb2 UAS (lanes 10–14) were incubated in the absence (lanes 3 and 10) or presence (lanes 4–7 and 11–14) of increasing concentrations of Ptr2 and subjected to •OH cleavage. Lanes 1 and 8 and lanes 2 and 9 show G and A+G chemical sequencing ladders, respectively. (C) Transcriptional activation at Prb2. DNA (base pairs −185 to +84 relative to the start site of rb2 transcription) was incubated in the absence (lane 1) or presence (lanes 2–5) of increasing amounts of Ptr2 and was used in a single-round transcription assay. (D) Levels of activated transcription relative to basal transcription. (E) Transcription elicited by Ptr2 on Prb2 templates containing the wild-type (lanes 2–4) and mutant (lanes 6–8) UAS are compared. (F) Primer extension analysis of Prb2 transcripts generated in vitro by using M. jannaschii genomic DNA, in the absence (lane 1) or in the presence (lane 2) of 1 μM Ptr2. The level of activation elicited by Ptr2 is indicated below lane 2.
Transcription experiments using total M. jannaschii genomic DNA confirmed that activation of transcription by Ptr2 at Prb2 is not confined to assays using short promoter segments but is also manifested in competition with diverse binding sites for all components of the in vitro system (Fig. 2F).
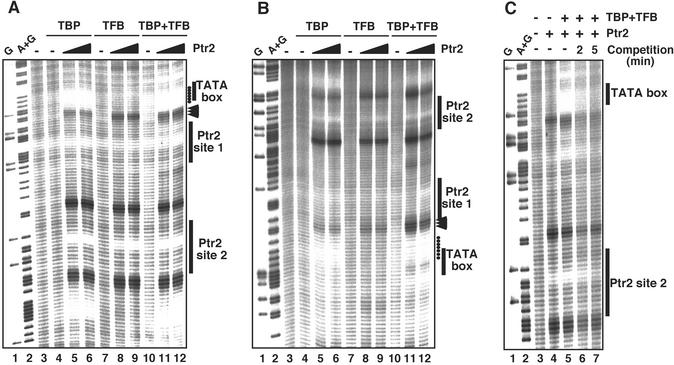
The importance of recruitment in transcriptional activation is underscored by diverse “activator-bypass” experiments in which high levels of transcription are achieved by artificially tethering a component of the core transcription machinery near the promoter (19–23). We used •OH footprinting to monitor recruitment of TBP and/or TFB to the rb2 promoter by Ptr2. TBP, at a concentration of 80 nM, did not significantly protect against •OH cleavage around the TATA box (Fig. 3 A and B, compare lanes 4 with lanes 3), allowing its Ptr2-mediated recruitment to be readily detected as protection against •OH cleavage around the TATA box (Fig. 3 A and B, compare lanes 5 and 6 with lanes 4). In the absence of Ptr2, TBP and TFB together generated incomplete protection around the TATA box, specifying only a small effect of TFB alone on promoter occupancy by TBP (Fig. 3 A and B, compare lanes 10 with lanes 4). Efficient recruitment of TBP and TFB by DNA-bound Ptr2 was manifested by increased protection as well as upstream and downstream extensions of the TATA-box footprint, with the downstream extension clearly TFB-dependent (compare lanes 11 and 12 with lanes 5 and 6 in each panel). TFB binding was not detected in the absence of TBP, and TFB was not recruited to the promoter by Ptr2 in the absence of TBP (lanes 7–9 in each panel). Taken together, these data suggest that Ptr2 conveys its stimulatory effect on the M. jannaschii transcriptional machinery, at least in part, through direct recruitment of TBP to promoter DNA.
Figure 3.
Ptr2-mediated recruitment of TBP to the rb2 promoter. The recruitment of TBP (80 nM; lanes 4–6), TFB (80 nM; lanes 7–9), or both factors (lanes 10–12) to Prb2 DNA preincubated in the absence (lanes 4, 7, and 10) or presence (lanes 5, 8, and 11) of 150 nM or 300 nM (lanes 6, 9, and 12) Ptr2, was monitored by •OH footprinting. Positions of strong protection by Ptr2-recruited TBP around the TATA box, on the nontranscribed (A) and transcribed (B) strand, are indicated by filled circles. Enhanced •OH reactivity between the TATA box and Ptr2 site 1 when TBP, TFB, and Ptr2 are all bound to DNA, especially prominent on the bottom strand, is indicated with arrowheads. Ptr2 also recruited TBP to an A+T-rich sequence located upstream of the Ptr2 site 2 (centered at base pair −97), but the functional significance of this, if any, has not been analyzed. (C) Stability of Ptr2–TBP–TFB–DNA complexes. rb2 promoter complexes were assembled in the presence of 300 nM Ptr2, 80 nM TBP, and 80 nM TFB, as in A, and were subjected to •OH cleavage either before (lane 5) or after (lanes 6 and 7) challenge with a 10-fold excess of unlabeled specific DNA for the indicated times.
The stability of Ptr2–TBP–TFB–DNA complexes formed at the rb2 promoter was examined by •OH footprinting and was found to be low (t1/2 ≈ 2–3 min), with the characteristic Ptr2 and TBP footprints quickly dissipating after challenge with excess specific competitor DNA (Fig. 3 C, compare lanes 6 and 7 with lane 5).
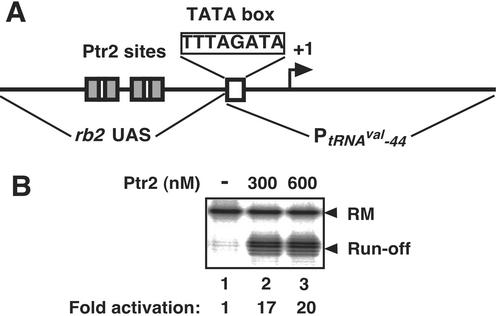
The rb2 gene TATA element (5′-TATATACC-3′; Fig. 2A) deviates from a proposed all-A/T consensus for methanogens (5′-TTTATATA-3′; ref. 1), at the downstream end. Single G substitutions at that end of the strong M. vannielii tRNAVal promoter substantially reduce transcription in a M. thermolithotrophicus in vitro system (24). To test whether Ptr2 might also restore transcription at a heterologous promoter with an artificially weakened TATA element, a hybrid construct was generated by inserting the entire upstream segment of Prb2 (base pairs −185 to −32) immediately upstream of the weakened TATA box of a variant tRNAVal promoter (tRNAVal-44: 5′-TTTAGATA-3′, ≈3% of wild-type activity; ref. 24; Fig. 4A). Ptr2 activated initiation of transcription at this hybrid promoter almost 20-fold in the M. jannaschii in vitro transcription system (Fig. 4B, lanes 2 and 3). This result indicates that Ptr2 can effectively rescue the transcriptional defect of a heterologous promoter, and that its binding sites near the rb2 promoter serve as a transportable cis-acting element for transcriptional activation.
Figure 4.
Ptr2-dependent activation of transcription at a heterologous promoter. (A) The hybrid rb2UAS/tRNAVal-44 promoter, generated by replacing the rb2 transcription unit (base pairs −31 to +84, relative to the start site of transcription) with the base pair −30/+85 segment of the M. vannielii tRNAVal-44 promoter. (B) The effect of Ptr2 on single rounds of transcription initiating at the rb2UAS/tRNAVal-44 promoter. Levels of activation elicited by Ptr2 are indicated below lanes 2 and 3.
Discussion
Our findings represent a direct (i.e., in vitro) demonstration of positive regulation of a eukaryote-like archaeal transcription apparatus by a cognate bacterial-type regulator, and also specify facilitated recruitment of TBP as a mechanism of transcriptional activation in archaea. There is an obvious correspondence with eukaryotic RNA polymerase II transcription, in which TBP association with promoters strongly correlates with PIC assembly, and transcriptional activation is similarly generated by increased recruitment of TBP and its associated factors to promoters by regulatory proteins (25–27). It remains to be seen whether TBP is the main basal component of the archaeal transcription machinery targeted by its bacterial-type positive regulators, and whether Ptr2 can also affect subsequent steps of the initiation process (promoter opening and/or promoter clearance). Ongoing analyses of the effects of various promoter architectures (e.g., Ptr2-site placement and polarity relative to the TATA element) on the transcriptional response to Ptr2 should provide further insight into the activation mechanism. The ability to readily detect Ptr2-mediated transcription activation on total M. jannaschii genomic DNA (Fig. 2F) also opens up the global exploration of the Ptr2 regulon by using runoff transcription/macroarray analyses (ROMA) (28).
In eukaryotes, transcriptional activation is generated through the stabilization of PICs, which effectively marks the promoter for rapid and preferential reinitiation (29–32). Activation of bacterial transcription, on the other hand, and to a large extent, does not depend on strong promoter marking and can be entirely independent of it. At Prb2, it appears that activation is generated by PICs whose transience is comparable to that of canonical bacterial activator–promoter complexes. The dynamic character of this activation raises the possibility that, although deploying eukaryal-type core components, archaeal transcription machineries have the potential to hew close to the bacterial norm.
Archaea, like bacteria, contain proteins that compact their chromosomes. The methanogenic euryarchaea, which include M. jannaschi, elicit special interest because they encode homologues of histones H3 and H4 that bind preferentially to nucleosome-localizing sequences (NLSs), just as eukaryal histone octamers and H3/H4 tetramers do, and can form regularly spaced histone arrays that resemble beads-on-a-string chromatin fibers on DNA containing regularly spaced NLSs (33–35). Archaeal histones can suppress basal transcription at strong promoters (36); it will be interesting to see whether they serve as “enforcers” of positive regulation at inherently weak promoters such as rb2 and fdxA. The recent in vitro assembly of M. jannaschii RNA polymerase from purified recombinant subunits opens up the exploration of subunit and subassembly requirements for basal promoter-specific transcription (37), and our work extends the reach of such an analysis to factor-activated transcription.
The three-dimensional structure of P. furiosus LrpA, the closest known Ptr2 homologue (55% amino acid identity, 82% similarity), has been determined recently (38), and it provides the structural basis for a detailed mapping of the TBP-interacting surface(s) of Ptr2. The high degree of sequence, structural, and functional conservation among archaeal TBPs also invites an exploration of Ptr2-mediated activation of heterologous archaeal transcription machineries (i.e., the euryarchaea M. thermolithotrophicus and P. furiosus, and the crenarchaea Sulfolobus acidocaldarius and Sulfolobus shibatae) in vitro.
Supplementary Material
Acknowledgments
We thank G. A. Kassavetis for helpful discussions; K. Adelman, R. R. Burgess, M. Kamali, G. A. Kassavetis, and L. B. Rothman-Denes for their critical reading of the manuscript; the National Institute of General Medical Sciences for support of this research at the University of California at San Diego; and the Deutsche Forschungsgemeinschaft for research support at Kiel, Germany.
Abbreviations
- TBP
TATA-binding protein
- TFB
transcription factor B
- SELEX
systematic evolution of ligands by exponential enrichment
- PIC
preinitiation complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Thomm M. FEMS Microbiol Rev. 1996;18:159–171. doi: 10.1111/j.1574-6976.1996.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi S A, Jackson S P. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 3.Bell S D, Kosa P L, Sigler P B, Jackson S P. Proc Natl Acad Sci USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanzelka B L, Darcy T J, Reeve J N. J Bacteriol. 2001;183:1813–1818. doi: 10.1128/JB.183.5.1813-1818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell S D, Brinkman A B, van der Oost J, Jackson S P. EMBO Rep. 2001;2:133–138. doi: 10.1093/embo-reports/kve021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aravind L, Koonin E V. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kyrpides N C, Ouzounis C A. Proc Natl Acad Sci USA. 1999;96:8545–8550. doi: 10.1073/pnas.96.15.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell S D, Cairns S S, Robson R L, Jackson S P. Mol Cell. 1999;4:971–982. doi: 10.1016/s1097-2765(00)80226-9. [DOI] [PubMed] [Google Scholar]
- 9.Brinkman A B, Dahlke I, Tuininga J E, Lammers T, Dumay V, de Heus E, Lebbink J H, Thomm M, de Vos W M, van der Oost J. J Biol Chem. 2000;275:38160–38169. doi: 10.1074/jbc.M005916200. [DOI] [PubMed] [Google Scholar]
- 10.Dahlke I, Thomm M. Nucleic Acids Res. 2002;30:701–710. doi: 10.1093/nar/30.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell S D, Jackson S P. J Biol Chem. 2000;275:31624–31629. doi: 10.1074/jbc.M005422200. [DOI] [PubMed] [Google Scholar]
- 12.Hausner W, Thomm M. J Biol Chem. 1993;268:24047–24052. [PubMed] [Google Scholar]
- 13.Ouhammouch M, Geiduschek E P. EMBO J. 2001;20:146–156. doi: 10.1093/emboj/20.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 15.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Wu J, Friedberg D, Plakto J, Calvo J M. J Bacteriol. 1994;176:1831–1839. doi: 10.1128/jb.176.7.1831-1839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer J. FEBS Lett. 2001;509:1–5. doi: 10.1016/s0014-5793(01)03049-6. [DOI] [PubMed] [Google Scholar]
- 18.Jafri S, Chen S, Calvo J M. J Bacteriol. 2002;184:5293–5300. doi: 10.1128/JB.184.19.5293-5300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee S, Struhl K. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 21.Klages N, Strubin M. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 22.Xiao H, Friesen J D, Lis J T. Mol Cell Biol. 1995;15:5757–5761. doi: 10.1128/mcb.15.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dove S L, Joung J K, Hochschild A. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 24.Hausner W, Frey G, Thomm M. J Mol Biol. 1991;222:495–508. doi: 10.1016/0022-2836(91)90492-o. [DOI] [PubMed] [Google Scholar]
- 25.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 26.Kuras L, Struhl K. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 27.Li X Y, Virbasius A, Zhu X, Green M R. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 28.Cao M, Kobel P A, Morshedi M M, Wu M F, Paddon C, Helmann J D. J Mol Biol. 2002;316:443–457. doi: 10.1006/jmbi.2001.5372. [DOI] [PubMed] [Google Scholar]
- 29.Trendelenburg M F, Gurdon J B. Nature. 1978;276:292–294. doi: 10.1038/276292a0. [DOI] [PubMed] [Google Scholar]
- 30.Bieker J J, Martin P L, Roeder R G. Cell. 1985;40:119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- 31.Yudkovsky N, Ranish J A, Hahn S. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 32.Bertolino E, Singh H. Mol Cell. 2002;10:397–407. doi: 10.1016/s1097-2765(02)00597-x. [DOI] [PubMed] [Google Scholar]
- 33.Bailey K A, Pereira S L, Widom J, Reeve J N. J Mol Biol. 2000;303:25–34. doi: 10.1006/jmbi.2000.4128. [DOI] [PubMed] [Google Scholar]
- 34.Sandman K, Reeve J N. Adv Appl Microbiol. 2001;50:75–99. doi: 10.1016/s0065-2164(01)50004-0. [DOI] [PubMed] [Google Scholar]
- 35.Tomschik M, Karymov M A, Zlatanova J, Leuba S H. Structure (London) 2001;9:1201–1211. doi: 10.1016/s0969-2126(01)00682-7. [DOI] [PubMed] [Google Scholar]
- 36.Soares D, Dahlke I, Li W T, Sandman K, Hethke C, Thomm M, Reeve J N. Extremophiles. 1998;2:75–81. doi: 10.1007/s007920050045. [DOI] [PubMed] [Google Scholar]
- 37.Werner F, Weinzierl R O. Mol Cell. 2002;10:635–646. doi: 10.1016/s1097-2765(02)00629-9. [DOI] [PubMed] [Google Scholar]
- 38.Leonard P M, Smits S H, Sedelnikova S E, Brinkman A B, de Vos W M, van der Oost J, Rice D W, Rafferty J B. EMBO J. 2001;20:990–997. doi: 10.1093/emboj/20.5.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.