Abstract
Carbohydrate-responsive element binding protein (ChREBP) is a transcription factor that activates lipogenic genes in liver in response to excess carbohydrate in the diet. ChREBP is regulated in a reciprocal manner by glucose and cAMP. cAMP-dependent protein kinase (protein kinase A) phosphorylates two physiologically important sites in ChREBP, Ser-196, which is located near nuclear localization signal sequence (NLS), and Thr-666, within the basic helix–loop–helix (bHLH) site, resulting in inactivation of nuclear translocation of ChREBP and of the DNA-binding activity, respectively. We demonstrate here that crude cytosolic extracts from livers of rats fed a high carbohydrate diet contained protein phosphatase (PPase) activity that dephosphorylated a peptide containing Ser-196, whereas a PPase in the nuclear extract catalyzed dephosphorylation of Ser-568 and Thr-666. All these PPases are activated specifically by xylulose 5-phosphate (Xu5P), but not by other sugar phosphates. Furthermore, addition of Xu5P elevated PPase activity to the level observed in extracts of fed liver cells. These partially purified PPases were characterized as PP2A-ABδC by immunoblotting with specific antibodies. These results suggest that (ia) Xu5P-dependent PPase is responsible for activation of transcription of the L-type pyruvate kinase gene and lipogenic enzyme genes, and (ii) Xu5P is the glucose signaling compound. Thus, we propose that the same Xu5P-activated PPase controls both acute and long-term regulation of glucose metabolism and fat synthesis.
Evolutionary pressures favor the ability to store nutrients as fat during times of abundant food supply as a safeguard against starvation during periods of food shortage in which energy intake is not sufficient to meet demands. In mammals the predominant storage form of energy is triglycerides synthesized from fatty acids derived from the diet or synthesized from carbohydrates through the intermediate acetyl-CoA. Glucose is the essential energy source. The pathways involved in the conversion of excess glucose to triglycerides include glycolysis and lipogenesis, which occur primarily in liver. Feeding a diet high in carbohydrate to rats causes rapid activation of a number of key enzymes in glycolysis and lipogenesis. These enzymes include fructose-6-P 2-kinase/fructose-2,6-P2 bisphosphatase, pyruvate kinase, fatty acid synthase, and acetyl-CoA carboxylase (reviewed in refs. 1 and 2). Fructose 2,6-bisphosphate (Fru-2,6-P2), the most potent activator of phosphofructokinase (PFK), increases rapidly in liver in response to high glucose. Fru-2,6-P2 is synthesized and degraded by a bifunctional enzyme termed 6-phosphofructo-2-kinase/Fru-2,6-P2 bisphosphatase, which is inhibited by phosphorylation catalyzed by cAMP-dependent protein kinase (PKA). Glucose activates the enzyme by dephosphorylation catalyzed by xylulose 5-phosphate (Xu5P)-activated protein phosphatase (PP2A) (3), resulting in increased Fru-2,6-P2. Thus, the glucose signaling mechanism in the activation of PFK and glycolysis is mediated by increased Xu5P, which acts as a glucose signaling compound and recruits and activates a specific protein serine/threonine phosphatase (PPase). As a long-term response, high-carbohydrate diet induces expression of many of the genes encoding the glycolytic and lipogenic enzymes, thereby promoting conversion of excess carbohydrate to storage fat in liver.
While insulin and glucagon play crucial roles in homeostasis of carbohydrate and fatty acids, glucose also has been implicated as an independent signal for activating the synthesis of glycolytic and lipogenic enzymes. The mechanism by which transcription is activated by high glucose feeding has been poorly understood (4–8). It is known that not glucose itself but its metabolites activate the gene transcription. Among the glucose metabolites, glucose 6-phosphate (9), a metabolite of pyruvate (10), and Xu5P (11) have been suggested as a glucose signal that regulates a glucose-responsive transcription factor. Because a mixture of the various intermediates is formed in vivo, it has been difficult to identify the true signaling compound, especially when the target glucose-responsive transcription factor is not known.
More recently, we purified and identified a previously uncharacterized hepatic transcription factor, designated carbohydrate-responsive element binding protein (ChREBP) (12). This factor meets a number of requirements for carbohydrate-responsive activation of transcription of the L-type pyruvate kinase (LPK) as well as the lipogenic enzyme genes such as acetyl-CoA carboxylase and fatty acid synthase.
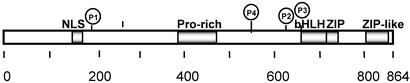
ChREBP is a member of the basic helix–loop–helix/leucine zipper (bHLH/ZIP) family of transcription factors with Mr = 94,600 and contains a nuclear localization signal (NLS), proline-rich stretches (PRO), a bHLH/ZIP, and a ZIP-like domain (Fig. 1). We demonstrated that the NLS site is inhibited by phosphorylation of Ser-196 by PKA and that ChREBP is localized in the cytosol of hepatocytes. However, when an animal is fed a diet high in carbohydrate, ChREBP is activated by being imported into the nucleus (13), and inside the nucleus the DNA-binding activity of ChREBP is activated by dephosphorylation of Thr-666, which is situated within the basic amino acids essential for DNA binding (Fig. 1). Thus, excess glucose stimulates ChREBP activity by two mechanisms: dephosphorylation and activation of the NLS in cytosol and DNA binding in the nucleus. In addition, fatty acid metabolism produces AMP, which activates AMP-activated protein kinase (AMPK). AMPK inhibits the DNA-binding activity of ChREBP by phosphorylation of Ser-568 of ChREBP (14).
Figure 1.
The domain structure of ChREBP. The locations of the NLS, proline-rich stretch, bHLH/leucine zipper (bHLH/ZIP), and ZIP-like domains are indicated. The locations of three phosphorylation sites of PKA are designated as P1, P2, and P3. The AMP-activated protein kinase (AMPK) site is indicated as P4.
The remaining questions are (i) how does glucose (or a glucose metabolite) activate ChREBP, and (ii) what is the glucose signaling compound? Here we find that high glucose activates ChREBP by dephosphorylation of ChREBP by PPases in the cytoplasm and the nuclei. These PPases are specifically activated by a metabolite in the extracts of cytoplasm and the metabolite was identified as Xu5P, suggesting that Xu5P is the glucose signaling compound. The PPases were partially purified and characterized as PP2A, the Xu5P-activated PPase described previously (15).
Materials and Methods
Materials.
[γ-32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq) was purchased from Amersham Pharmacia Biosciences. The PKA catalytic subunit was from New England Biolabs. AMPK was purchased from Upstate Biotechnology (Lake Placid, NY). Okadaic acid and other inhibitors were from Calbiochem. DEAE-cellulose (DE52) was purchased from Whatman, and Mono Q columns were from Amersham Pharmacia Biosciences. All other chemicals were reagent grade from commercial sources.
Animals.
Male Sprague–Dawley rats weighing 200–250 g were purchased from Sasco (Omaha, NE). Rats were starved for 48 h and then fed 24 h with the National Institutes of Health high-sucrose diet lab chow (by weight: 20% casein, 60% sucrose, 15% cellulose, 2.5% minerals, and 2.5% vitamins).
Preparation of Cytosolic and Nuclear Extracts of Liver.
Liver was homogenized in a motor-driven Potter–Elvehjem homogenizer in 1 vol of extraction buffer containing 10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.74 mM spermidine, 0.25 M sucrose, 50 mM imidazole, 1 mM DTT, 4 mM phenylmethanesulfonyl fluoride, and 1 mM benzamidine, and the extract was centrifuged at 10,000 × g for 30 min.
Liver nuclear extracts were prepared according to Hattori et al. (16), modified to include the PEG precipitation described previously (8). Nuclei were suspended in nuclear lysis buffer (10% glycerol/10 mM Hepes, pH 7.9/0.1 mM EDTA/1 mM DTT/4 mM phenylmethanesulfonyl fluoride/1 mM benzamidine) and centrifuged at 27,000 × g for 25 min. Protein concentration was determined by the Bradford method (17).
Preparation of Phosphorylated Substrates and Protein Phosphatase Assay.
Synthetic peptides corresponding to amino acids 187–201 (YYKKRLRKSSREGDF; P1 peptide), 556–573 (STVPSTLLRPPESPDAVP, P4 peptide), and 655–674 (VDNNKMENRRITHISAEQKR; P3 peptide) of ChREBP were prepared by the Peptide Synthesis Service of the University of Texas Southwestern Medical Center. These peptides were phosphorylated by 10 units of PKA for 6 h at 30°C. P4 peptide corresponding to residues 556–573 (STVPSTLLRPPESPDAVP) was phosphorylated by 10 units of AMPK for 5 h at 30°C. The reaction mixtures were passed through a Dowex AG1X8 column (0.5 cm × 5 cm) that had been equilibrated with 30% (wt/vol) acetic acid to remove residual [32P]ATP. The labeled peptides in the pass-through fraction were lyophilized.
Protein phosphatase activity was determined by the method of Donella-Deana et al. (18). The reaction mixture contained in a final volume of 50 μl, 20 mM Mops at pH 7.0, 1 mM DTT, 25 μg of BSA, and 32P-labeled substrate (100 μM) and was incubated for 15 min at 30°C. The reaction was terminated by the addition of 10% trichloroacetic acid. Phosphate released was converted to a phosphomolybdic complex, which was extracted by addition of equal volumes of organic solvent (isobutyl alcohol/heptane, 1:1). The reaction mixture was mixed thoroughly and centrifuged. An aliquot of the top layer was then added to 5 ml of scintillation fluid and the radioactivity was measured. One unit of protein phosphatase is defined as that amount which catalyzes the release of 1.0 μmol of 32P per min at 30°C.
Construction of Plasmids.
Full-length rat ChREBP cDNA (ref. 14, GenBank accession no. AB074517) was ligated into the mammalian expression vector pcDNA3 (Invitrogen) or the pEGFP-N3 vector encoding enhanced green fluorescent protein (GFP; CLONTECH), as previously described (13).
Primary Hepatocyte Culture and Transfection.
Primary hepatocytes were prepared from male Sprague–Dawley rats (250–300 g) by using the collagenase perfusion method (13) and plated in collagen-coated 35-mm tissue culture plates (Primaria Falcon/Franklin Lakes, NJ) at a density of 1.0 × 106 cells per well in glucose-free DMEM (Life Technologies) supplemented with 10 nM dexamethasone, 0.1 unit/ml insulin, 100 units/ml penicillin, 100 μg/ml streptomycin, 10% dialyzed FBS, and 5.5 mM glucose. After a 6-h attachment period, hepatocytes were transfected by using synthetic liposomes (Lipofectamine 2000 from Life Technologies) in Opti-MEM I reduced-serum medium as described (13) and incubated for 12 h. The media containing the liposome–DNA complex was removed and replaced with glucose-free DMEM containing 27.5 mM glucose.
Determination of Subcellular Localization of ChREBP.
Subcellular localization of GFP-fused ChREBP was determined by using a confocal laser scanning microscope (Bio Rad) and nuclear localization of ChREBP was quantitated as described previously (13).
Transcription of LPK Assayed by Using a Luciferase Reporter.
Transfected cells were cultured for 12 h in medium containing 27.5 mM glucose, washed twice with 2 ml of Ca2+- and Mg2+-free PBS, and lysed with Passive Lysis Buffer (Promega). Luciferase activity assay was performed as described previously (13). After measuring the firefly luciferase signal (LAF) and the Renilla (sea pansy) luciferase signal (LAR), the relative luciferase activity (RLA) was calculated as RLA = LAF/LAR, where RLA was calculated as a percentage, i.e., % RLA = RLA/(RLA)max. To compare the RLA in one cell line with another, an index of RLA was calculated as IRLA = RLA/RLASV40, where RLASV40 is the ratio of firefly luciferase signal with a simian virus 40 promoter in pGL3 divided by the Renilla luciferase signal in pRL-TK.
Metabolite Measurements.
HClO4 extracts of freeze-clamped livers were prepared as described (19). Xu5P was assayed by the method of Casazza and Veech (20), except the formation of fructose 6-phosphate coupled to NADPH formation was measured fluorimetrically by adding phosphoglucose isomerase and glucose-6-phosphate dehydrogenase with excitation and emission wavelengths at 354 and 452 nm, respectively. All other metabolites were assayed spectrophotometrically by enzymatic methods (21).
Results
ChREBP contains three phosphorylation sites, P1, P3, and P4, as shown in Fig. 1, and this nomenclature will be used in the following results and discussion. P1 and P3 are phosphorylated by PKA, whereas P4 is phosphorylated by AMPK, all resulting in inactivation of nuclear translocation or DNA binding activity. The P2 site affects the DNA-binding activity but it does not appear to be important in regulation by cAMP and glucose in hepatocytes (13). We proposed that glucose activates specific PPases, which dephosphorylate phosphorylated ChREBP.
Dephosphorylation of P1 Site and Activation of Nuclear Translocation.
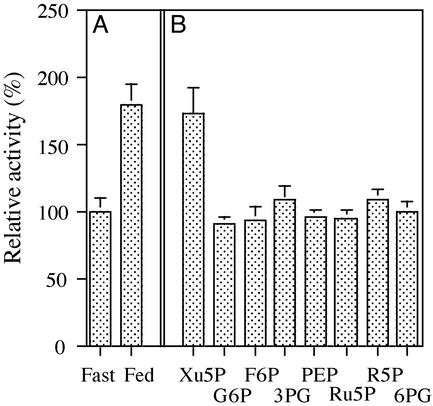
The first step in glucose activation of ChREBP is the dephosphorylation of Ser-196 located near the NLS site. To investigate this reaction a synthetic peptide containing amino acids 187–201 (P1 peptide in Fig. 1) of ChREBP was phosphorylated with a catalytic subunit of PKA and used as a substrate to search for a PPase and a metabolite that activated the PPase. The cytosolic fraction of liver extract of rats fed a high-carbohydrate diet was used as a source of the PPase and metabolite. These extracts were compared with a corresponding liver extract from fasted rats. The PPase activity in the high-carbohydrate liver extract was ≈1.8 times higher than that of the fasted liver extract (Fig. 2A).
Figure 2.
Dephosphorylation of P1 site peptide by PPase in cytosol. (A) PPase activity in the cytosolic extracts of livers from fasted rats vs. high-carbohydrate-fed rats. The PPase activity was determined with the assay method described in Materials and Methods with the P1 site peptide as a substrate. Crude cytosolic extract (5 μg) was added to the reaction mixture, incubated for 15 min at 30°C, and assayed for [32P]phosphate released. The PPase activity was expressed relative to that in the fasted liver extract, which was 100%. (B) Activation of desalted crude extracts by various metabolites. To remove endogenous metabolites the crude extracts were filtered through Sephadex G50 by centrifugation, and the filtered extract (5 μg) was assayed for the PPase activity in the presence of 50 μM sugar phosphates. G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Ru5P, ribulose 5-phosphate; R5P; ribose 5-phosphate; 6PG, 6-phosphogluconate.
To demonstrate the possible presence of a metabolite that could serve as an activator of the PPase, the fed-liver extracts were desalted by passing through Sephadex G50 to remove endogenous metabolites. Various intermediates of glycolysis and the pentose shunt pathways were examined for a possible activator of the PPase. The results showed that among those glucose metabolites examined, only Xu5P (50 μM) was able to activate the dephosphorylation reaction catalyzed by PPase (Fig. 2B). Similarly, Xu5P activated the extract from the starved animals. The extent of the activation was close to that observed with the crude extract, suggesting that Xu5P could account completely for the activation in the extract.
Characterization of the Cytosolic PPase.
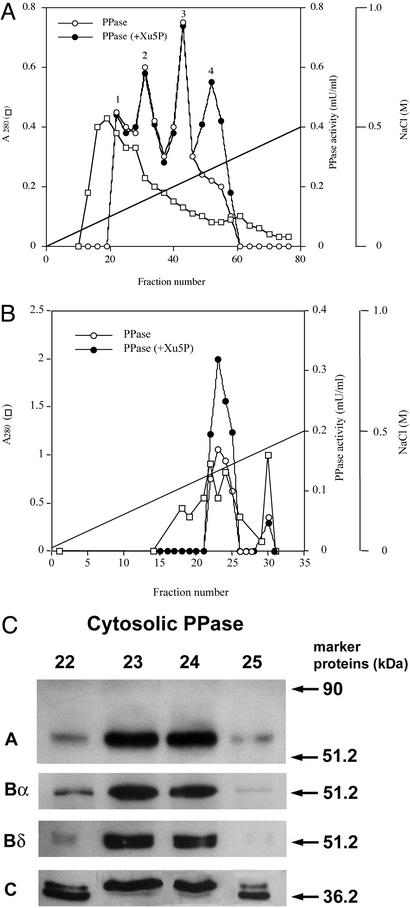
To characterize the cytosolic Xu5P-activated PPase responsible for dephosphorylation of the P1 site of ChREBP, the cytosolic extract was chromatographed on a DEAE-cellulose column as described previously (3). At least three peaks of PPase activity were detectable (Fig. 3A, peaks 1–3), but none of these PPases was activated by Xu5P. The Xu5P-activated PPase was eluted at 0.35 M NaCl (peak 4) after the three major PPases had been eluted, and this PPase was detectable only in the presence of Xu5P (Fig. 3A).
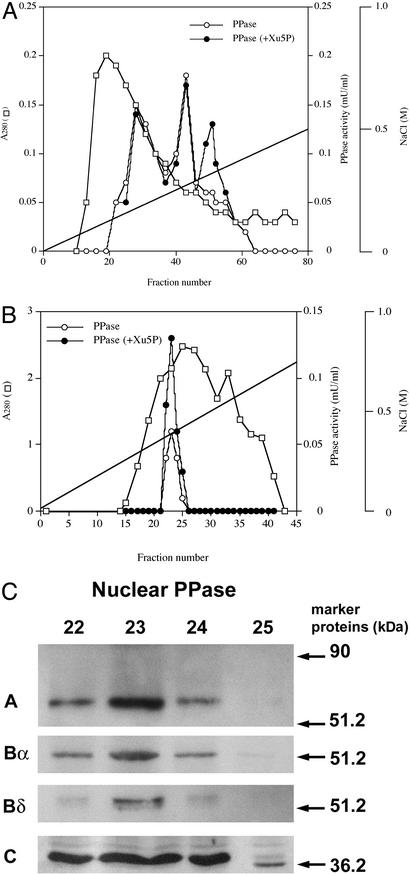
Figure 3.
(A) DEAE-cellulose chromatography of Xu5P-activated PPase in cytosol. The cytosolic extract (1,500 mg) from rat liver was chromatographed on a DEAE-cellulose column (3 cm × 10 cm) as described previously (15). The PPase activity was assayed with P1 peptide as a substrate in the absence (○) or presence (●) of 50 μM Xu5P. □, A280. (B) Mono Q chromatography of the Xu5P-activated PPase eluted from the DEAE-cellulose column in A. The fractions (48–56) were pooled and loaded on a Mono Q column (0.5 cm × 5 cm). PPase was eluted with a linear gradient of 0–0.5 M NaCl in 50 mM Tris⋅HCl/0.1 mM EDTA/1 mM DTT. (C) Immunoblot of Xu5P-activated PPase eluted from the Mono Q column. The fractions (22–25, 15 μl each) were subjected to PAGE, electroblotted onto a nitrocellulose filter, and incubated with primary antibodies against PP2A A, Bα, Bδ, and C.
To characterize the PPase further, the fractions containing the Xu5P-activated PPase from the DEAE-cellulose chromatography were pooled and further purified by chromatography on a Mono Q column. Both a major and a minor peak of PPase activity were eluted from the column (Fig. 3B). The major peak, representing >90% of the PPase activity, was activated by Xu5P, but the minor peak, <10%, was insensitive to the pentose-phosphate activation. Xu5P activated equally all of the PPase in the eluted fractions, suggesting that the major peak contained one type of Xu5P-activated PPase free of any other PPases. Immunoblots of these fractions with antibodies against PP2A subunits A, Bα, Bδ, and C (22–25) indicated that the Xu5P-activated PPase was a trimer consisted of A, Bδ, and C subunits (Fig. 3C). Furthermore, the Bδ peak as well as the A and the C peaks corresponded exactly with the Xu5P-activated peak. The antibodies against Bδ are specific for this isoform, but the antibodies against Bα reacted with both Bα and Bδ. The immunoreactivity indicated that the regulatory subunit is likely Bδ (25), and the estimated molecular weights of the subunits were identical to those we reported previously (15). Furthermore, the relative composition of these subunits in these PPase fractions appear to be the same, suggesting the presence of the same PPase in these fractions.
To determine the type of the PPase, various inhibitors of PPases (Mono Q eluate) were examined. Okadaic acid, a potent inhibitor of PP2A (IC50 = 1 nM; reviewed in refs. 22–24), inhibited ≈65% and 50% in the absence and presence of Xu5P (data not shown). At higher concentration (10 nM), okadaic acid inhibited completely in the absence and presence, respectively, of Xu5P. Cantharidic acid (500 nM), an indiscriminate inhibitor at this concentration, inhibited the Xu5P-activated PPase completely. However, deltamethrin (300 nM), a PP2B inhibitor, did not affect the activity. These results as well as the similarity of this Xu5P-activated PPase to the previously found liver PPase (22–25) and the above immunoreactivity suggested strongly that this PPase was type 2A.
Dephosphorylation of P3 Site and Activation of the DNA-Binding Activity of ChREBP in Nuclear Extract.
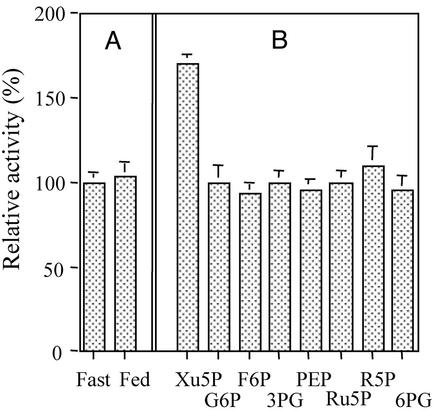
The DNA-binding activity of ChREBP is inhibited by the reversible phosphorylation of Thr-666 (P3 site peptide, Fig. 1), which is located within a bHLH (13). Glucose activation of the DNA-binding activity would be expected to occur by dephosphorylation of the P3 and P4 sites and should occur in the nucleus. Thus we searched for a specific PPase catalyzing the dephosphorylation reaction in the nuclei of liver from rats fed a high-carbohydrate diet. Synthetic peptides of the bHLH region corresponding to amino acids 655–674 and 556–573 containing the target Thr-666 (P3) and Ser-568 (P4), respectively, were prepared and phosphorylated by PKA and used as a substrate. As shown in Fig. 4A (with P3), PPase activity was identical in the crude nuclear extracts from both fed and starved rat liver, in contrast to the cytosolic extracts shown in Fig. 2A. The nuclear PPase was activated by added Xu5P, which suggested that the metabolites were absent from the nuclear extract (Fig. 4B). Similar to the cytosolic PPase, Xu5P was the only metabolite able to activate the nuclear PPase. The absence of the metabolites in the nuclear extract was probably due to loss of metabolites by diffusion during the isolation of the nuclei by gradient centrifugation. The lack of difference between the nuclear extracts of fed and fasted livers and full recovery of the enzyme activity with addition of Xu5P supported the idea that this pentose phosphate was the only factor necessary for the full activation of the PPase, and no other activator appeared to be required.
Figure 4.
PPase activity in nuclear extract. P3 peptide was used as substrate. (A) PPase activity in crude nuclear extracts (5 μg) from fasted and high-carbohydrate-fed rat livers was assayed as in Fig. 2. (B) Various sugar phosphates (50 μM) were added to the assay mixture and PPase activity was determined as in Fig. 2B. The same results were obtained with P4 peptide as a substrate (data not shown).
Characterization of the Nuclear Xu5P-Activated PPases.
To characterize the nuclear Xu5P-activated PPase, crude nuclear extract was chromatographed on DEAE-cellulose as shown in Fig. 5A. The elution pattern was identical to that of the cytosolic PPases, and the nuclear Xu5P-activated PPase was eluted at the same concentration (0.35 M) of NaCl as the cytosolic Xu5P-activated PPase. The elution pattern of the nuclear PPase from the Mono Q column was also identical to that of the cytosolic PPase (Fig. 5B). Furthermore, the immunoblots of the Mono Q eluates (Fig. 5C) were essentially identical to those of the cytosolic PPase (Fig. 3C).
Figure 5.
(A and B) DEAE-cellulose and Mono Q chromatography of Xu5P-activated PPase in nuclear extract. Crude nuclear extract (800 mg) was subjected to DEAE-cellulose (A) followed by Mono Q (B) chromatography as in Fig. 3A. The PPase activity was assayed with P3 as a substrate in the absence (○) or presence (●) of 50 μM Xu5P. (C) Immunoblots of PPase eluted from the Mono Q column as in Fig. 3C. Fractions 22–25 (15 μl) were subjected to PAGE, electroblotting, and immunoreaction as above.
Xu5P-activated PPase in the nuclear extract (the Mono Q eluate) was inhibited by 1 nM okadaic acid 70% and 63%, respectively, in the presence and absence of Xu5P, and >95% by 10 nM okadaic acid. The nuclear PPase also was completely inhibited by 500 nM cantharidic acid, but was not affected by 300 nM deltamethrin (data not shown). The same results were obtained with the PPase acting on the P4 peptide as the substrate.
Substrate specificities of the cytosolic and the nuclear Xu5P-activated PPases toward P1, P3, and P4 peptides were compared with the Mono-Q-purified enzymes. Both PPases dephosphorylated all peptide sites, and they showed essentially the same activity with respect to P1 and P2 peptides but somewhat lower activity with P3. Furthermore, both enzymes were activated by Xu5P to the same extent (1.8-fold). All these data suggested that the cytosolic and nuclear PPases activated by Xu5P were similar if not identical PPases. This conclusion was supported further by the results of immunoreactivity assays using the specific antibodies against PP2A-Bα and PP2A-Bδ. Both the cytosolic and the nuclear Xu5P-activated PPase demonstrated immunoreactivity with Bα and more specific Bδ, suggesting that these PP2As could be the PP2ABδC isoform.
Glucose-Induced Kinetic Changes in Xu5P Level, ChREBP Translocation, and Transcription Activity in Hepatocytes.
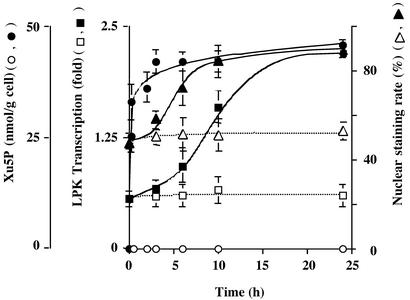
We proposed previously that for the glucose activation of LPK transcription, ChREBP must first be translocated from cytosol to nucleus by dephosphorylation of the P1 site near the NLS, and translocation is followed by activation of the DNA-binding activity by dephosphorylation of the P3 and/or P4 sites near or within the bHLH domain. If Xu5P is indeed the glucose signaling compound essential for the activation of ChREBP and LPK transcription, then the formation of Xu5P must precede the nuclear import, which is followed by transcription. We have examined the kinetics of these three events in primary hepatocytes transfected with GFP-fused ChREBP or ChREBP and incubated in high or low concentration of glucose. As shown in Fig. 6, the formation of Xu5P was quite rapid, taking <30 min to reach 80% of the maximum and remaining at the maximum concentration of 43 nmol/g of cell for 24 h under high glucose. The nuclear translocation began in ≈3 h and reached half maximum at ≈5 h and maximum at 10 h. The LPK transcription also commenced slowly after 3 h and reached a half maximum in ≈10 h and the maximum after ≈15 h. However, in low-glucose medium, the Xu5P formation, ChREBP translocation, and the LPK transcription remained constant (Fig. 6). These results are consistent with the proposed sequence of reactions in the glucose-stimulated pathway of ChREBP activation.
Figure 6.
Kinetics of Xu5P formation, nuclear import, and the LPK transcription activity in primary hepatocytes. The hepatocytes transfected with either GFP-ChREBP or ChREBP were incubated in low (open symbols, 5.5 mM) and high (filled symbols, 27.5 mM) concentrations of glucose. At indicated time intervals, the hepatocytes were harvested. An aliquot was immediately treated with HClO4 for metabolite determination, and the remainder was used for assay of translocation and LPK transcription, as described in Materials and Methods. ●, Xu5P formation; ▴, nuclear import; ■, LPK transcription.
Discussion
Glucose and cAMP are opposing factors in the regulation of hepatocytic gene expression. Previous studies from this laboratory demonstrated that cAMP-activated PKA inhibits ChREBP at two levels, the nuclear import by phosphorylation of Ser-196 of ChREBP and the DNA-binding activity by phosphorylation of Thr-666, located within the bHLH domain (13) (Fig. 1). We also demonstrated that dephosphorylation of these sites of the expressed C terminus of ChREBP containing the bHLH with the catalytic subunit of PP2A resulted in activation in vitro. In addition, high-fat diet also inhibits glucose activation of lipogenic enzyme synthesis. This inhibition is the result of phosphorylation/inactivation of Ser-568 of ChREBP by AMPK (14), and dephosphorylation of the site results in activation of transcription of the lipogenesis enzymes. These results suggest that the simplest mechanism for glucose action is mediated by activation of protein phosphatase. In this report we described the identification and characterization of protein phosphatases in cytoplasm and nucleus, which catalyzed dephosphorylation/activation of ChREBP in response to high glucose and, in turn, activated transcription of lipogenic enzyme genes. Furthermore, we demonstrated the existence of a glucose signaling compound in the fed livers and identified it as Xu5P.
We have shown that a protein phosphatase(s) was present in the cytosolic fraction of livers from rats fed a high-carbohydrate diet, and this phosphatase catalyzed the dephosphorylation of Ser-196, the P1 site. We also found a PPase in nuclei that dephosphorylated Thr-666, the P3 site, as well as Ser-568, the P4 site. The chromatographic elution patterns, the inhibition by various PPase inhibitors, the substrate specificity, and activation by Xu5P suggested that the cytosolic and the nuclear PPases are very similar, if not identical. Moreover, we determined previously the amino acid sequences of seven peptides derived from the Xu5P-activated PP2A that dephosphorylates 6-phosphofructo-2-kinase/Fru-2,6-P2ase (15) and concluded that the PPase was similar to PP2A Bα, since PP2ABδ was not known at that time. Subsequently, Strack et al. (25) cloned the Bδ subunit. A comparison of the amino acid sequences of Xu5P-activated PP2A B subunit showed that although five peptides were identical to Bδ, two others showed one or three amino acid differences. These minor differences could be due to errors in the previous sequence analysis, or these enzymes could consist of another B subunit.
We report here the nuclear PP2A that is activated by Xu5P. The previously reported Xu5P PP2A is clearly cytosolic, and is responsible for increased fructose-2,6-P2 synthesis by activation of fructose-6-P 2-kinase/Fru-2,6-P2ase in response to excess glucose, resulting in activation of PFK and glycolysis (3, 26). The nuclear PP2A has characteristics similar to those of the cytosolic PP2A as far as we could determine. Thus, we believe the same Xu5P-activated PP2A appeared to activate the inactive phosphorylated ChREBP as well as the bifunctional enzyme. PP2A in mammalian cells has a variety of diverse functions in signaling pathways. Among the known PP2As, the Xu5P-activated PP2A is unique in that it is activated by a small molecular weight substance. The mechanism of the activation by Xu5P, however, is not known at present.
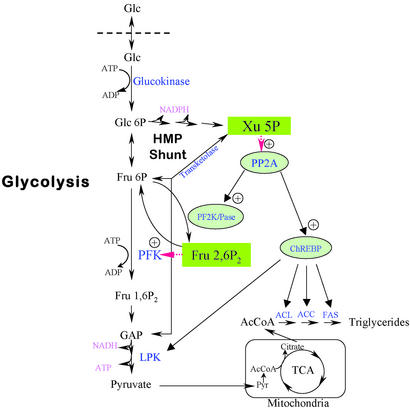
The results presented here demonstrated that the glucose signaling compound is likely Xu5P. Furthermore, Xu5P is the only activator necessary to account for the glucose activation in liver. Xu5P is an intermediate of the nonoxidative part of the pentose shunt pathway, and it could form directly from two glycolytic intermediates, glyceraldehyde 3-phosphate (GAP) and fructose-6-P (F6P) by a transketolase reaction (Fig. 7). Both GAP and F6P are linked to the activation of PFK, which depends on a potent activator, Fru-2,6-P2, and Fru-2,6-P2 formation in turn depends on the Xu5P-activated PP2A, as demonstrated in this laboratory (15, 26). Immediately after ingestion of excess glucose, F6P in the liver rises, followed by increased Xu5P, resulting in the activation of PFK caused by increased Fru-2,6-P2. The rise in Xu5P then not only activates Fru-2,6-P2 synthesis but also activates ChREBP to stimulate transcription of lipogenic enzyme genes. The transcription can be turned off with decreasing Xu5P when PFK activity and glycolysis slow down. Thus the entire carbohydrate metabolism and lipogenesis in acute and long-term regulation could be controlled exquisitely by Xu5P. Furthermore, Xu5P/transketolase could be termed a “glucose sensing” system; however, strictly speaking, Xu5P/transketolase is not sensing the glucose level at all but is sensing the activity of PFK and glycolytic rates (Fig. 7). Therefore, as we pointed out previously (15, 26), both glycolysis and pentose shunt pathways are coordinately regulated by the second messengers, Xu5P and Fru-2,6-P2, in response to glucose and hormonal changes, respectively. Furthermore, a bigger picture emerges that these two messengers determine the redox states, NAD/NADH and NADP/NADPH ratios, and the energy potential [ATP/(ADP + Pi)] in liver.
Figure 7.
Abbreviated glycolysis, gluconeogenesis, and pentose shunt pathways and roles of Xu5P and Fru-2,6-P2 in activation of PP2A and PFK, respectively. The scheme illustrates the formation of Xu5P from fructose 6-phosphate (F6P) and glyceraldehyde 3-phosphate (GAP) by transketolase, which activates PP2A. Xu5P PP2A then activates 6-phosphofructo-2-kinase/Fru-2,6-P2 phosphatase (PF2K/Pase) to increase Fru-2,6-P2, resulting in activation of PFK. The same PP2A also activates ChREBP in the activation of lipogenic gene transcription, including ATP citrate lyase (ACL), acetyl-CoA carboxylase (ACC), and fatty acid synthase (FAS). Glu, glucose; HMP, hexose monophosphate; TCA, tricarboxylic acid cycle.
If the same Xu5P-dependent PP2A activates both acute glucose metabolism and long-term transcription, the question may be raised as to whether the concentration of Xu5P remains high enough to function in the long term. For example, induction of LPK transcription takes several hours, but Xu5P forms within a few minutes upon feeding high amounts of glucose (3). The kinetic results presented here (Fig. 6) demonstrated that Xu5P formation in hepatocytes under high glucose was rapid and remained high during the activation of transcription, indicating that there was sufficient Xu5P present for the duration of transcription.
In summary, we conclude that ChREBP contains multiple phosphorylation sites that are regulated exclusively by a phosphorylation/dephosphorylation mechanism (Fig. 1). The phosphorylation of ChREBP, which is catalyzed by PKA (that was activated by glucagon/cAMP) and AMPK (activated by AMP under high-fat diet) inactivates nuclear translocation and the DNA-binding activity. We propose that the intracellular signaling mechanism that is responsible for the glucose activation is as follows. Excess glucose raises the Xu5P level, resulting in activation of Xu5P-activated PP2A, which in turn activates the inactive phosphorylated ChREBP by dephosphorylation of the multiple sites. The activity of ChREBP, therefore, is determined by the relative activities of the protein kinases and PP2A, which are regulated by cAMP, AMP, and Xu5P concentrations, sensors of nutritional and hormonal status of liver, and play vital roles in regulation of short and long-term carbohydrate and energy metabolism as well as fat synthesis.
Acknowledgments
We are grateful to Dr. Marc Mumby (University of Texas Southwestern Medical School) for valuable advice and providing antibodies against PP2A-C subunit. We thank Dr. Rick Bruick (University of Texas Southwestern Medical School) and Dr. Sarah McIntire (Texas Women's University) for critical review of the paper. This work was supported by Department of Veterans Affairs Merit Review and National Institutes of Health Grant DK54914.
Abbreviations
- AMPK
AMP-activated protein kinase
- bHLH
basic helix–loop–helix
- ChRE
carbohydrate-responsive element
- ChREBP
ChRE-binding protein
- Fru-2,6-P2
fructose 2,6-bisphosphate
- LPK
L-type pyruvate kinase
- NLS
nuclear localization signal
- PFK
phosphofructokinase
- PKA
cAMP-dependent protein kinase
- PP2A
xylulose 5-phosphate-activated protein phosphatase
- PPase
protein serine/threonine phosphatase
- Xu5P
xylulose 5-phosphate
References
- 1.Girard J, Ferre P, Foufelle F. Annu Rev Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 2.Towle H C, Kaytor E N, Shih H M. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 3.Nishimura M, Fedorov S, Uyeda K. J Biol Chem. 1994;269:26100–26106. [PubMed] [Google Scholar]
- 4.Towle H C. J Biol Chem. 1995;270:23235–23238. doi: 10.1074/jbc.270.40.23235. [DOI] [PubMed] [Google Scholar]
- 5.Vaulont S, Vasseur-Cognet M, Kahn A. J Biol Chem. 2000;275:31555–31558. doi: 10.1074/jbc.R000016200. [DOI] [PubMed] [Google Scholar]
- 6.Cuif M H, Porteu A, Kahn A, Vaulont S. J Biol Chem. 1993;268:13769–13772. [PubMed] [Google Scholar]
- 7.Thompson K S, Towle H C. J Biol Chem. 1991;266:8679–8682. [PubMed] [Google Scholar]
- 8.Hasegawa J, Osatomi K, Wu R F, Uyeda K. J Biol Chem. 1999;274:1100–1107. doi: 10.1074/jbc.274.2.1100. [DOI] [PubMed] [Google Scholar]
- 9.Prip-Buus C, Perdereau D, Foufelle F, Maury J, Ferre P, Girard J. Eur J Biochem. 1995;230:309–315. doi: 10.1111/j.1432-1033.1995.0309i.x. [DOI] [PubMed] [Google Scholar]
- 10.Kang R, Yamada K, Tanaka T, Lu T, Noguchi T. J Biochem (Tokyo) 1996;119:162–166. doi: 10.1093/oxfordjournals.jbchem.a021203. [DOI] [PubMed] [Google Scholar]
- 11.Doiron B, Cuif M H, Chen R, Kahn A. J Biol Chem. 1996;271:5321–5324. doi: 10.1074/jbc.271.10.5321. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita H, Takenoshita M, Sakurai M, Bruick R K, Hanzel W J, Shillinglaw W, Arnot D, Uyeda K. Proc Natl Acad Sci USA. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Proc Natl Acad Sci USA. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura M, Uyeda K. J Biol Chem. 1995;270:26341–26346. doi: 10.1074/jbc.270.44.26341. [DOI] [PubMed] [Google Scholar]
- 16.Hattori M, Tugores A, Veloz L, Karin M, Brenner D A. DNA Cell Biol. 1990;9:777–781. doi: 10.1089/dna.1990.9.777. [DOI] [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Donella-Deana A D, Varro A, Dockray G J, Pinna L A. Biochim Biophys Acta. 1990;1095:75–77. doi: 10.1016/0167-4889(91)90046-z. [DOI] [PubMed] [Google Scholar]
- 19.Veech R L, Guynn R, Veloso D. Biochem J. 1972;127:387–397. doi: 10.1042/bj1270387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casazza J P, Veech R L. J Biol Chem. 1986;261:690–698. [PubMed] [Google Scholar]
- 21.Lowry O H, Passonneau J V. A Flexible System of Enzymatic Analysis. New York: Academic; 1982. pp. 146–218. [Google Scholar]
- 22.Mumby M C, Walter G. Physiol Rev. 1993;73:673–699. doi: 10.1152/physrev.1993.73.4.673. [DOI] [PubMed] [Google Scholar]
- 23.Shenolikar S. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- 24.Wera S, Hemmings B A. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strack S, Chang D, Zaucha J A, Colbran R J, Wadzinski B E. FEBS Lett. 1999;460:462–466. doi: 10.1016/s0014-5793(99)01377-0. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y Q, Uyeda K. J Biol Chem. 1996;271:8824–8830. doi: 10.1074/jbc.271.15.8824. [DOI] [PubMed] [Google Scholar]
- 27.Pallas D C, Weller W, Jaspers S, Miller T B, Lane W S, Roberts T M. J Virol. 1992;66:886–893. doi: 10.1128/jvi.66.2.886-893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitoh Y, Yamamoto H, Ushio Y, Miyamoto E. Brain Res. 1989;489:291–301. doi: 10.1016/0006-8993(89)90862-7. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama N, Shima H, Hatano Y, Osawa Y, Sugimura T, Nagao M. Eur J Biochem. 1995;230:766–772. doi: 10.1111/j.1432-1033.1995.tb20619.x. [DOI] [PubMed] [Google Scholar]