Abstract
Antiandrogens given to antagonize androgen receptor (AR) activity gradually lose their efficacy as antagonists and eventually function as agonists to promote (instead of block) AR-mediated growth of prostate cancer cells. The mechanisms of how antiandrogens acquire this agonist activity during hormonal therapy are largely unknown. Here, we report that expression of a dominant-negative AR-associated protein 55 (dARA55) coregulator, inhibits AR transcriptional activity and reduces the agonist activity of antiandrogens. Inducibly expressed dARA55 inhibits prostate-specific antigen and cell growth in prostate cancer cells. Further dissection of the molecular mechanism shows dARA55 can selectively suppress endogenous AR-associated protein 55 (ARA55) enhanced AR transactivation by means of interruption of dimerization between ARA55 and ARA55. These results were confirmed by using RNA interference-mediated silencing of the ARA55 gene. These results therefore provide evidence that AR function could be suppressed without mutation or change in AR itself. Taken together, these findings not only demonstrate the important roles of the ARA55 coregulator in the AR-mediated growth of prostate cancer, they also may provide a critical target for developing therapeutic agents for the antiandrogen therapy that almost always fails in the treatment of hormone-refractory prostate cancer.
Prostate cancer is the most frequently diagnosed malignancy in aging men and the second leading cause of cancer-related deaths, claiming ≈40,000 men each year in the United States (1). Androgens and androgen receptor (AR) play important roles in this malignancy, and for the past 50 years androgen ablation has remained a major therapeutic option for the treatment of locally advanced or metastatic prostate cancer. Being androgen-dependent, prostate tumors quickly regress upon androgen ablation therapy as carried out by surgical or pharmacological castration (2, 3). Although complete disappearances of symptoms are achieved in a few cases, in most of the cases the response does not imply cure for the disease. Tumor progression and therapy failures are very common when treatment is continued for many months or years (4). A major contributory factor for such therapy failures may be the presence of a substantial amount of adrenal androgens that are readily convertible to 5α-dihydrotestosterone (DHT) in the prostate tissue (5, 6). Recently, ablation therapy has been supplemented with antiandrogens to produce the effect that is called a complete androgen blockade (6, 7). Although this rational use of ablation and antiandrogens was expected to completely block AR-mediated growth and progression of prostate cancer, this combined therapy also failed to show any significant improvement in the 5-year survival of the prostate cancer patients (8). Although 80–90% of patients initially respond favorably, this therapy fails to sustain its efficacy when continued beyond 1–2 years. Tumors gradually become resistant, androgen dependency is lost, and, eventually, the majority of the patients develop androgen-independent progression of the disease. This condition is called “hormone-refractory” prostate cancer, which is not curable by any present-day therapy. The AR loses its specificity and becomes activatable by antiandrogens or other nonandrogenic molecules, which has been a major problem in treating these patients. The molecular events that result in antiandrogens losing their efficacy as antagonists and/or transform them into androgenic molecules to promote AR-mediated growth are mostly unknown. One of the hallmarks of hormone-refractory prostate cancer is that growth still depends largely on activation of the AR (9). All possible androgen-independent pathways so far identified eventually promote cell growth through the activation of AR signaling (10, 11). A majority of androgen-independent tumors and metastases retain a functional AR-signaling pathway, giving rise to wide speculation that modulation of the AR-signaling pathway may be necessary for hormone-refractory growth of prostate cancer cells. Apart from mutations in AR itself as reported (12–14), we believe an aberrant modulation of AR transcriptional activity by AR coregulators may contribute to such an aberrant AR-mediated growth and, in particular, to agonist activity of antiandrogens in prostate cancer cells.
AR-associated protein 55 (ARA55), one of the AR coregulators, has been characterized as having the capacity to modulate AR specificity in response to agonists and antagonists (15). This AR coactivator has been reported to enhance AR transcriptional activity in response not only to normally weak adrenal androgens but also to the antiandrogens hydroxyflutamide (HF) and other nonandrogenic steroids, including 17β-estradiol (E2) (16–18). A frequently occurring AR mutation (mtART877A) in prostate cancer patients promotes hypersensitivity to ARA55 in the up-regulation of androgen-regulated genes. The commonly used antiandrogens, including HF, E2, cyproterone acetate, and bicalutamide, promote (instead of block) AR transcriptional activity in the presence of ARA55 in prostate cancer cells (18). ARA55 also may be involved in the “superactivation” of AR signaling by HER2/Neu in the presence of very low levels of androgens in prostate cancer cells (19, 20). In addition, expression levels of ARA55 were higher in tissue samples from hormone-refractory prostate cancers than in those from androgen-dependent cancers (21).
In the present study, using an in vitro mutagenesis and a yeast two-hybrid screening technique, we have identified a dominant-negative ARA55 mutant (dARA55) that can inhibit AR transcriptional activity by disrupting the normal function of endogenous ARA55 in the cells. Ectopic expression of this dARA55 abolishes the agonist activity of antiandrogens in prostate cancer cells. Our findings provide direct evidence for the contribution of ARA55 in the acquired agonist activity of antagonists and other nonandrogenic molecules and in making prostate cancer cells resistant to ablation and/or antiandrogen therapy.
Materials and Methods
Chemicals and Plasmids.
E2, DHT, and hydroxylamine were purchased from Sigma, and HF was a gift from Schering. The plasmid pACT2-ARA55 fused with GAL4 activation domain was the clone originally identified from the prostate cDNA library, and pAS2-AR containing the C terminus of the AR ligand-binding domain from wild-type AR fused to GAL4 DNA-binding domain, pCMX-GAL4-AR containing AR ligand-binding domain fused with GAL4 DNA-binding domain and pCMX-VP16-ARA55, and dARA55 fused to the activation domain of VP16 were constructed as described (15). The plasmid pSG5-dARA55 was constructed by subcloning EcoRI fragment from yeast vector pACT2-ARA55 into pSG5 expression vector, and pSG5-dARA55 also was re-created by site-directed mutagenesis by using the Quick-change mutagenesis kit (Invitrogen). Plasmids pSG5-AR, pSG5-ARA55, pSG5-ARA70, pSG5-ARA54, and pSG5-SRC-1 were constructed as described (15, 18). The plasmids pBIG-ARA55 and pBIG-dARA55 were constructed by inserting EcoRI fragment into pBIG2i expression vector as described (22). AR antibody (NH27) was from A. Mizokami, ARA55 antibody was from PharMingen, and the prostate-specific antigen (PSA) antibody was obtained from DAKO.
Generation of Mutant ARA55 Library.
The ARA55 mutant library was made by incubating 100 μg of the plasmid DNA pACT2-ARA55 in 5 ml of 1 M hydroxylamine for 1 h at 70°C. The plasmid DNA was extracted with phenol/chloroform (1:1) and recovered by precipitation with ethanol.
Yeast Two-Hybrid System.
The plasmid pAS2-AR expressing AR ligand-binding domain first was transformed into yeast strain Y190. The pAS2-AR transformants were selected on plates containing synthetic dropout (SD) medium lacking tryptophan (SD/−Trp). The hydroxylamine-mutated pACT2-ARA55 library then was transformed into the selected Y190 yeast cells expressing AR (previously transformed as mentioned above). The transformants then were plated on SD medium lacking tryptophan and leucine (SD/−Trp/−Leu) in the presence of DHT. A pool of white colonies indicating no interaction between AR and ARA55 mutants was selected by the β-galactosidase colony-lift filter assay. The plasmid DNA of ARA55 mutants was isolated from yeast cells that had spontaneously lost the cycloheximide-sensitive pAS2-AR plasmid upon plating the white colonies on SD/−Leu in presence of 10 μg/ml cycloheximide (Sigma). This pool of noninteracting mutants was transformed further into yeast strain Y190 expressing both AR and ARA55 (previously generated by a sequential transformation). A number of mutants that generated more than 80% white colonies on β-galactosidase assay were selected as potential dominant-negative mutants, because these mutants not only had lost their ability to interact with AR on their own, but also could prevent interaction between wtARA55 and AR.
Construction of DNA Vector-Based RNA Interference (RNAi) Plasmids.
The 19-nt coding sequences corresponding to the target sequences, followed by a 5-nt spacer, an inverted repeat of the coding sequences plus 5 Ts, were subcloned into plasmid BS/U6 (23) at the ApaI (blunt)/EcoRI site. For example, the DNA template (sense) for RNAi-B (corresponding to nucleotides 327–345) is GGA GGA CCA GTC TGA ATT CCG TTC TTC AGA CTG GTC CTC CTT TTT. The selection of coding sequences was determined empirically and was analyzed by blast search to avoid any significant sequence homology with other genes. Using primer extension, we first generated the double-stranded DNA with an ApaI (blunt) and EcoRI site, which then was cloned into BS/U6 plasmid after enzyme digestion. The construction of RNAi-A, RNAi-C, and RNAi-D was also done in a similar manner.
Cell Culture and Transient Transfection.
The PC-3 and DU145 cells were routinely maintained in DMEM at 37°C under 5% CO2/95% air. The CWR22-R and LNCaP cells were maintained in RPMI medium 1640 (Life Technologies, Rockville, MD). All media were supplemented with 100 units/ml penicillin G sodium and 100 μg/ml streptomycin sulfate, supplemented with 10% (vol/vol) FCS. Cells were seeded in the culture medium on 12-well Petri dishes at a density of 1 × 106 cells per dish for 24 h before transfection. One hour before transfection, the medium was replaced with 10% charcoal/dextran-treated FBS medium for transfection using SuperFect transfection reagent as described by the manufacturer (Qiagen). After 2 h, the medium was replaced with fresh charcoal/dextran-treated FBS medium, and cells were treated with ligand or ethanol and incubated for an additional 24 h.
Luciferase Assay.
Transfected and treated cells were washed two times with PBS, lysed by adding 200 μl of passive lysis buffer, and harvested by scrapping, and luciferase activity was measured by the Dual Luciferase Assay Kit (Promega). Another reporter gene, encoding Renilla luciferase, was used as an internal control to confirm transfection efficiency.
Thiazolyl Blue Assay.
The thiazolyl blue assay is a quantitative colorimetric assay for mammalian cell survival and proliferation. LNCaP413T55/55fl cells were grown in six-well plates in RPMI medium 1640 with 5% charcoal/dextran FCS. Cells were treated with DHT, E2, or HF in the presence or absence of doxycycline. After each treatment period, 200 μl of thiazolyl blue (5 mg/ml; Sigma) was added into each plate with 1 ml of medium for 3 h at 37°C. After incubation, 2 ml of 0.04 M HCl in isopropyl alcohol was added into each well. After several rounds of pipetting and 5 min of incubation at room temperature, the absorbency was read at a wavelength of 570 nm.
Mammalian Two-Hybrid System.
Protein–protein interactions in mammalian two-hybrid systems were characterized in DU145 cells by measuring luciferase activity. AR, dARA55, and ARA55 were fused either to GAL4 DNA-binding domain in pCMX mammalian expression vector or to VP16 activation domain, generating pCMX-GAL4-AR, pCMX-GAL4-dARA55, VP16-ARA55, and VP16-dARA55 constructs. The plasmids were transfected into DU145 cells, and the luciferase activity was measured as described above.
Inducible and Stable Transfection.
LNCaP cells were transfected with pBIG-dARA55 or pBIG vector. The pBIG2i vector contains all of the elements for tetracycline-responsive gene expression and a selective marker conferring resistance to hygromycin B for the selection of stable cell lines (24). The transfected cells were incubated for 24 h in RPMI medium 1640 supplemented with 10% FCS. The medium then was supplemented with 100 μg/ml hygromycin B (GIBCO/BRL) and incubated for 2 weeks. The resulting resistant clones were selected and cultured in the presence of doxycycline and examined for the expression of dARA55 by Western blotting. The selected pBIG-dARA55-expressing cells then were stably transfected with pCDNA3-His-wtARA55 plasmid, and neomycin-resistant clones were selected and verified for expression of His-ARA55. We thus generated an inducible (for dARA55) and a stable (for His-wtARA55) LNCaP cell line (LNCaP413T55/55fl).
Western Blot Analysis.
Total protein of cell extracts (doxycycline or control-treated) obtained from LNCaP413T55/55fl cells was determined by using the Bio-Rad protein assay. Equal amounts of proteins were electrophoresed on an SDS/polyacrylamide gel and blotted onto nitrocellulose membrane by using a standard protocol. The detection of the specific antibodies binding to the ARA55, AR, PSA, or β-actin proteins was accomplished by using the enhanced chemiluminescence Western blotting detection kit (Amersham Pharmacia).
Results
Identification of dARA55 Mutants.
By using hydroxylamine-mediated mutagenesis, a library of ARA55 mutants was generated by engineering random point mutations in ARA55. The mutated library then was screened by yeast two-hybrid system for those mutants that did not interact with AR. The screening system was extended to screen out those noninteracting mutants that could effectively interfere with the interaction between ARA55 and AR in a dominant-negative fashion. A pool of potential dominant-negative mutants was thus generated by this double-negative selection in the yeast cells.
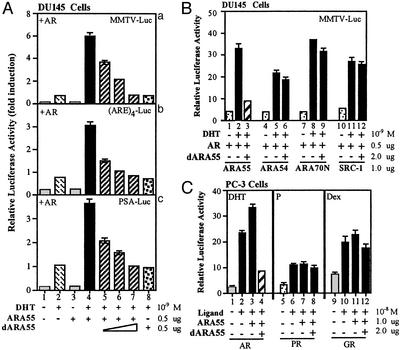
The selected mutants then were subcloned into the mammalian expression vector pSG5 and assayed for their dominant-negative effects in DU145 cells that do not express ARA55 or AR. We cotransfected ARA55 mutants and ARA55 along with AR and assayed for AR transcriptional activity with a mouse mammary tumor virus–luciferase (MMTV-Luc) reporter gene. As shown in Fig. 1Aa, cotransfection of ARA55 enhances AR transactivation (lane 2 vs. 4). The addition of a select ARA55 mutant significantly suppresses the ARA55-enhanced AR transactivation in a dose-dependent manner (lanes 5–7). When MMTV-Luc was replaced by PSA-Luc containing a natural AR promoter or by a synthetic (ARE)4-Luc, similar results were obtained (Fig. 1A b and c, respectively). Because DU145 cells do not express endogenous ARA55, we notice that the addition of dARA55 into this ARA55-negative cell shows little suppression effect for AR transactivation (lanes 2 vs. 8), suggesting dARA55 may go through ARA55 to suppress AR transactivation. To verify its specificity, we then cotransfected this mutant in the presence of either ARA55 or other related AR coregulators, including ARA54, ARA70, and SRC-1, in DU145 cells and assayed for AR transactivation. As expected, this mutant selectively inhibited ARA55-enhanced AR transactivation without any significant effect on other related coregulators (Fig. 1B). The inhibition was also relatively selective for AR, because the mutant showed little or no effect on other related steroid receptors, including progesterone receptor and glucocorticoid receptor in PC-3 cells that express endogenous ARA55 (Fig. 1C). We selected this mutant as dominant-negative ARA55 mutant and designated it as dARA55. Sequence analysis revealed a single point mutation (G-to-A transition) at the first position of codon 413, resulting in an alanine-to-threonine substitution.
Figure 1.
A select ARA55 mutant (dARA55) functions as a dominant-negative mutant to inhibit ARA55-enhanced AR transactivation in mammalian cells. (A) DU145 cells were transfected with ARA55, mutant ARA55 (dARA55) alone, or in combination as indicated, along with a fixed amount of AR (0.5 μg) and the reporter gene MMTV-Luc (0.5 μg). After 24 h of treatment with 1 nM DHT or vehicle, Luc activity was measured as described in Materials and Methods. The experiment was repeated with two other reporter genes, PSA-Luc and (ARE)4-Luc. (B) DU145 cells were transfected with AR (0.5 μg), MMTV-Luc (0.5 μg), and AR coactivators (1 μg each) as indicated either in the presence or in the absence of dARA55 (2 μg). After 24 h of 1 nM DHT treatment, the Luc activity was measured as before. (C) PC-3 cells that express endogenous ARA55 were transfected with AR, progesterone receptor, or glucocorticoid receptor expression plasmids alone or in the presence of ARA55 or dARA55. After 24 h of DHT, progesterone (P), or dexamethasone (Dex) treatment, receptor activity was measured by using an MMTV-Luc reporter gene assay.
dARA55 Inhibits Endogenous ARA55 to Suppress AR Transcriptional Activity in Prostate Cancer Cells.
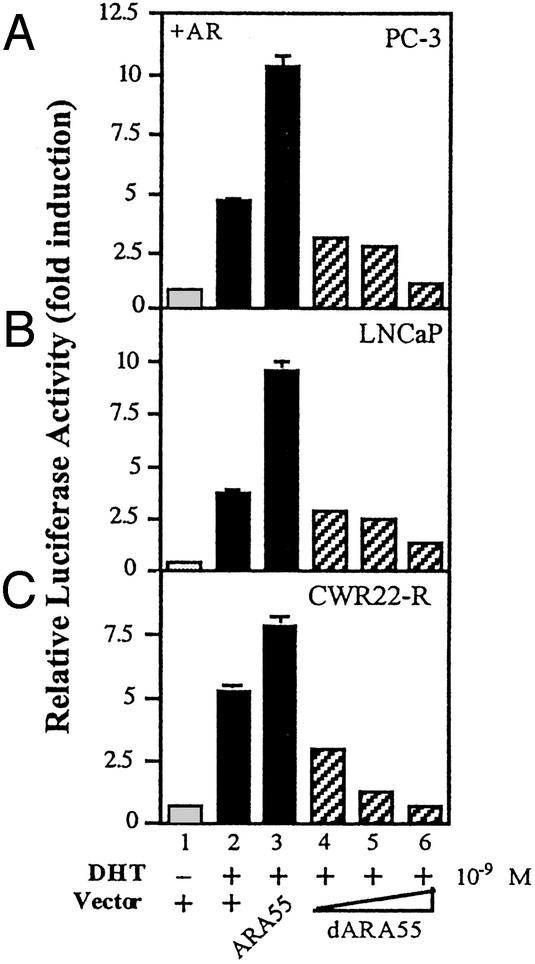
To test whether dARA55 inactivates endogenous ARA55, PC-3 cells that express endogenous ARA55 were transfected with ARA55 or dARA55 along with AR and an MMTV-Luc reporter gene. After 24-h androgen treatment, cells were harvested and assayed for Luc activity as a function of AR transcriptional activity. As depicted in Fig. 2A, 1 nM DHT treatment stimulated AR transactivation ≈15-fold compared with mock-treated cells. Although PC-3 cells express endogenous ARA55, transfection of ARA55 further increased AR transactivation (lanes 2 vs. 3). As expected, transfection of dARA55 significantly inhibited AR transactivation in a dose-dependent manner (lanes 4–6). This inhibition by dARA55 was confirmed in two other prostate cancer cell lines, CWR22-R and LNCaP (Fig. 2 B and C, respectively), which express endogenous ARA55 as well as AR. The CWR22-R expresses a mutant AR and represents a hormone-refractory state of prostate cancer (25, 26). The LNCaP cells also express a mutant AR, which frequently is found in advanced prostate cancer patients. These results clearly demonstrate that dARA55 can selectively suppress endogenous ARA55-mediated AR transactivation in prostate cancer cells.
Figure 2.
dARA55 inhibits endogenous ARA55 to suppress AR transcriptional activity in prostate cancer cells. PC-3 cells expressing endogenous ARA55 were transfected with ARA55, dARA55, or pSG5 control vector as indicated. After 24-h treatment of 1 nM DHT or vehicle, the luciferase activity was measured (A). The experiment was repeated with two other prostate cancer cell lines including LNCaP (B) and CWR22-R (C), which express endogenous ARA55 as well as AR. After 24-h ligand treatment, AR transcriptional activities were measured and presented as above. Values represent the mean ± SD of at least three independent determinations and are presented as the ratio of MMTV-Luc vs. Renilla-Luc activity for each sample.
dARA55 Reduces the Agonist Activity of Antiandrogens in Prostate Cancer Cells.
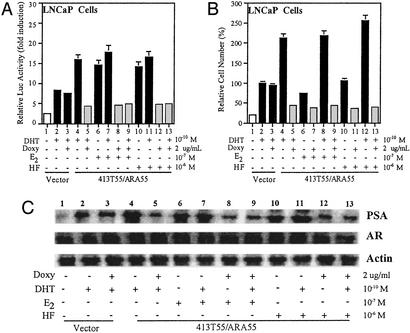
Antiandrogens given to antagonize residual androgen action during maximal ablation therapy lose their efficacy as antagonists and eventually function as agonists to promote AR-mediated growth of prostate cancer cells (13–15). To test whether ARA55 plays any role in this agonist activity of antiandrogens, we designed experimental conditions that closely mimicked conditions of maximal androgen ablation therapy. Prostate cancer patients may have minimal concentrations of androgens (0.1 nM) and a relatively high level of antiandrogens (1–5 μM) during ablation therapy. We used a modified LNCaP cell line (LNCaP413T55/55fl), which expresses both wtARA55 (stable expression) and dARA55 (inducible upon doxycycline treatment). We introduced this modification in our laboratory and confirmed the expression levels of ARA55 and dARA55 by Western blot analysis (data not shown). We transfected the cells with MMTV-Luc reporter gene and, after 48 h of androgen and/or antiandrogen treatment in the above conditions in either the presence or the absence of doxycycline, harvested the cells, and assayed for AR transcriptional activity by using the luciferase assay. As depicted in Fig. 3A, the antiandrogen HF and nonandrogenic molecule E2 increased AR activity, functioning as an agonist in ARA55-expressing LNCaP413T55/55fl cells (lanes 6 and 10). In conditions of androgen ablation therapy, these molecules failed to antagonize or block androgen actions in the cells (lanes 7 and 11). However, expression of dARA55 induced by doxycycline treatment significantly reduced this agonist activity of these antiandrogens in the cells (lanes 8–9 and 12–13).
Figure 3.
dARA55 abolishes the agonist activity of antiandrogens in prostate cancer cells. (A) LNCaP cells expressing both ARA55 (stable His-55fl) and dARA55 [inducible upon doxycycline (Doxy) treatment] were transfected with control vector and MMTV-Luc plasmids in the presence or absence of doxycycline as indicated. After 48-h treatment of DHT or antiandrogens alone or in combination as indicated, cells were harvested and assayed for AR transcriptional activity as described earlier. The transfection efficiency of the samples was normalized by using Renilla-Luc as an internal control. Values represent the mean ± SD of at least three independent determinations. (B) LNCaP cells (413T55/ARA55fl) as in A were applied to thiazolyl blue assay for the relative cell number (compared with treated vector alone, set as 100% in lane 3) determination. (C) LNCaP cells (LNCaP413T55/55fl) were cultured in the presence or absence of doxycycline. After 48-h treatment of DHT or antiandrogens alone or in combination as indicated, cells were harvested and lysed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and analyzed for PSA expression by Western blotting using PSA antibody as described above. The expression of AR and β-actin is shown in the middle and bottom blot, respectively. In all blots, LNCaP cells expressing pBIG vector were used as control.
Using the same LNCaP413T55/55fl cells, we then tested the effect of dARA55 on cell proliferation in similar treatment conditions. As shown in Fig. 3B, the expression of ARA55 (−doxycycline) increased cell growth, whereas dARA55 (+doxycycline-treated) resulted in a significant decrease in cell growth in response to DHT, E2, or HF alone or when combined as indicated. As a control, we also used vector-expressed LNCaP cells that did not show any significant differences in either the presence or the absence of doxycycline (lanes 2 and 3, respectively).
The inhibition of AR transactivation as well as cell growth by the dARA55 was well reflected in the diminished expression of PSA levels in the cells as measured by Western blot analysis. The expression of PSA, an AR target gene, has been used as the most useful diagnostic marker to monitor progression of prostate cancer (27, 28). As depicted in Fig. 3C, both E2 and HF by themselves significantly increased PSA expression (lanes 6 and 10) in untreated LNCaP413T55/55fl cells, whereas cells treated with doxycycline showed significantly diminished expression of PSA by these drugs (lanes 8 and 12). In conditions of maximal androgen ablation therapy, instead of blocking androgen action, these drugs increased PSA expression in the untreated cells (lanes 7 and 11). In contrast, both E2 and HF were able to reduce the androgen action and inhibited PSA expression in treated cells (lanes 9 and 13). The dARA55, thus by inactivating ARA55, reduces the agonist activity of antiandrogens in prostate cancer cells. These findings suggest that the activation of AR by antiandrogens or other nonandrogenic molecules may be dependent on the expression of ARA55 in the cells.
RNAi-Mediated Silencing of ARA55 Gene Inhibits AR Transactivation and Reduces the Agonist Activity of Antiandrogens in LNCaP Cells.
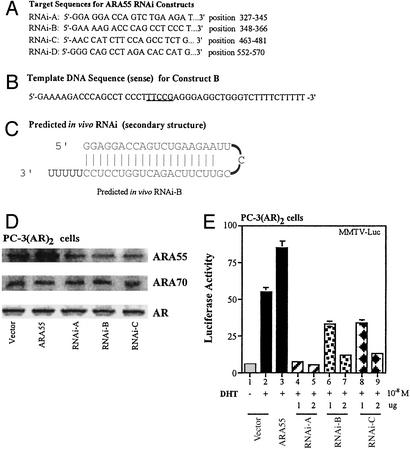
To confirm these roles of ARA55 on AR transactivation and on the agonist activity of antiandrogens, we investigated the effect of RNAi-mediated ARA55 gene silencing in LNCaP cells as shown in Fig. 4. We constructed four RNAi constructs as described in Materials and Methods. We first tested their effect on AR transactivation in DU145 cancer cells. Whereas construct D had no effect, constructs A, B, and C were highly effective in inhibiting exogenously transfected, ARA55-enhanced AR transactivation (data not shown). To see whether these three constructs also were able to inhibit endogenous ARA55 and thereby able to inhibit endogenous AR function, we transfected construct A, B, or C into PC-3(AR)2 cells, which express both ARA55 and AR. As depicted in Fig. 4E, whereas all three RNAi-constructs were able to inhibit endogenous AR function in a dose-dependent manner (lanes 4–9), construct A seemed to inhibit basal AR activity in high doses (lane 5). Western blot analysis using cell lysates from PC-3(AR)2 transfected with each of these RNAi constructs confirmed the silencing of ARA55 gene in the cells (Fig. 4D). The silencing was specific for ARA55 only, because the expression of a related AR coactivator, ARA70, or AR protein was not affected by any of these RNAi constructs.
Figure 4.
RNAi-mediated silencing of ARA55 gene inhibits AR transactivation in prostate cancer cells. (A) Target DNA sequences for constructs RNAi-A, RNAi-B, RNAi-C, and RNAi-D. (B) Template DNA sequences (sense) for RNAi-B. (C) Secondary structure for RNAi-B as predicted to be generated in vivo. (D) PC-3(AR)2 cells were transfected with empty vector, ARA55, or RNAi constructs as before. After 36-h incubation in normal serum (10% FCS), cells were harvested, lysed with RIPA buffer, and analyzed for expression of ARA55, ARA70, or AR by Western blotting, using specific antibodies for ARA55, ARA70, or AR (top, middle, and bottom blots, respectively). (E) PC-3(AR)2 cells were transfected with empty vector, ARA55, or RNAi constructs as indicated, along with a reporter gene, MMTV-Luc. After 24-h treatment of 1 nM DHT or vehicle, luciferase activity was measured.
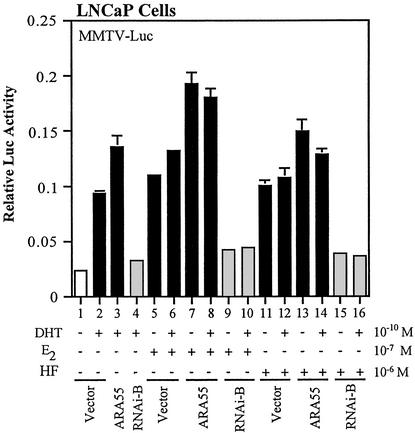
To test whether the reducing effect of antiandrogens by the dARA55 can be reproduced by RNAi-mediated gene silencing of ARA55, we transfected LNCaP cells with construct B and measured AR transactivation in conditions of androgen ablation therapy as described earlier. As depicted in Fig. 5, although both E2 and HF were able to increase AR transactivation (lanes 5, 6, 11, and 12), transfection of RNAi-B significantly reduced the agonist activity of these antiandrogens in either the absence or the presence of DHT. These findings are well consistent with our previous findings with dARA55 mutants. Taken together, our study strongly suggests a role of ARA55 in the acquired agonist activity of antiandrogens in LNCaP prostate cancer cells.
Figure 5.
RNAi-mediated inactivation of ARA55 reduces the agonist activity of antiandrogens in LNCaP cells. LNCaP cells were transfected with ARA55, RNAi-B, or control vector along with a reporter gene, MMTV-Luc. After 24-h treatment of DHT or antiandrogens alone or in combination as indicated, cells were harvested and assayed for AR transcriptional activity. Values represent the mean ± SD of at least three independent determinations and are presented as the ratio of MMTV-Luc to Renilla-Luc activity for each sample.
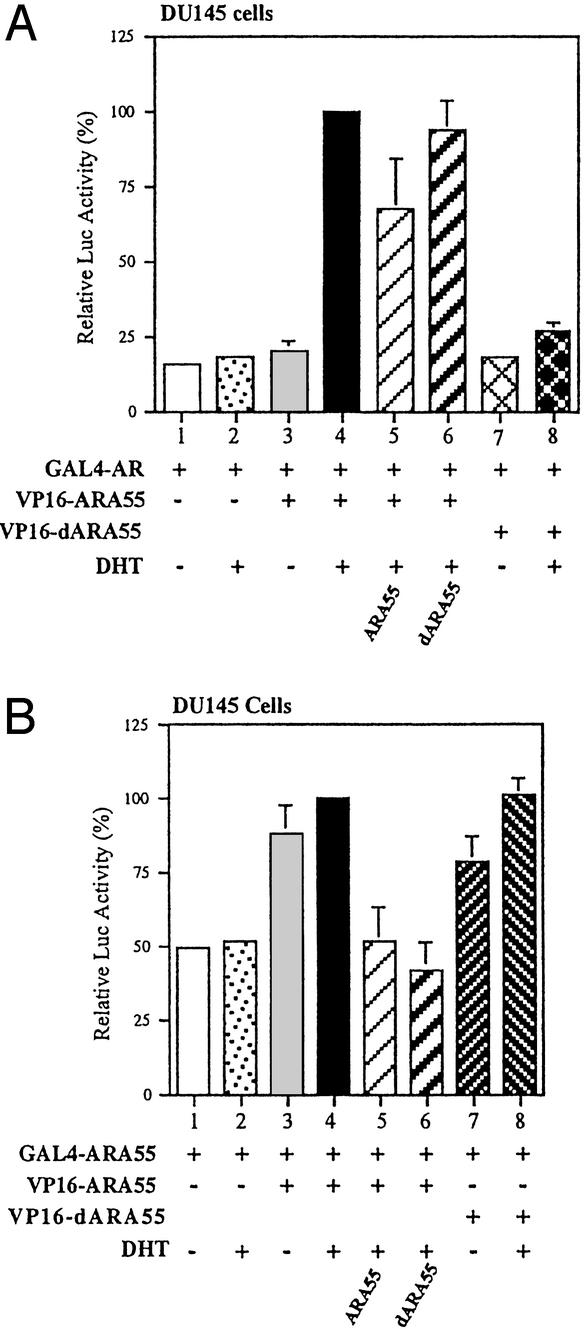
dARA55 Forms a Nonfunctional Homomer with ARA55.
The dARA55 was identified on the assumption that the noninteracting mtARA55 might form a nonfunctional heteromer with ARA55, which may or may not interact with AR, but will interrupt AR transcriptional activity. We used mammalian two-hybrid assay to verify this phenomenon. The dARA55, ARA55, and AR were fused to GAL4 DNA-binding domain or VP16 activation domain, and plasmids were transfected into DU145 cells. The interactions between expressed proteins were analyzed by using pG5-Luc reporter gene. As shown in Fig. 6A, compared with ARA55, the dARA55 lost its ability to interact with AR (lanes 4 vs. 8), which is consistent with our yeast two-hybrid screening results. The dARA55 could form a heteromer with ARA55, similar to ARA55-ARA55 dimerizations (Fig. 6B, lanes 3 and 4 vs. lanes 7 and 8). The addition of dARA55 substantially disrupts the interaction between ARA55 and ARA55 (Fig. 6B, lane 6). Although dARA55 could form a heteromer with ARA55, it did not prevent ARA55 interaction with AR (Fig. 6A, lane 6). These findings suggest that dARA55 may form a nonfunctional heteromer with ARA55 that does not support AR transcriptional activity in the cells.
Figure 6.
The effect of dARA55 on the interaction between AR-ARA55 and ARA55-ARA55 homodimer. DU145 cells were transfected with 2.5 μg of GAL4-hybrid expression plasmids pGAL4-AR (A) and GAL4-ARA55 (B). Also included in both A and B were 2.5 μg of pCMX-VP16-ARA55 or pCMX-VP16-dARA55 and 2.5 μg of pG5-Luc reporter gene with or without pSG5-ARA55 or pSG5-dARA55. Each Luc activity is presented relative to that of lane 4 in each graph (filled bars, set as 100%). Values represent the mean ± SD of at least three independent determinations.
Discussion
Since the cloning of its complementary DNA structure in 1988 (29), the AR-signaling pathway has been studied extensively to elucidate its role in the development and progression of prostate cancer. Substantial evidence indicated that ligand-bound AR regulates target genes by a mechanism involving coregulators such as ARA55. We investigated whether interruption of the AR coregulator function could lead to down-regulation of AR activity in the cells, which may have critical implications in regulating prostate cancer progression. Using in vitro mutagenesis and a double-negative selection in yeast two-hybrid screening, we have identified a dominant-negative AR coactivator, dARA55, which inhibits ARA55-enhanced AR transcriptional activity in prostate cancer cells. The strategy for identification of a dominant-negative ARA55 mutant was based on the rationale that any mutant that loses its interaction with AR but still forms a nonfunctional heteromer with wtARA55, which may or may not form a complex with AR, might be able to interrupt the AR transcriptional activity. We created an ARA55 library by using hydroxylamine-mediated mutagenesis, which induces random point mutations. Hydroxylamine reacts with double-stranded target DNA to create N4-hydroxycytosine and N2-hydroxyguanine, which can base-pair with adenine and thymine, respectively, resulting in both C-to-T and G-to-A transition mutations and, thereby, generating an assortment of random mutations in the DNA (30). The dominant-negative mutant then was isolated by two-hybrid screening of the mutant library in the yeast and confirmed by reporter gene assay in mammalian cells. A single point mutation (G-to-A transition) generated an alanine-to-threonine substitution at amino acid 413, transforming a potent AR coactivator (ARA55) into a strong inhibitor (ARA55A413T) of AR transcriptional activity. Whereas ARA55 in an oligomeric form interacts with AR and enhances its transcriptional activity, the dARA55 lacking AR interaction forms a nonfunctional heteromer with ARA55 and inhibits ARA55-enhanced AR activity. Ectopic expression of the dominant-negative mutant not only inhibits androgen-dependent AR transcriptional activity but also reduces the agonist activity of antiandrogens or nonandrogenic molecules in prostate cancer cells. Identical results obtained by using RNAi-mediated silencing of ARA55 gene further strengthen the role of ARA55 in the acquired agonist activity of antiandrogens in LNCaP cells.
Androgens and AR play a vital role both in normal prostate development and in the progression of prostate cancer. Therefore, blocking the AR-signaling pathway is critical for controlling prostate cancer. Effectiveness of therapeutic agents, including androgen ablation and/or antiandrogens, depends on sustained blockade of AR-mediated growth of prostate cancer cells. Maximum ablation therapy used in the treatment of prostate cancer, although initially effective, fails to sustain its efficacy when continued beyond 1–2 years. Tumor cells become resistant to ablation therapy and AR loses its specificity and becomes activatable by antiandrogens or other antiandrogenic therapeutic agents, the mechanisms of which are largely unknown. Previously, a possible role of AR mutations has been reported by a number of groups (12–14). The abrogation of the agonist activity of antiandrogens in conditions of androgen ablation therapy by a dominant-negative ARA55 mutant in the present study establishes a role of ARA55 in the process. The LNCaP cells used in this study express a mutant AR (T877A), which also is found in prostate cancer patients, that can change the AR specificity in response to agonists and antagonists. The inhibition of AR transcriptional activity and/or the agonist activity of antiandrogens in these cells by a dominant-negative ARA55 mutant, and also by a RNAi-construct against ARA55, clearly indicates a probable dominant role of ARA55 over AR mutation in the process.
The interruption of AR–ARA55 interaction thus offers a target for developing new therapeutic agents that can reduce the agonist activity of antiandrogens and inhibit AR-mediated growth of prostate cancer cells. The dominant-negative mutant itself may be evaluated as a gene therapeutic reagent. However, the effects of dARA55 on tumorigenesis in mice need to be examined to see whether the mutant can rescue the function of antiandrogens or whether it will be sufficiently effective to suppress the progress of prostate tumor growth in vivo. In addition, the role of other AR coregulators needs to be evaluated to the same extent.
Acknowledgments
We thank Karen Wolf for manuscript preparation. This work was supported by the George Whipple Professorship Endowment and National Institutes of Health Grants CA-71570 and DK-60905.
Abbreviations
- AR
androgen receptor
- ARA55
AR-associated protein 55
- dARA55
dominant-negative ARA55
- RNAi
RNA interference
- DHT
5α-dihydrotestosterone
- HF
hydroxyflutamide
- PSA
prostate-specific antigen
- E2
17β-estradiol
- SD
synthetic dropout
- MMTV-Luc
mouse mammary tumor virus–luciferase
References
- 1.Jemal A, Thomas H, Murray T, Thun M. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Stevens R, Hodges C V. Arch Surg. 1941;43:209–233. [Google Scholar]
- 3.Westin P, Stattin P, Damber J-E, Bergh A. Am J Pathol. 1995;146:1368–1375. [PMC free article] [PubMed] [Google Scholar]
- 4.Gittes R F. N Engl J Med. 1991;324:1892–1893. doi: 10.1056/NEJM199101243240406. [DOI] [PubMed] [Google Scholar]
- 5.Dennis L J, Griffiths K E. Semin Surg Oncol. 1993;18:52–74. doi: 10.1002/(sici)1098-2388(200001/02)18:1<52::aid-ssu8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Labrie F, Belanger A, Dupont A, Luu-The V, Simard J, Labrie C. Clin Invest Med. 1993;16:475–492. [PubMed] [Google Scholar]
- 7.Goktas S, Crawford E D. Semin Oncol. 1999;26:162–173. [PubMed] [Google Scholar]
- 8.Prostate Cancer Trialist's Collaborative Group. Lancet. 2000;355:1491–1498. [PubMed] [Google Scholar]
- 9.Feldman B J, Feldman D. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 10.Darne C, Veyssiere G, Jean C. Eur J Biochem. 1998;256:541–549. doi: 10.1046/j.1432-1327.1998.2560541.x. [DOI] [PubMed] [Google Scholar]
- 11.Culig Z, Hobisch A, Cronauer M V, Radmayr C, Trapman J, Hittmair A, Bartsch G K. Cancer Res. 1994;54:5474–5478. [PubMed] [Google Scholar]
- 12.Taplin M E, Bubley G J, Shuster T D, Frantz M E, Spooner A E, Ogata G K, Keer H N, Balk S P. N Engl J Med. 1995;332:1393–1398. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 13.Taplin M E, Bubley G J, Ko Y J, Small E J, Upton M, Rajeshkumar B, Balk S P. Cancer Res. 1999;59:2511–2515. [PubMed] [Google Scholar]
- 14.Culig Z, Hobisch A, Cronauer M V, Cato A C, Hittmair A, Radmayr C, Eberle J, Bartsch G, Klocker H. Mol Endocrinol. 1993;7:1541–1550. doi: 10.1210/mend.7.12.8145761. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto N, Yeh S, Kang H Y, Inui S, Chang H C, Mizokami A, Chang C. J Biol Chem. 1999;274:8316–8321. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- 16.Yeh S, Miyamoto H, Chang C. Lancet. 1997;349:852–853. doi: 10.1016/S0140-6736(05)61756-4. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto H, Yeh S, Wilding G, Chang C. Proc Natl Acad Sci USA. 1998;95:7379–7384. doi: 10.1073/pnas.95.13.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh S, Kang H Y, Miyamoto H, Nishimura K, Chang H C, Ting H J, Rahman M, Lin H K, Fujimoto N, Hu Y C, et al. Endocrine. 1999;11:195–202. doi: 10.1385/endo:11:2:195. [DOI] [PubMed] [Google Scholar]
- 19.Craft N, Shostak Y, Carey M, Sawyears C. Nat Med. 1999;5:280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 20.Yeh S, Lin H-K, Kang H-Y, Thin T H, Lin M-F, Chang C. Proc Natl Acad Sci USA. 1999;96:5458–5463. doi: 10.1073/pnas.96.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto N, Mizokami A, Harada S, Matsumoto T. Urology. 2001;58:289–294. doi: 10.1016/s0090-4295(01)01117-7. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Yang Y, Guo X, Sampson E R, Hsu C L, Tsai M Y, Yeh S, Wu G, Guo Y, Chang C. J Biol Chem. 2002;277:15426–15431. doi: 10.1074/jbc.M111218200. [DOI] [PubMed] [Google Scholar]
- 23.Sui G, Soohoo C, Affar E B, Gay F, Shi Y, Forrester W C, Shi Y. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strathdee C A, McLeod M R, Hall J R. Gene. 1999;229:21–29. doi: 10.1016/s0378-1119(99)00045-1. [DOI] [PubMed] [Google Scholar]
- 25.Nagabhushan M, Miller C M, Pretlow T P, Giaconia J M, Edgehouse N L, Schwartz S, Kung H J, de Vere White R W, Gumerlock P H, Resnick M I, et al. Cancer Res. 1996;56:3042–3046. [PubMed] [Google Scholar]
- 26.Wainstein M A, He F, Robinson D, Kung H J, Schwartz S, Giaconia J M, Edgehouse N L, Pretlow T P, Bodner D R, Kursh E D, et al. Cancer Res. 1994;54:6049–6052. [PubMed] [Google Scholar]
- 27.Palmberg C, Koivisto P, Visakorpi T, Tammela T L. Eur Urol. 1999;36:191–196. doi: 10.1159/000067996. [DOI] [PubMed] [Google Scholar]
- 28.Kelly W K, Scher H I, Mazumdar M, Vlamis V, Schwartz M, Fossa S D. J Clin Oncol. 1993;11:607–615. doi: 10.1200/JCO.1993.11.4.607. [DOI] [PubMed] [Google Scholar]
- 29.Chang C, Kokontis J, Liao S. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R S, Boeke J D. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]