Abstract
All nonsteroidal antiinflammatory drugs (NSAIDs) inhibit the cyclooxygenase (COX) isozymes to different extents, which accounts for their anti-inflammatory and analgesic activities and their gastrointestinal side effects. We have exploited biochemical differences between the two COX enzymes to identify a strategy for converting carboxylate-containing NSAIDs into selective COX-2 inhibitors. Derivatization of the carboxylate moiety in moderately selective COX-1 inhibitors, such as 5,8,11,14-eicosatetraynoic acid (ETYA) and arylacetic and fenamic acid NSAIDs, exemplified by indomethacin and meclofenamic acid, respectively, generated potent and selective COX-2 inhibitors. In the indomethacin series, esters and primary and secondary amides are superior to tertiary amides as selective inhibitors. Only the amide derivatives of ETYA and meclofenamic acid inhibit COX-2; the esters are either inactive or nonselective. Inhibition kinetics reveal that indomethacin amides behave as slow, tight-binding inhibitors of COX-2 and that selectivity is a function of the time-dependent step. Site-directed mutagenesis of murine COX-2 indicates that the molecular basis for selectivity differs from the parent NSAIDs and from diarylheterocycles. Selectivity arises from novel interactions at the opening and at the apex of the substrate-binding site. Lead compounds in the present study are potent inhibitors of COX-2 activity in cultured inflammatory cells. Furthermore, indomethacin amides are orally active, nonulcerogenic, anti-inflammatory agents in an in vivo model of acute inflammation. Expansion of this approach can be envisioned for the modification of all carboxylic acid-containing NSAIDs into selective COX-2 inhibitors.
Cyclooxygenase (COX; prostaglandin endoperoxide synthase, EC 1.14.99.1) metabolizes arachidonic acid to prostaglandin (PG) H2, which serves as the precursor for the biosynthesis of various PGs, thromboxanes, and prostacyclin (1). COX activity originates from two distinct and independently regulated isozymes, COX-1 and COX-2 (2). COX-1 is a constitutive enzyme, whereas COX-2 is inducible and short-lived. COX-2 is the product of an immediate-early gene, and its expression is stimulated by a host of growth factors, cytokines, and mitogens (3). COX-1 appears responsible for the biosynthesis of PGs in the gastric mucosa and in the kidney, whereas COX-2 appears responsible for biosynthesis in inflammatory cells and the central nervous system (4). Nonsteroidal anti-inflammatory drugs (NSAIDs) inhibit the two isoforms to different extents, and this feature accounts for their shared therapeutic properties and side effects (5). The differential tissue distribution of the COX isozymes has provided a rationale for the development of COX-2-selective inhibitors as nonulcerogenic, anti-inflammatory, and analgesic agents (6).
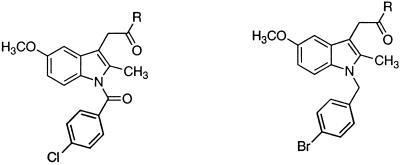
Most selective COX-2 inhibitors, including the recently approved drugs celecoxib (7) and rofecoxib (8), belong to the diarylheterocycle class of compounds (9–11). Diarylheterocycles have been investigated extensively as COX-2 inhibitors since the description of the 2,3-diarylthiophene, DuP 697, as a nonulcerogenic anti-inflammatory agent (12). In contrast, relatively few reports document structural modifications of NSAIDs into selective COX-2 inhibitors. Indomethacin (13, 14), zomepirac (15), aspirin (16, 17), and flurbiprofen (18) are the only examples of NSAIDs that have been transformed successfully into COX-2-selective inhibitors. However, the methodologies used in their modifications are not general and required extensive structure–activity relationship (SAR) studies on individual compounds. For instance, replacement of the 4-chlorobenzoyl group in indomethacin with a 4-bromobenzyl moiety generates a COX-2-selective inhibitor (13). In contrast, exchanging the carboxylate moiety in aspirin with alkyl sulfide functionalities affords specific COX-2 inhibitors (16, 17).
We have exploited biochemical differences between the COX isoforms to improve upon the selectivity of carboxylate-containing NSAIDs as COX-2 inhibitors. Cocrystallization and site-directed mutagenesis studies have confirmed that ion pairing of the carboxylic acid group in NSAIDs and arachidonate to the positively charged R120 residue in COX-1 is a major contributor to both inhibition and catalysis (19–22). However, recent studies reveal that ion pairing of the carboxylate moiety of arachidonate with R120 is not as important a determinant of catalysis by COX-2 (23). In addition, anandamide, the ethanolamide derivative of arachidonic acid, is reported to be a selective COX-2 substrate (24). Based on these observations, we hypothesized that derivatization of the carboxylate moiety in NSAIDs would eliminate their ability to inhibit COX-1 without significantly affecting their COX-2 inhibitory properties. Because many NSAIDs contain a carboxylic acid group, this would represent a general strategy for the conversion of nonselective NSAIDs into selective COX-2 inhibitors.
We report the successful transformation of a substrate analog inhibitor and NSAIDs from the arylacetic acid and fenamic acid classes into selective COX-2 inhibitors. The facile nature of this approach features a single chemical derivatization (amidation or esterification) of the carboxylate moiety in NSAIDs, which generates an impressive array of potent and highly selective COX-2 inhibitors. Lead compounds in this series exhibit anti-inflammatory activity after oral administration but do not induce gastric lesions. Site-directed mutagenesis identifies regions in the COX-2 protein that are responsible for the selectivity of inhibition exhibited by this novel class of inhibitors.
Materials and Methods
Materials.
[1-14C]Arachidonic acid (≈55–57 mCi/mmol) was purchased from DuPont/NEN or American Radiolabeled Chemicals (St. Louis). Hematin was purchased from Sigma. COX-1 was purified from ram seminal vesicles (Oxford Biomedical Research, Oxford, MI) as described (25). The specific activity of the protein was 20 μmol O2/min·mg, and the percentage of holoprotein was 13.5%. ApoCOX-1 was prepared as outlined (26). Apoenzyme was reconstituted by the addition of hematin to the assay mixtures. Human COX-2 was expressed in SF-9 insect cells by using the pVL1393 expression vector (PharMingen) (27). ETYA (5,8,11,14-eicosatetraynoic acid), indomethacin, and meclofenamic acid were purchased from Sigma. All other chemicals were obtained from Aldrich. Ester and amide derivatives were synthesized by coupling the carboxylic acid with appropriate alcohols or amines in the presence of dicyclohexyldicarbodiimide, bis-(2-oxo-3-oxazolidinyl)phosphonic chloride, or 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride. All new compounds were characterized by 1H-NMR, 13C-NMR, and MS. Detailed SAR studies and synthetic procedures will be published elsewhere.
Time-Dependent and Competitive Inhibition Assays.
For the time-dependent inhibition studies, recombinant human COX-2 (66 nM) or ovine COX-1 (44 nM) in 100 mM Tris⋅HCl buffer (pH 8.0) containing 500 μM phenol was incubated with appropriate test compounds in DMSO (0–66 μM) at 25°C for 20 min and then analyzed for remaining COX activity by treatment with [1-14C]arachidonic acid (50 μM, 57 mCi/mmol) for 30 sec at 37°C. Competitive inhibition by the test compounds was studied by adding COX-1 (2 nM) or COX-2 (2 nM) to an incubation mixture containing [1-14C]arachidonate (2 μM) and inhibitor (0–20 μM). Isolation and quantification of radio-labeled prostanoid products has been described (16). All IC50 values are average determinations from two independent experiments.
Inhibition of COX-2 Activity in Activated RAW264.7 Cells.
Protocols for COX-2 inhibition in cultured cells have been described (17). Briefly, cells (6.2 × 106 cells per T25 flask) were activated with lipopolysaccharide (1 μg/ml) and IFN-γ (10 units/ml) in serum-free DMEM for 7 hr and then treated with inhibitor (0–5 μM) for 30 min at 37°C. Exogenous arachidonate metabolism was determined by adding [1-14C]arachidonic acid (20 μM) for 15 min at 37°C. IC50 values are the average of two independent determinations.
Generation and Inhibition of Wild-Type and Mutant Murine COX-2.
Site-directed mutagenesis was performed as described (27). Wild-type and mutant murine COX-2s were expressed in SF-9 insect cells from baculovirus vectors (PharMingen) and purified by ion-exchange chromatography and gel filtration. All of the purified proteins were shown by densitometric scanning of a 7.5% SDS/PAGE gel to be >80% pure. The peroxidase activity of all purified proteins was measured by using the guaiacol peroxidase assay. Inhibition assays in triplicate were performed by preincubating enzyme (60–80 nM) and inhibitors (0–5 μM) for 20 min at 25°C followed by the addition of [1-14C]arachidonic acid (50–120 μM) for 30 sec at 37°C.
Carrageenan-Induced Rat Foot Paw Edema Assay.
All procedures were performed according to approved animal protocols (M/98/251; Vanderbilt University Animal Care Committee). Male Sprague–Dawley rats (Harlan–Sprague–Dawley) (150–175 g) were fasted for 18 hr and then injected with λ-carrageenan (0.1 ml of a 1% suspension in 0.85% saline; Fluka) into the right hind footpad. After 1 hr, 90 μl of DMSO or 90 μl of inhibitor in DMSO was added to 6 ml of corn oil, and the rats were gavaged with 0.5 ml of the corn oil suspension containing DMSO or inhibitor. The ipsilateral footpad volume was measured with a water displacement plethysmometer (Ugo Basile, Varese, Italy, distributed by Stoelting) at 3 hr postinjection and compared with the initial preinjection paw volume. Inhibitor concentrations were varied with six animals per group. Each experiment was performed in duplicate.
Microsomal Metabolism Experiments.
Liver microsomes were prepared from Sprague–Dawley rats by differential centrifugation (28) and stored in 10 mM Tris buffer, pH 7.4, containing 1 mM EDTA and 20% glycerol at −80°C. Human liver microsomes were a gift from F. P. Guengerich (Vanderbilt University). Protein content was determined by a bicinchoninic acid protein assay (Pierce), and cytochrome P450 content was determined from the carbon monoxide difference spectrum (29). Compound 19 (10 μM) was incubated with microsomes for 2 min at 37°C in 50 mM phosphate buffer, pH 7.4, and metabolism was initiated by addition of NADPH. Reaction was terminated by addition of an equal volume of ice-cold acetonitrile, and the samples were placed immediately on ice. Samples were frozen at −80°C before solid-phase extraction and analysis by HPLC.
Gastrointestinal Ulcerogenicity.
Male Sprague–Dawley rats (150–175 g) were fasted for 18 hr with free access to water. DMSO or inhibitor in DMSO was mixed with 60 μl of absolute ethanol and 60 μl of Tween 80 before addition of 5.82 ml of PBS. The animals (n = 6 per group) received an oral gavage of 0.5 ml at the indicated dosages at t = 0 hr and food at t = 8 hr, followed by euthanization with carbon dioxide at t = 24 hr. The stomach and 3 cm of the adjoining duodenum were excised, opened along the greater curvature, and rinsed with cold PBS. The antrum of the stomach and the first 3 cm of the duodenum were examined for lesions with a dissecting microscope. The incidence is reported as the combined number of rats in duplicate experiments that contained one or more major lesions (1-mm length or greater).
Results
Selective COX-2 Inhibition by Amide Derivatives of ETYA.
The selective COX-2 substrate properties of anandamide prompted us to prepare the ethanolamide derivative 1 of the COX-1-selective inhibitor, ETYA (30), as a potential COX-2-selective inhibitor. Although 1 was inactive against the COX isoforms, the 3-phenylpropyl amide analog 2 exhibited modest COX-2-selective inhibition, compared with ETYA (IC50 COX-1/COX-2 = 4 vs. 0.1, respectively). Substituted aromatic amides, such as the 3-(4-methoxyphenyl)propylamide 3, also exhibited modest, but selective, COX-2 inhibition (IC50 COX-1/COX-2 = 2). Although we did not undertake an extensive SAR analysis in the ETYA series, these studies demonstrated a “proof-of-concept” that modification of carboxylate groups in relatively COX-1-selective inhibitors can alter the selectivity toward the two isoforms.
Selective COX-2 Inhibition by Amide and Ester Derivatives of Indomethacin.
Our preliminary results with ETYA encouraged us to analyze NSAIDs as targets for derivatization. Indomethacin was chosen from the arylacetic acid category of NSAIDs. Like ETYA, indomethacin is a slow, tight-binding, relatively COX-1-selective inhibitor (31), but conversion of its carboxylate to a methyl amide (4), methyl ester (5), or an ethanolamide (6) led to compounds that exhibited potent and selective COX-2 inhibition (Table 1). Apart from alkyl derivatives, many aromatic analogs also displayed COX-2-selective inhibition as illustrated by aromatic amide and ester derivatives 7 and 8, respectively. COX-2 selectivity, particularly in the aromatic ester series, was extremely sensitive to the nature and position of substituents on the phenyl ring. For example, the 4-methylmercaptophenyl ester 9 was only 8-fold selective as a COX-2 inhibitor, whereas the 2-methylmercaptophenyl isomer 10 was >1,100-fold selective as a COX-2 inhibitor. Interestingly, replacement of the 4-methylmercapto group in 9 with a 4-methoxy substituent (compound 8) also regenerated selectivity. As observed with 9, the 4-fluorophenyl (11) and the 3-pyridyl (12) esters exhibited dramatic losses in selectivity. COX-2 selectivity was regained by simply exchanging the ester linkage with an amide group (compounds 13-15). Interestingly, tertiary amides exemplified by 16 and 17 revealed little or no COX inhibition at the concentration range studied. However, primary amide analog 18 revealed potency and selectivity similar to many of the secondary amides. A crucial determinant of inhibitory potency was the presence of the 4-chlorobenzoyl group on the indole ring. Its replacement in amide 19 or ester 20 with a 4-bromobenzyl group generated inactive compounds (analogs 21 and 22) (see Table 1). To compare the inhibitory potency of indomethacin amide and ester derivatives with that of the previously reported COX-2 inhibitor 23, we synthesized 23 (13) and evaluated its inhibitory potency against human COX-2 and ovine COX-1. Although 23 displayed selective COX-2 inhibition, its COX-2 inhibitory potency was much less than most of the potent amide or ester analogs in the series. For example, the phenethyl amide derivative 19 was ≈40-fold more potent as a COX-2 inhibitor and ≈50- to 100-fold more selective than 23.
Table 1.
SAR studies on the selective COX-2 inhibition by indomethacin amides and esters
| Compound | R | IC50*
|
IC50 (COX-1)/ IC50 (COX-2)† | |
|---|---|---|---|---|
| oCOX-1 | hCOX-2 | |||
| Indomethacin | OH | 0.050 | 0.75 | 0.070 |
| 4 | HNCH3 | >66 | 0.70 | >90 |
| 5 | OCH3 | 33 | 0.25 | 130 |
| 6 | HN(CH2)2OH | >66 | 0.25 | >287 |
| 7 | HNC6H5(4-NHCOCH3) | >66 | 0.12 | >600 |
| 8 | OC6H5(4-OCH3) | >66 | 0.040 | >1,700 |
| 9 | OC6H5(4-SCH3) | 2.6 | 0.30 | 8.7 |
| 10 | OC6H5(2-SCH3) | >66 | 0.060 | >1,100 |
| 11 | OC6H5(4-F) | 3.0 | 0.075 | 40 |
| 12 | O(3-C5H4N) | 2.5 | 0.050 | 50 |
| 13 | HNC6H5(4-SCH3) | >66 | 0.12 | >600 |
| 14 | HNC6H5(4-F) | >66 | 0.060 | >1,100 |
| 15 | HN(3-C5H4N) | >66 | 0.050 | >1,300 |
| 16 | NC5H10 | >66 | >16.5 | — |
| 17 | N(CH3)(CH2)2C6H5 | >66 | >16.5 | — |
| 18 | NH2 | >20 | 0.70 | >29 |
| 19 | HN(CH2)2C6H5 | >66 | 0.060 | >1,100 |
| 20 | O(CH2)2C6H5 | >66 | 0.050 | >1,320 |
| 21 | ‡ | >66 | >66 | — |
| 22 | ‡ | >66 | >66 | — |
| 23 | ‡ | >66 | 2.5 | >26 |
IC50 values in μM represent time-dependent COX inhibition and are average values from duplicate experiments.
†COX-2 selectivity ratio.
‡ Contains p-bromobenzyl group on the indole nitrogen. The R group in compounds 21, 22, and 23 are phenethyl amide, phenethyl ester, and free acid, respectively.
Selective COX-2 Inhibition by Amide Derivatives of Meclofenamic Acid.
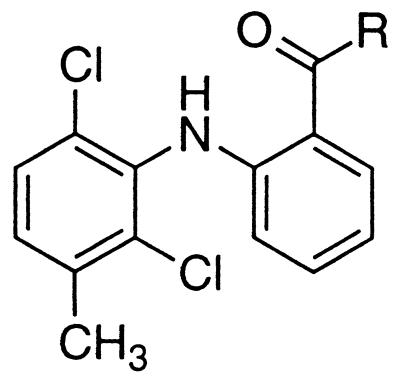
This methodology was expanded further to include meclofenamic acid, as an example from the fenamic acid class of NSAIDs. Meclofenamic acid is a nonselective inhibitor of the COX isoforms (32). The corresponding methyl amide 24, however, exhibited some COX-2-selective inhibition (Table 2). As evidenced in the ETYA series, alkyl or aryl esters were either nonselective or inactive as COX inhibitors. Therefore, further SAR analysis was limited to amides only. Increments in the alkyl chain length of 24 increased potency and incorporation of a terminal halogen in the alkyl chain increased selectivity as shown with the 3-chloropropyl amide 25. Introduction of a terminal hydroxyl group in the alkyl chain, however, led to significant losses in potency and selectivity as seen with the ethanolamide 26. Potency and selectivity were regained by functionalization of the hydroxyl group in 26 as illustrated with the phenoxy analog 27. Compound 27 was the most selective inhibitor in the meclofenamate series, with a COX-2 selectivity ratio of 440. Apart from 27, certain O-(substituted)hydroxamate derivatives (compounds 28 and 29) also demonstrated good COX-2-selective inhibitory properties. Replacement of the oxygen in the hydroxamate moiety in 28 with a methylene generated 30, which exhibited loss of potency and selectivity. A striking feature in the SAR analysis was the inhibitory profile of meclofenamic acid–amino acid conjugates. Thus, derivatives in which the carboxylate moiety in amino acid conjugates was esterified (compound 31) displayed potent and modestly selective COX-2 inhibition, whereas compounds such as 32, which possessed a free carboxylate, revealed nonselective COX inhibition.
Table 2.
SAR studies on the selective COX-2 inhibition by meclofenamic acid amides
| Compound | R | IC50*
|
IC50 (COX-1)/ IC50 (COX-2)† | |
|---|---|---|---|---|
| oCOX-1 | hCOX-2 | |||
| Meclofenamic acid | OH | 0.040 | 0.050 | 0.72 |
| 24 | HNCH3 | 16.5 | 5.5 | 3.0 |
| 25 | HN(CH2)3Cl | 2.4 | 0.060 | 40 |
| 26 | HN(CH2)2OH | 0.90 | 0.60 | 1.4 |
| 27 | HN(CH2)2OC6H5 | 66 | 0.15 | 440 |
| 28 | HNOCH2C6H5 | 66 | 1.0 | 66 |
| 29 | HNOCH2C6H5(4-NO2) | 60 | 0.20 | 273 |
| 30 | HN(CH2)2C6H5 | 4.0 | 4.5 | 0.90 |
| 31 | HNCH2CO2CH3 | 1.2 | 0.070 | 17 |
| 32 | HNCH2CO2H | 0.30 | 0.40 | 0.75 |
IC50 values in μM represent time-dependent COX inhibition and are average values from duplicate experiments.
†COX-2 selectivity ratio.
Kinetics and Molecular Basis of Selective COX-2 Inhibition.
The kinetics of selective COX-2 inhibition were probed with the indomethacin phenethyl amide derivative 19. Like indomethacin, 19 behaved as a slow, tight-binding inhibitor of COX-2. However, the time course of COX-2 inhibition was much slower than that by indomethacin or diarylheterocycles such as 2-(2-chlorophenyl)-4-[4-(methylsulfonyl)phenyl]-5-[4-(methoxy)phenyl]thiazole (SC-58092; ref. 33). For instance, maximal inhibition of COX-2 was achieved within 1–2 min with a 2-fold excess of SC-58092, whereas optimal COX-2 inhibition with a 2-fold excess of amide 19 required 10–15 min. No time-dependent inhibition of COX-1 by 19 was observed even at very high concentrations (>80 μM). Furthermore, 19 did not display any significant competitive inhibition of COX-1 or COX-2 at concentrations 10-fold greater than that of arachidonate, whereas SC-58092 was an excellent competitive inhibitor of both isoforms at low inhibitor concentrations (0.05–0.50 μM). Therefore, COX-2 inhibition kinetics of indomethacin amides are different from those observed with other COX-2-selective inhibitor classes such as the diarylheterocycles and the acidic sulfonamides (34).
Site-directed mutagenesis provided an opportunity to probe the molecular basis for the interaction of NSAID analogs with murine COX-2 (Table 3). Several site-directed mutants, including the R120Q and R120A mutants, were constructed as representatives of active-site regions important for the binding of indomethacin and meclofenamic acid (15–18). Although the R120Q mutant was resistant to inhibition by indomethacin, it was potently inhibited by ester 8 and amide 19. Furthermore, the indomethacin analog 23, which contains a free carboxylate group, was much less active than indomethacin esters and amides against wild-type protein and was unable to inhibit the R120Q mutant (see Table 3). Surprisingly, meclofenamic acid, which also contains a free carboxylate moiety, inhibited the R120A mutant at a potency comparable to the wild-type enzyme. Likewise, the IC50 values for the inhibition of wild type and the R120A COX-2 mutant by the phenoxyethyl amide derivative of meclofenamic acid (compound 27) were similar (Table 3). These results confirm our hypothesis that the R120 interaction is not essential for selective COX-2 inhibition by NSAID amides and esters.
Table 3.
Inhibition of murine COX-2 mutants by NSAID ester and amide derivatives
| Mutant | IC50, μM*
|
|||||
|---|---|---|---|---|---|---|
| Indomethacin | Ester 8 | Amide 19 | Bromobenzyl analog 23 | Meclofenamic acid | Amide 27 | |
| Wild-type mouse COX-2 | 0.21 | 0.050 | 0.26 | 3.8 | 0.13 | 0.40 |
| R120Q | >2.5 (0% inhibition) | 0.070 | 0.17 | |||
| R120A | >2.5 (20% inhibition) | 0.060 | >2.5 | >5.0 (0% inhibition) | 0.42 | 0.55 |
| Y355F | >5.0 (8% inhibition) | >5.0 (10% inhibition) | >5.0 (8% inhibition) | >3.0 (0% inhibition) | >3.0 (30% inhibition) | |
| E524L | 0.28 | >5.0 (22% inhibition) | >5.0 (25% inhibition) | 0.37 | 0.90 | |
| L503F | 0.53 | 0.36 | >2.5 (35% inhibition) | >5.0 (8% inhibition) | 0.13 | 0.5 |
| S530A | 0.22 | 0.080 | 0.70 | 0.13 | 0.13 | |
| V523I | 0.45 | 0.20 | 0.98 | |||
| V523IR513HV434I | 2.1 | 0.17 | 1.1 | 0.30 | 0.30 | |
IC50 values in μM represent time-dependent inhibition and average determinations from three experiments. The amino acid designations are based on the COX-1 amino acid sequence.
Together with R120, Y355 forms a constriction at the mouth of the substrate access channel and is an important determinant of NSAID binding (15, 19, 35). Indomethacin and both 8 and 19 were incapable of inhibiting the Y355F mutant. Likewise, no inhibition of the Y355F mutant was discernible with meclofenamic acid and the corresponding amide derivative 27. The E524L mutant, a residue positioned at the mouth of the COX channel that forms a salt bridge with R120, was potently inhibited by indomethacin but not by 8 or 19. Meclofenamic acid as well as the corresponding amide 27 inhibited the E524L mutant, albeit at a slightly lower potency. The possibility that S530, the site of aspirin acetylation (36, 37), could influence COX inhibition by indomethacin, meclofenamic acid, and their corresponding derivatives also was examined. IC50 values for the S530A inhibition by indomethacin, meclofenamic acid, 8, 19, and 27 were similar to those observed with the wild-type enzyme. Furthermore, the parent NSAIDs and their derivatives, 8, 19, and 27, were potent inhibitors of the V523I and V434I/R513H/V523I mutants, residues that are responsible for the COX-2 selectivity of diarylheterocycles (38–40).
Comparison of the COX structures reveals that the conserved L384 is oriented differentially in the COX-1 and -2 active sites, because of the effects of a residue at position 503. In COX-1, the presence of a phenylalanine at position 503 results in the placement of the L384 side chain into the active site. In COX-2, a smaller leucine at position 503 allows the L384 side chain to orient away from the active site and generates a solvent accessible space in the apex of the COX-2 active site. To test the possibility that this region contributed to the selectivity of the esters and amides, we assessed our inhibitors and the parent NSAID against the L503F mutant. IC50 values for the inhibition of this mutant and wild-type enzyme by indomethacin were comparable. However, ester 8 and amide 19 were significantly less potent as inhibitors (see Table 3). Likewise, indomethacin analog 23 did not inhibit this mutant, suggesting some similarity in the molecular basis for selective COX-2 inhibition. However, meclofenamic acid and its corresponding amide 27 were able to inhibit the L503F mutant with similar potency as observed with wild-type enzyme. Overall, these results establish that the COX-2 selectivity by indomethacin analogs arises from a combination of novel interactions at the mouth of the COX-2 active site with Y355 and E524 and at the apex of the active site with L503. These results also suggest subtle differences between the interactions of the esters and amides at the COX-2 active site. No apparent differences in the inhibitory profile of meclofenamic acid and amide 27 against the various mutants are discernible from this study. Both the free acid and the amide 27 are inactive against the conserved Y355F residue. Therefore, the COX-2 selectivity of 27 must arise from interactions elsewhere in the active site.
Inhibition of COX-2 Activity in Intact Cells.
The ability of indomethacin ester and amide derivatives to inhibit COX-2 in intact cells was assayed in RAW264.7 macrophages in which COX-2 activity was induced by pathologic stimuli. The macrophages were exposed to lipopolysaccharide and IFN-γ to induce COX-2 and then treated with several concentrations of ester 8 or amides 7 or 19. The IC50 values for inhibition of PGD2 production by 7, 8, and 19 were 0.009, 0.2, and 0.04 μM, respectively. Meclofenamate amides 25 and 31 also inhibited COX-2 activity in these macrophages with IC50 values of 0.2 and 0.12 μM, respectively. The IC50 values for inhibition of PGD2 synthesis by indomethacin and meclofenamic acid were 0.01 and 0.06 μM, respectively. Thus, the COX-2-selective derivatives of indomethacin and meclofenamic acid were comparable to the parent NSAIDs as inhibitors of COX-2 activity in cultured inflammatory cells.
Selective COX-2 Inhibition by Indomethacin Amides Is Anti-Inflammatory and Nonulcerogenic.
To test the hypothesis that selective COX-2 inhibition by indomethacin amides is anti-inflammatory but nonulcerogenic, we analyzed the in vivo anti-inflammatory properties of two indomethacin amides (compounds 7 and 19) in the carrageenan-induced rat footpad edema model after oral administration. Maximal edematous response in the carrageenan-injected right hind paw (0.7–0.9 ml) in control animals (n = 6) occurred 3 hr postinjection, and administration of indomethacin 1 hr after carrageenan reduced paw volume with an ED50 value of 2 mg/kg. This value is comparable to the literature value (41). Both 7 and 19 displayed anti-inflammatory effects with ED50 values of 0.8 mg/kg and 1.5 mg/kg, respectively. As shown in Table 4, oral administration of indomethacin at 10 mg/kg (5-fold higher than the ED50 value) produced gastric lesions in six of 12 rats. In contrast, oral administration of 7 and 19 at 50 mg/kg (≈50-fold higher than the ED50 value) was nonulcerogenic. These data are consistent with the lack of COX-1 inhibitory activity of compounds 7 and 19 and suggest that their anti-inflammatory activities are not caused by their hydrolysis to indomethacin.
Table 4.
Antral ulcers in fasted/fed rats with oral administration of indomethacin and amide derivatives 7 and 19
| Inhibitor | Dosage, mg/kg | Incidence (n = 12)* |
|---|---|---|
| Vehicle | 0 | 0/12 |
| Indomethacin | 10 | 6/12 |
| 7 | 50 | 0/12 |
| 19 | 50 | 0/12 |
The incidence is reported as the number of rats that contained one or more major lesions (1-mm length or greater).
The plasma stability of amide 19 was assessed by incubating 20 μg with 500 μl of human plasma for 18 hr at 37°C. After organic extraction, the formation of indomethacin in plasma was monitored by tandem MS with electrospray ionization. Examination of parent ions yielding a daughter ion at m/z 140 (4-chlorobenzoyl moiety common in indomethacin and 19) upon collision-induced dissociation (CID) revealed a single molecular ion at m/z 461 with a CID spectrum identical to a synthetic standard of 19. In contrast, when plasma samples treated with indomethacin and 19 (50 μg and 20 μg each) were analyzed, two molecular ions at m/z 358 and 461 generated fragments at m/z 140 upon CID. These molecular ions generated CID identical to indomethacin and 19, respectively. Thus, indomethacin amides such as 19 do not appear to be hydrolytically labile in plasma.
Finally, compound 19 was incubated for varying times with rat liver microsomes or human liver microsomes in the presence or absence of NADPH. The incubation mixtures were quenched with acetonitrile and analyzed by HPLC. No products that coeluted with indomethacin were formed under any conditions, even when significant metabolism of compound 19 was detected. All of the in vivo and in vitro data suggest that 19 and, by inference, the other amide derivatives are not converted metabolically to indomethacin.
Discussion
The findings in this report validate our hypothesis that neutralization of the carboxylate of NSAIDs can generate COX-2-selective inhibitors. As predicted from the differential effect of R120 mutations in COX-1 and COX-2 on arachidonate oxygenation, esterification or amidation of NSAIDs abolishes COX-1 inhibitory activity while maintaining COX-2 inhibitory activity. SAR analysis reveals that structurally diverse functionalities can serve as part of the ester/amide linkage in indomethacin, resulting in highly selective COX-2 inhibitors. Based on these observations and the array of readily available amines and alcohols, one can envision the development of a clinical candidate with an “ideal” pharmacological profile, solely based on the indomethacin template.
Preliminary biochemical analysis of indomethacin amides as COX-2 inhibitors reveals that they conform to the two-step kinetic mechanism, typical of slow, tight-binding NSAIDs. However, the time course of COX-2 inhibition is much slower for indomethacin amides than diarylheterocycles. These results presumably reflect a slow on-rate for the initial association of the amides with COX-2. A useful consequence of this slow on-rate is the complete absence of competitive inhibition of either COX-2 or COX-1 by the indomethacin amides. Competitive inhibition of COX-1 has been observed for some diarylheterocycles and presents a practical limitation to the in vivo selectivity of this class of inhibitors (34, 42).
As predicted, COX-2 inhibition by indomethacin amides and esters is unaffected by mutations of R120. This differs from the effect of this mutation on inhibition by indomethacin, zomepirac, and several of their analogs. COX-2 inhibition by indomethacin amides and esters depends on Y355 and E524. Furthermore, indomethacin does not inhibit the Y355F mutant. The latter result is consistent with the observation that the Y355F COX-1 mutant is resistant to the inhibitory effects of indomethacin (20). For reasons that are unclear, our results on the lack of inhibition of the Y355F COX-2 mutant by indomethacin are opposite to those obtained by Swinney and colleagues (43), who reported that indomethacin was a better time-dependent inhibitor of the Y355F human COX-2 mutant.
Residues that account for the COX-2 selectivity by diarylheterocycles are not important for the selectivity demonstrated by NSAID amides and esters. However, the “leucine tickle region” in COX-2 (15), which results from the presence of a leucine at position 503 instead of a phenylalanine as in COX-1, is important for COX-2 potency and selectivity by NSAID analogs, as well as indomethacin analog 23. The inhibitory potency of the parent NSAIDs, however, is not affected by mutations in this region of the protein. Thus, NSAID amides and esters interact with unique combinations of regions in the COX-2 protein; the substituted amide or ester linkage interacts with Y355 and E524 at the mouth of the active site, whereas the 4-chlorobenzoyl moiety interacts with the leucine tickle region.
As with the indomethacin derivatives, meclofenamate amides do not require R120 for COX-2 inhibition. Unlike indomethacin, which is inactive against the R120A mutant, meclofenamic acid is an inhibitor of this mutant, albeit with a slightly higher IC50 value. This result is consistent with that obtained by several other groups (21, 22, 44). However, the reason(s) for the inhibitory effects of a free carboxylate-containing NSAID against R120A is unclear. The general consensus that all carboxylate-containing NSAIDs bind to R120 therefore must be viewed with caution. Except for the Y355F mutant, which was resistant to the inhibitory effects of meclofenamic acid and its corresponding amide 27, all other mutants were inhibited potently by 27 and the parent NSAID. Because this residue is conserved in the two isozymes, COX-2-selective inhibition by 27 must arise from interaction(s) of its phenoxyethyl group with residue(s) below the constriction site formed by R120 and Y355.
Indomethacin amides and esters inhibit COX-2 activity in macrophages and are potent anti-inflammatory agents in the rat footpad carrageenan-induced edema model. The ED50 values for oral activity of the candidate amides (1–1.5 mg/kg) are similar to that of indomethacin (2 mg/kg). Unlike indomethacin, the corresponding amides were nonulcerogenic even at doses well exceeding their therapeutic efficacy. The similarity in anti-inflammatory activity between 7, 19, and indomethacin and the lack of ulcerogenic activity in 7 and 19 suggest that their anti-inflammatory activity is not caused by hydrolysis to indomethacin. This is supported by plasma stability studies and in vitro experiments with rat liver and human liver microsomes. No conversion of compound 19 to indomethacin was detected under any conditions.
Although amidation of meclofenamic acid also results in many potent and selective COX-2 inhibitors, further optimization may be necessary to improve the potency and selectivity of the current derivatives. The strategy outlined in this report should be general for all carboxylic acid-containing NSAIDs although different amide or ester substituents may be required for each NSAID. Because amides and esters are generated in a single step from the parent NSAID, combinatorial chemistry should facilitate the optimization process.
Acknowledgments
This research was funded by a research grant and center grants from the National Institutes of Health (CA47479, ES00267, and CA68485). S.W.R. is the recipient of a National Health and Medical Research Council of Australia C. J. Martin Fellowship.
Abbreviations
- COX
cyclooxygenase
- PG
prostaglandin
- NSAID
nonsteroidal anti-inflammatory drug
- SAR
structure–activity relationship
- ETYA
5,8,11,14-eicosatetraynoic acid
- CID
collision-induced dissociation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hamberg M, Samuelsson B. Proc Natl Acad Sci USA. 1973;70:899–903. doi: 10.1073/pnas.70.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith W L, Garavito R M, DeWitt D L. J Biol Chem. 1996;271:33157–33160. doi: 10.1074/jbc.271.52.33157. [DOI] [PubMed] [Google Scholar]
- 3.Herschman H R. Biochim Biophys Acta Lipids Lipid Metab. 1996;1299:125–140. doi: 10.1016/0005-2760(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 4.Needleman P, Isakson P C. J Rheumatol. 1997;24, Suppl 49:6–8. [PubMed] [Google Scholar]
- 5.Vane J R, Bakhle Y S, Botting R M. Annu Rev Pharmacol Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 6.Seibert K, Zhang Y, Leahy K, Hauser S, Masferrer J, Isakson P. Adv Exp Med Biol. 1997;400A:167–170. doi: 10.1007/978-1-4615-5325-0_24. [DOI] [PubMed] [Google Scholar]
- 7.Simon L S, Lanza F L, Lipsky P E, Hubbard R C, Talwalker S, Schwartz B D, Isakson P C, Geis G S. Arthritis Rheum. 1998;41:1591–1602. doi: 10.1002/1529-0131(199809)41:9<1591::AID-ART9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Ehrich E W, Dallob A, De Lepeleire I, Van Hecken A, Riendeau D, Yuan W Y, Porras A, Wittreich J, Seibold J R, DeSchepp P, et al. Clin Pharmacol Ther. 1999;65:336–347. doi: 10.1016/S0009-9236(99)70113-X. [DOI] [PubMed] [Google Scholar]
- 9.Prasit P, Riendeau D. Annu Rep Med Chem. 1997;32:211–220. [Google Scholar]
- 10.Talley J J. Prog Med Chem. 1999;36:201–234. doi: 10.1016/s0079-6468(08)70048-1. [DOI] [PubMed] [Google Scholar]
- 11.Marnett L J, Kalgutkar A S. Curr Opin Chem Biol. 1998;2:482–490. doi: 10.1016/s1367-5931(98)80124-5. [DOI] [PubMed] [Google Scholar]
- 12.Gans K R, Galbraith W, Roman R J, Haber S B, Kerr J S, Schmidt W K, Smith C, Hewes W E, Ackerman N R. J Pharmacol Exp Ther. 1990;254:180–187. [PubMed] [Google Scholar]
- 13.Black W C, Bayly C, Belley M, Chan C-C, Charleson S, Denis D, Gauthier J Y, Gordon R, Guay D, Kargman S, et al. Bioorg Med Chem Lett. 1996;6:725–730. [Google Scholar]
- 14.Leblanc Y, Black W C, Chan C C, Charleson S, Delorme D, Denis D, Gauthier J Y, Grimm E L, Gordon R, Guay D, et al. Bioorg Med Chem Lett. 1996;6:731–736. [Google Scholar]
- 15.Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner M F. Nat Struct Biol. 1996;3:927–933. doi: 10.1038/nsb1196-927. [DOI] [PubMed] [Google Scholar]
- 16.Kalgutkar A S, Crews B C, Rowlinson S W, Garner C, Seibert K, Marnett L J. Science. 1998;280:1268–1270. doi: 10.1126/science.280.5367.1268. [DOI] [PubMed] [Google Scholar]
- 17.Kalgutkar A S, Kozak K R, Crews B C, Hochgesang G P, Jr, Marnett L J. J Med Chem. 1998;41:4800–4818. doi: 10.1021/jm980303s. [DOI] [PubMed] [Google Scholar]
- 18.Bayly C I, Black W C, Léger S, Ouimet N, Ouellet M, Percival M D. Bioorg Med Chem Lett. 1999;9:307–312. doi: 10.1016/s0960-894x(98)00717-3. [DOI] [PubMed] [Google Scholar]
- 19.Picot D, Loll P J, Garavito R M. Nature (London) 1994;367:243–249. doi: 10.1038/367243a0. [DOI] [PubMed] [Google Scholar]
- 20.Loll P J, Picot D, Ekabo O, Garavito R M. Biochemistry. 1996;35:7330–7340. doi: 10.1021/bi952776w. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya D K, Lecomte M, Rieke C J, Garavito R M, Smith W L. J Biol Chem. 1996;271:2179–2184. doi: 10.1074/jbc.271.4.2179. [DOI] [PubMed] [Google Scholar]
- 22.Mancini J A, Riendeau D, Falgueyret J-P, Vickers P J, O'Neill G P. J Biol Chem. 1995;270:29372–29377. doi: 10.1074/jbc.270.49.29372. [DOI] [PubMed] [Google Scholar]
- 23.Rieke C J, Mulichak A M, Garavito R M, Smith W L. J Biol Chem. 1999;274:17109–17114. doi: 10.1074/jbc.274.24.17109. [DOI] [PubMed] [Google Scholar]
- 24.Yu M, Ives D, Ramesha C S. J Biol Chem. 1997;272:21181–21186. doi: 10.1074/jbc.272.34.21181. [DOI] [PubMed] [Google Scholar]
- 25.Marnett L J, Siedlik P H, Ochs R C, Pagels W D, Das M, Honn K V, Warnock R H, Tainer B E, Eling T E. Mol Pharmacol. 1984;26:328–335. [PubMed] [Google Scholar]
- 26.Odenwaller R, Chen Y-N P, Marnett L J. Methods Enzymol. 1990;187:479–485. doi: 10.1016/0076-6879(90)87054-7. [DOI] [PubMed] [Google Scholar]
- 27.Rowlinson S W, Crews B C, Lanzo C A, Marnett L J. J Biol Chem. 1999;274:23305–23310. doi: 10.1074/jbc.274.33.23305. [DOI] [PubMed] [Google Scholar]
- 28.Guengerich F P. In: Principles and Methods of Toxicology. Hayes A W, editor. New York: Raven; 1994. pp. 1259–1313. [Google Scholar]
- 29.Omura T, Sato R. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 30.Ahern D G, Downing D T. Biochim Biophys Acta. 1970;210:456–461. doi: 10.1016/0005-2760(70)90042-1. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell J A, Akarasereenont P, Thiemermann C, Flower R J, Vane J R. Proc Natl Acad Sci USA. 1993;90:11693–11697. doi: 10.1073/pnas.90.24.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meade E A, Smith W L, DeWitt D L. J Biol Chem. 1993;268:6610–6614. [PubMed] [Google Scholar]
- 33.Kalgutkar A S, Crews B C, Marnett L J. Biochemistry. 1996;35:9076–9082. doi: 10.1021/bi9605752. [DOI] [PubMed] [Google Scholar]
- 34.Copeland R A, Williams J M, Giannaras J, Nurnberg S, Covington M, Pinto D, Pick S, Trzaskos J M. Proc Natl Acad Sci USA. 1994;91:11202–11206. doi: 10.1073/pnas.91.23.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurumbail R G, Stevens A M, Gierse J K, McDonald J J, Stegeman R A, Pak J Y, Gildehaus D, Miyashiro J M, Penning T D, Seibert K, et al. Nature (London) 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 36.Van Der Ouderaa F J, Buytenhek M, Nugteren D H, Van Dorp D A. Eur J Biochem. 1980;109:1–8. doi: 10.1111/j.1432-1033.1980.tb04760.x. [DOI] [PubMed] [Google Scholar]
- 37.DeWitt D L, El-Harith E A, Kraemer S A, Andrews M J, Yao E F, Armstrong R L, Smith W L. J Biol Chem. 1990;265:5192–5198. [PubMed] [Google Scholar]
- 38.Gierse J K, McDonald J J, Hauser S D, Rangwala S H, Koboldt C M, Seibert K. J Biol Chem. 1996;271:15810–15814. doi: 10.1074/jbc.271.26.15810. [DOI] [PubMed] [Google Scholar]
- 39.Guo Q, Wang L-H, Ruan K-H, Kulmacz R J. J Biol Chem. 1996;271:19134–19139. doi: 10.1074/jbc.271.32.19134. [DOI] [PubMed] [Google Scholar]
- 40.Wong E, Bayly C, Waterman H L, Riendeau D, Mancini J A. J Biol Chem. 1997;272:9280–9286. doi: 10.1074/jbc.272.14.9280. [DOI] [PubMed] [Google Scholar]
- 41.Mukherjee A, Hale V, Borga O, Stein R. Inflamm Res. 1996;45:531–540. doi: 10.1007/BF02342223. [DOI] [PubMed] [Google Scholar]
- 42.Gierse J K, Koboldt C M, Walker M C, Seibert K, Isakson P C. Biochem J. 1999;339:607–614. [PMC free article] [PubMed] [Google Scholar]
- 43.So O-Y, Scarafia L E, Mak A Y, Callan O H, Swinney D C. J Biol Chem. 1998;273:5801–5807. doi: 10.1074/jbc.273.10.5801. [DOI] [PubMed] [Google Scholar]
- 44.Greig G M, Francis D A, Falgueyret J P, Ouellet M, Percival M D, Roy P, Bayly C, Mancini J A, O'Neill G P. Mol Pharmacol. 1997;52:829–838. doi: 10.1124/mol.52.5.829. [DOI] [PubMed] [Google Scholar]