Abstract
Activin, nodal, Vg1, and growth and differentiation factor 1 are members of the transforming growth factor β superfamily and signal via the activin type II (ActRII/IIB) and type I (ALK4) serine/threonine kinase receptors. Unlike activins, however, signaling by nodal, Vg1, and growth and differentiation factor 1 requires a coreceptor from the epidermal growth factor-Cripto-FRL1-Cryptic protein family such as Cripto. Cripto has important roles during development and oncogenesis and binds nodal or related ligands and ALK4 to facilitate assembly of type I and type II receptor signaling complexes. Because Cripto mediates signaling via activin receptors and binds directly to ALK4, we tested whether transfection with Cripto would affect the ability of activin to signal and/or interact with its receptors. Here we show that Cripto can form a complex with activin and ActRII/IIB. We were unable to detect activin binding to Cripto in the absence of ActRII/IIB, indicating that unlike nodal, activin requires type II receptors to bind Cripto. If cotransfected with ActRII/IIB and ALK4, Cripto inhibited crosslinking of activin to ALK4 and the association of ALK4 with ActRII/IIB. In addition, Cripto blocked activin signaling when transfected into either HepG2 cells or 293T cells. We have also shown that under conditions in which Cripto facilitates nodal signaling, it antagonizes activin. Inhibition of activin signaling provides an additional example of a Cripto effect on the regulation of signaling by transforming growth factor-β superfamily members. Because activin is a potent inhibitor of cell growth in multiple cell types, these results provide a mechanism that may partially explain the oncogenic action of Cripto.
Activins are members of the transforming growth factor β (TGF-β) superfamily (1, 2) that also includes the TGF-β, bone morphogenetic protein (BMP), growth and differentiation factor (GDF), and nodal-related families. The structurally related polypeptides of this superfamily control diverse cellular processes ranging from tissue patterning during embryogenesis to the control of homeostasis, cell growth, and differentiation in multiple adult tissues. Disruption or dysregulation of activin signaling is associated with multiple pathological states including reproductive disorders and carcinogenesis (3, 4). Activins are dimers consisting of two polypeptide β chains covalently linked by a disulfide bond. Although there are several β subunit genes and an extensive array of possible β–β dimers (5), only βA–βA (activin-A), βA–βB (activin-AB), and βB–βB (activin-B) have been isolated as dimeric proteins and shown to be biologically active.
Similar to other members of the TGF-β superfamily, activins exert their biological effects by interacting with two types of transmembrane receptors (types I and II) with intrinsic serine/threonine kinase activities. The initial step in signaling involves the binding of activin to a type II receptor, ActRII or ActRIIB (6–8), and the subsequent recruitment of the activin type I receptor activin-like kinase 4 (ALK4) (ActRIB) (9, 10). In this complex, the ActRII/IIB kinase phosphorylates ALK4 within a glycine- and serine-rich region called the GS domain, and this phosphorylation event activates the ALK4 kinase (1, 11). ALK4 subsequently phosphorylates cytoplasmic Smad proteins that assemble Smad4 and migrate to the nucleus to regulate transcription of activin-responsive genes (12, 13).
Members of the nodal family (14) and GDF-1/Vg1 (15) have also been shown to signal via ActRII/IIB and ALK4. Unlike activins, however, these TGF-β superfamily members require additional coreceptors from the epidermal growth factor (EGF)-Cripto-FRL1-Cryptic (CFC) protein family to assemble type II and type I receptors and generate signals (14, 15). The EGF-CFC family consists of small, extracellular signaling proteins including human and mouse Cripto and cryptic, Xenopus FRL1, and zebrafish one-eyed pinhead (16, 17). EGF-CFC proteins are known to act as anchored cell-surface coreceptors, but they also have activity when expressed as soluble proteins (17–20) or when they are secreted from the cell surface after enzymatic cleavage of their glycosylphosphatidylinositol anchor (21). Genetic studies in zebrafish and mice have shown that EGF-CFC proteins are required for mesoderm and endoderm formation as well as the establishment of left/right asymmetry during development (14). Cripto knockout mouse embryos lack a primitive streak and fail to form embryonic mesoderm (22). This phenotype is very similar to that observed in ActRIIA−/−;ActRIIB−/− (23), ALK4−/− (24), and Nodal−/− mice (25, 26), consistent with a requirement for nodal signaling via activin receptors and Cripto to initiate primitive streak elongation and mesoderm formation (14). It has been shown that Cripto independently binds nodal via its EGF-like domain and ALK4 via its CFC domain (27). Furthermore, selected point mutations in Cripto that block nodal binding or ALK4 binding disrupt nodal signaling (21, 27). Substantial biochemical evidence indicates that nodal and Vg1/GDF1 form a complex with activin receptors only in the presence of EGF-CFC proteins (15, 19, 21, 27, 28).
Because Cripto binds ALK4 and activin-related ligands to facilitate their signaling, we tested whether Cripto also binds activin to affect its ability to interact with its receptors and/or signal. Here we present evidence that Cripto indeed can form a complex with activin and ActRII/IIB that seems to be mutually exclusive with the formation of an activin–ActRII/IIB–ALK4 complex, and we further show that transfecting cells with Cripto can inhibit activin signaling.
Materials and Methods
Materials.
NuPAGE gels and molecular weight markers were obtained from Invitrogen. Recombinant human activin-A was generated by using a stable activin-expressing cell line generously provided by J. Mather (Genentech) and was purified by Wolfgang Fischer (Peptide Biology Laboratories, The Salk Institute). Activin-B and BMP-7 were purchased from R & D Systems. [125I]Activin-A and [125I]activin-B were prepared by using the chloramine T method as described (29). Anti-myc (9E10) monoclonal antibody and protein G agarose were purchased from Calbiochem. Polyclonal antibodies directed against ActRIIB (30) and ALK4 (10) have been described. Horseradish peroxidase-linked anti-rabbit IgG, anti-mouse IgG, and chemiluminescent substrate (Supersignal) were obtained from Pierce.
Expression Constructs.
Mouse Cripto constructs, each containing three C-terminal FLAG epitope tag sequences, have been described (27) and were gifts from Malcolm Whitman (Department of Cell Biology, Harvard Medical School, Boston). Mouse nodal was a gift from Michael Shen (Center for Advanced Biotechnology and Medicine, University of Medicine and Dentistry of New Jersey, Piscataway). Cripto-FLAG constructs, nodal, ActRII-myc, ActRIIB, and ALK4 were subcloned into pcDNA3 (Invitrogen) for mammalian expression.
Transfection of 293T and HepG2 Cells.
293T cells were grown in complete DMEM, and HepG2 cells were grown in complete α-MEM (DMEM and α-MEM were supplemented with 10% bovine calf serum, penicillin, streptomycin, and l-glutamine). Cells were grown in 5% CO2 to ≈40–60% confluence and then transfected by using Perfectin (Gene Therapy Systems, San Diego) for 293T cells or GenePorter 2 (Gene Therapy Systems) for HepG2 cells according to manufacturer instructions.
Covalent Crosslinking and Western Blotting.
293T cells were plated on six-well plates coated with poly-D-lysine at a density of 400,000 cells per well. Approximately 24 h later, cells were transfected with 2 μg of DNA per well (1 μg of RII/IIB/0.5 μg of ALK4/0.5 μg of Cripto) and then incubated an additional 48 h before harvesting. Covalent crosslinking was performed by first washing cells in Hepes dissociation buffer (HDB) (12.5 mM Hepes, pH 7.4/140 mM NaCl/5 mM KCl) and then incubating cells with [125I]activin-A in binding buffer (HDB containing 0.1% BSA/5 mM MgSO4/1.5 mM CaCl2) at room temperature for 4 h. Cells were washed in HDB, resuspended at 0.5 mM disuccinimidyl suberate in HDB, and incubated 30 min on ice. Crosslinking reactions were quenched with TBS (50 mM Tris⋅HCl, pH 7.5/150 mM NaCl), and cells were solubilized in lysis buffer (TBS containing 1% Nonidet P-40/0.5% deoxycholate/2 mM EDTA) and subjected to immunoprecipitation by using anti-myc, anti-ActRIIB, or anti-ALK4 antibodies. Immune complexes were analyzed by SDS/PAGE and autoradiography. For Western blotting, cells were solubilized in 200 μl of lysis buffer, and SDS/PAGE and electrotransfer to nitrocellulose were carried out by using NuPAGE gels and a NOVEX X-cell II apparatus as described (31).
Luciferase Assays in HepG2 and 293T Cells.
HepG2 cells were plated in 24-well plates at 150,000 cells per well and then transfected in triplicate ≈24 h later with 1 μg of DNA per well (800 ng of Cripto/100 ng of 3TP-lux or BRE-luc/100 ng of cytomegalovirus-β-galactosidase). Cells were treated with activin-A or BMP-7 ≈30 h posttransfection and then harvested ≈16 h after treatment. Cells were solubilized in 1% TX-100 solubilization buffer (1% Triton X-100/25 mM glycylglycine, pH 7.8/15 mM MgSO4/4 mM EGTA/1 mM DTT), and luciferase reporter activity was measured and normalized relative to β-galactosidase activities. 293T cells were plated on poly-D-lysine-treated 24-well plates at 150,000 cells per well and transfected in triplicate 24 h later with 0.5 μg of DNA per well (200 ng of Cripto/200 ng of nodal/50 ng of FAST2/25 ng of A3-lux/25 ng of cytomegalovirus-β-galactosidase). Cells were treated 6–8 h posttransfection with activin and then harvested 16 h after treatment. Luciferase assays were performed as described above.
Results
Structure of Cripto Constructs Tested for Effects on Activin Binding and Signaling.
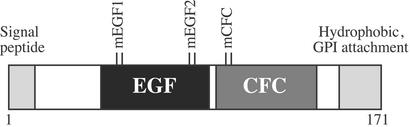
The domain structure of mouse Cripto (171 aa) is shown in Fig. 1 (reviewed in refs. 16 and 17). The mouse Cripto core protein has a molecular mass of ≈18 kDa and includes an N-terminal signal peptide, conserved EGF-like and CFC domains, and a hydrophobic C-terminal region that contains a glycosylphosphatidylinositol attachment site. Cripto proteins undergo a variety of processing and modification steps including removal of the N-terminal signal peptide, glycosylphosphatidylinositol attachment, and N- and O-linked glycosylation. Cripto isoforms with apparent molecular masses ranging from 14 to 60 kDa have been observed after SDS/PAGE (17). We tested the behavior of three Cripto mutants (Fig. 1) in this study that were shown by Yeo and Whitman (27) to be unable to mediate nodal signaling. These mutants are (i) Cripto mCFC (H104G and W107G), which has two point mutations within the CFC domain and does not bind ALK4 (27), (ii) Cripto ΔEGF, which has the entire EGF domain deleted and is therefore unable to bind nodal (21, 27), and (iii) Cripto EGF1⋅2mCFC (N69G, T72A, R88G, E91G, H104G, and W107G), which incorporates the mEGF1, mEGF2, and mCFC tandem point mutations (Fig. 1) and is defective in both ALK4 and nodal binding (21, 27).
Figure 1.
Domain structure of mouse Cripto. Conserved domains of Cripto are shown including the N-terminal signal peptide, EGF-like domain (EGF), CFC domain, and C-terminal hydrophobic region containing the site of glycosylphosphatidylinositol (GPI) attachment. The sites of three tandem point mutations are also indicated (mEGF1, N69G and T72A; mEGF2, R88G and E91G; mCFC, H104G and W107G).
Cripto Can Bind Activin in the Presence of ActRII/IIB and Block Activin Crosslinking to ALK4.
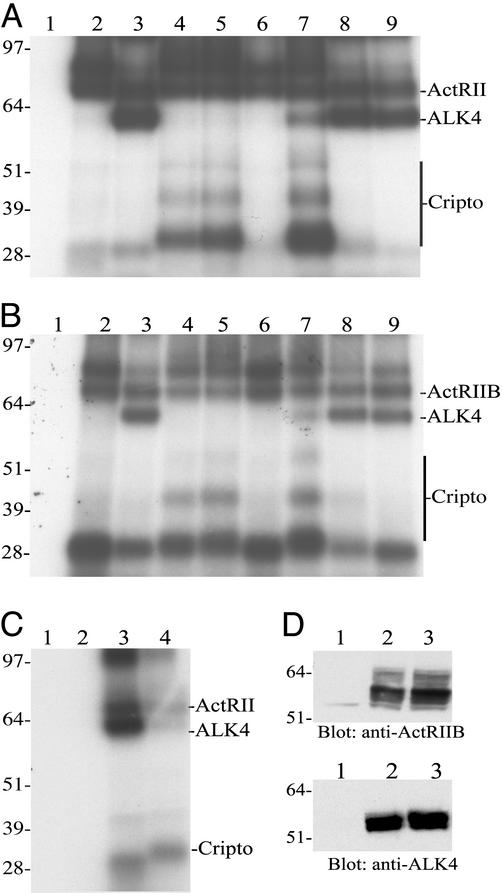
We have tested the ability of [125I]activin-A to form crosslinked complexes with Cripto in the presence or absence of activin receptors. Fig. 2 shows that when 293T cells were transfected with ActRII (Fig. 2A, lane 2) and then subjected to labeling and crosslinking with [125I]activin-A followed by immunoprecipitation with an antibody directed against ActRII, an activin–ActRII crosslinked complex of ≈80 kDa was evident, consistent with previous crosslinking results (30). The appearance of two ActRII–activin bands is routinely observed (31) and is likely the result of differential glycosylation of ActRII. Cotransfection of ActRII with ALK4 (Fig. 2A, lane 3) results in crosslinking of [125I]activin-A to both receptor types as indicated by the appearance of the activin–ALK4 crosslinked complex at ≈60 kDa. We have been unable to detect binding of [125I]activin-A to Cripto in the absence of activin type II receptors (Fig. 2C and data not shown). However, when ActRII was cotransfected with Cripto, activin-crosslinked complexes of ≈32, 45, and 52 kDa were observed (Fig. 2A, lane 4). These complexes are not present in samples in which Cripto was not transfected (Fig. 2A, lanes 1–3; the ≈28-kDa band represents the crosslinked [125I]activin-A dimer) and indicate the presence of Cripto species of ≈18, 31, and 38 kDa (the activin βA monomer is ≈14 kDa, and the gels were run under reducing conditions). The ≈18-, 31-, and 38-kDa forms likely have differential glycosylation and/or other modifications (17). The presence of [125I]activin-A–Cripto bands indicates the formation of stable activin–ActRII–Cripto complexes, because an antibody directed against ActRII was used in the immunoprecipitation. Activin–ActRII and activin–Cripto crosslinked bands were also evident when 293T cells were cotransfected with ActRII and Cripto and then subjected to immunoprecipitation by using an antibody directed against Cripto (data not shown). When the Cripto mCFC mutant or the Cripto ΔEGF mutant were cotransfected with ActRII, [125I]activin-A formed a crosslinked complex with Cripto mCFC (Fig. 2A, lane 5) but not Cripto ΔEGF (Fig. 2A, lane 6), indicating that the EGF-like domain of Cripto is required for activin binding.
Figure 2.
Covalent crosslinking of [125I]activin-A to type II activin receptors, Cripto, and ALK4. (A) 293T cells were transfected with the following constructs: lane 1, vector; lane 2, ActRII-myc; lane 3, ActRII-myc + ALK4; lane 4, ActRII-myc + Cripto; lane 5, ActRII-myc + Cripto mCFC; lane 6, ActRII-myc + Cripto ΔEGF; lane 7, ActRII-myc + ALK4 + Cripto; lane 8, ActRII-myc + ALK4 + Cripto mCFC; lane 9, ActRII-myc + ALK4 + Cripto ΔEGF. (B) 293T cells were transfected with the same constructs as described for A but with ActRIIB instead of ActRII-myc. (C) 293T cells were transfected with vector (lane 1), ALK4 + Cripto (lane 2), ActRII + ALK4 (lane 3), or ActRII + ALK4 + Cripto (lane 4). Cells were subjected to crosslinking with [125I]activin-A as described in Materials and Methods. Crosslinked complexes were isolated by immunoprecipitation by using an anti-myc antibody (targeting ActRII-myc) (A), an ActRIIB antibody (B), or an anti-ALK4 antibody (C). Immunoprecipitated proteins were resolved by SDS/PAGE and visualized by autoradiography as described in Materials and Methods. (D) 293T cells transfected with vector (lane 1), ActRIIB + ALK4 (lane 2), or ActRIIB + ALK4 + Cripto (lane 3) were solubilized and subjected to SDS/PAGE and Western blot analysis as described in Materials and Methods.
We further tested the effects of cotransfecting 293T cells with the three Cripto constructs and ActRII together with ALK4. When wild-type Cripto was transfected with ActRII and ALK4 (Fig. 2A, lane 7), [125I]activin-A formed a crosslinked complex with ActRII and Cripto, whereas crosslinking to ALK4 was greatly decreased relative to crosslinking in the absence of Cripto (Fig. 2A, compare lanes 3 and 7). Cotransfection with Cripto did not decrease expression of ALK4 as shown by Western blot (Fig. 2D and data not shown). Cripto mCFC did not block activin crosslinking to ALK4, but rather ALK4 prevented activin crosslinking to this mutant (Fig. 2A, compare lanes 5 and 8). This result is consistent with a competition between Cripto mCFC and ALK4 for binding the activin–ActRII complex and a reduced affinity of the mCFC mutant for the activin–ActRII complex relative to wild-type Cripto. Cripto ΔEGF was also unable to block crosslinking of activin to ALK4 in the presence of ActRII (Fig. 2A, lane 9). We performed parallel crosslinking studies by using ActRIIB instead of ActRII and immunoprecipitated crosslinked complexes with an antibody directed against ActRIIB. The results obtained with ActRIIB were very similar to those obtained by using ActRII (Fig. 2 A and B). We also tested the effects of Cripto on activin–ActRII–ALK4 complex formation as assessed after immunoprecipitation with an antibody directed against ALK4. Fig. 2C shows that when 293T cells were transfected with vector (Fig. 2C, lane 1) or cotransfected with ALK4 and Cripto (Fig. 2C, lane 2) and then subjected to crosslinking with [125I]activin-A, an ALK4 antibody failed to isolate labeled complexes. This is consistent with the inability of either Cripto or ALK4 to bind [125I]activin-A in the absence of type II receptors. When ActRII and ALK4 were coexpressed, the anti-ALK4 antibody precipitated a complex in which both ActRII and ALK4 were labeled (Fig. 2C, lane 3). However, cotransfection of Cripto with ActRII and ALK4 substantially blocked the appearance of these bands (Fig. 2C, lane 4) consistent with its ability to block crosslinking of activin to ALK4 and the association of ALK4 with ActRII.
Cripto Blocks Activin-A Signaling in HepG2 Cells.
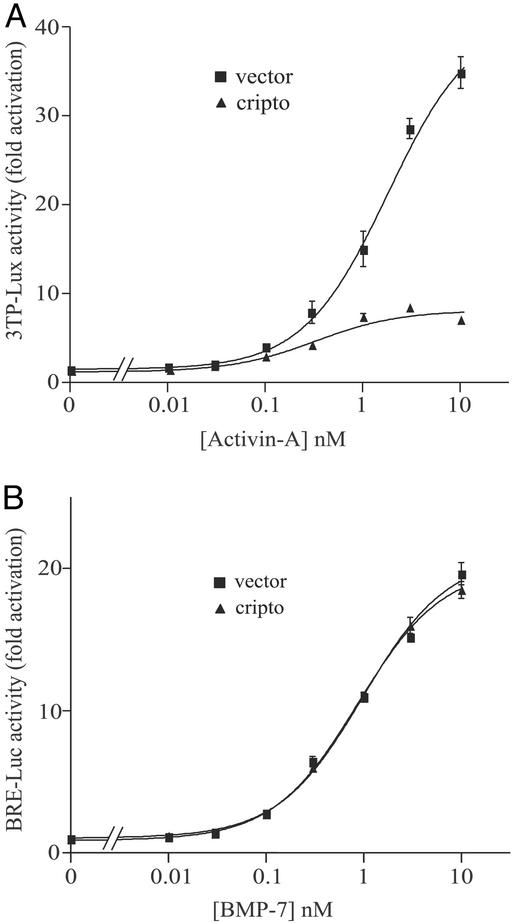
HepG2 cells do not express Cripto and require transfected Cripto to respond to nodal signals (32). Therefore, we tested the effects of transfected Cripto on activin signaling in this cell line. Cripto and the activin/TGF-β responsive luciferase reporter construct 3TP-lux (33) were transfected into HepG2 cells, and the effect of Cripto on activin-A-induced luciferase expression was measured. As shown in Fig. 3A, activin-A caused a dose-dependent increase in luciferase expression that was inhibited by Cripto. At maximal doses of activin-A there was an ≈4-fold reduction in activin-A signaling. As a control, we tested the effect of Cripto on the ability of the activin-A paralog BMP-7 to induce luciferase expression using the BMP-selective reporter BRE-luc (34). As shown in Fig. 3B, BMP-7 induced luciferase expression in HepG2 cells in a dose-dependent manner, but Cripto did not affect this induction, indicating that the effects of Cripto are selective for the activin pathway.
Figure 3.
Effects of Cripto on activin-A and BMP-7 signaling in HepG2 cells. HepG2 cells were transfected with either empty vector or Cripto as described in Materials and Methods and then treated with the indicated doses of either activin A (A) or BMP-7 (B). Luciferase activities were normalized relative to β-galactosidase activities, and data are presented as the fold increase in luciferase activity of cells treated with activin-A or BMP-7 relative to untreated cells.
Effects of Wild-Type and Mutant Cripto Constructs on Activin Signaling in 293T Cells.
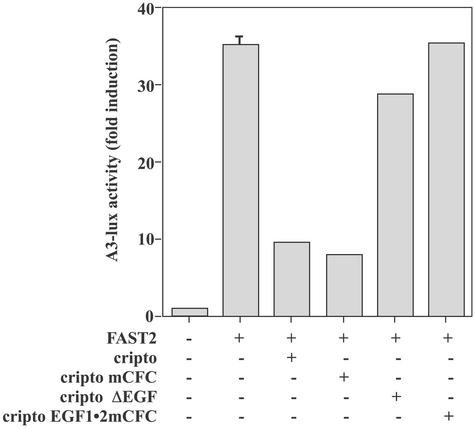
We further tested the effects of wild-type and mutant forms of Cripto on activin signaling in 293T cells. Similar to HepG2 cells, these cells do not express endogenous Cripto and have been used to characterize the effects of transfected EGF-CFC constructs on nodal signaling (21). It has been shown (21) that the transcription factor FAST2 (35) is required for induction of the activin-responsive A3-lux luciferase reporter (36) in 293T cells. We similarly showed that in the presence of FAST2, activin-A treatment caused a 30- to 40-fold induction of luciferase expression relative to untreated cells (Fig. 4, lane 2). Consistent with results in HepG2 cells, wild-type Cripto blocked activin signaling in 293T cells (Fig. 4, lane 3). The ability of Cripto to block activin-B signaling was similar to its ability to block activin-A signaling in these cells (data not shown). Similar to wild type Cripto, the Cripto mCFC mutant blocked activin-A signaling in these cells (Fig. 4, lane 4). In contrast, neither the Cripto ΔEGF mutant (Fig. 4, lane 5) nor the EGF1⋅2mCFC mutant (Fig. 4, lane 6; also see Fig. 1) were able to block activin-A signaling, consistent with data showing that these mutants do not block activin crosslinking to ALK4 (Fig. 2 and data not shown).
Figure 4.
Effects of wild-type Cripto and Cripto mutants on activin-A signaling in 293T cells. 293T cells were transfected with the indicated constructs as described in Materials and Methods and then treated with vehicle or 1 nM activin-A. Luciferase activities were normalized to β-galactosidase activities, and data are presented as the fold increase in luciferase activities relative to untreated cells.
Cripto Has Opposing Effects on Activin and Nodal Signaling in 293T Cells.
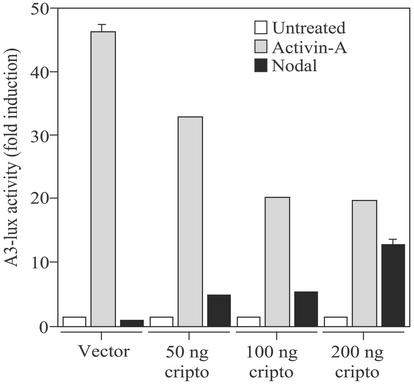
The results presented thus far indicate that Cripto has opposite effects on activin and nodal signaling. Therefore, we compared the effects of Cripto on activin-A and nodal signaling within the same system. It has been shown that transfection of nodal and Cripto into 293T cells resulted in secretion of processed nodal protein that generated signals in the cells producing it (21). We transfected 293T cells with FAST2, the A3-lux reporter plasmid, and various amounts of Cripto DNA and then treated the cells with activin-A or cotransfected them with the mouse nodal cDNA. Fig. 5 shows that in the absence of Cripto, activin-A treatment induced luciferase expression ≈45-fold relative to untreated cells and that cotransfection with increasing amounts of Cripto DNA caused a dose-dependent blockade of activin-A signaling. Conversely, nodal did not generate a detectable signal in the absence of Cripto, but its signaling increased as the amount of Cripto DNA transfected into the cells was increased (Fig. 5). Therefore, Cripto can have opposing effects on activin and nodal signaling despite the fact that both ligands use the same signaling receptors.
Figure 5.
Effects of Cripto on activin-A and nodal signaling in 293T cells. 293T cells were transfected with either empty vector or nodal and the indicated amount of Cripto DNA as described in Materials and Methods and then treated where indicated with 1 nM activin-A. Luciferase values were normalized to β-galactosidase activities, and data are presented as the fold increase in luciferase activities relative to untreated cells.
Model of the Mechanism of Action of Cripto.
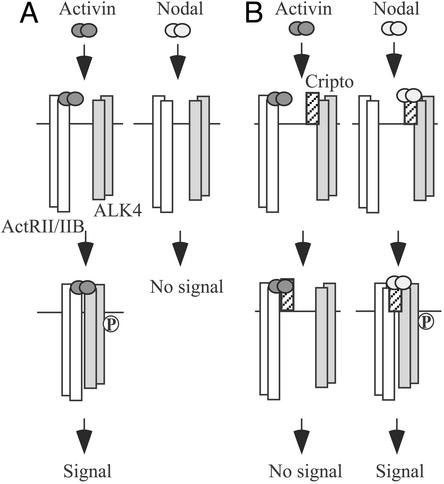
Our data are consistent with a model (Fig. 6) in which activin and nodal have distinct mechanisms for signaling via activin receptors. In the absence of Cripto (Fig. 6A), activin first binds ActRII/IIB to form a complex that can recruit ALK4 and then generate signals. Nodal, on the other hand, does not bind and assemble ActRII/IIB and ALK4 in the absence of Cripto (or a related EGF-CFC protein). In the presence of Cripto (Fig. 6B), activin binds ActRII/IIB and can then form a complex with Cripto. Cripto prevents binding of the activin–ActRII/IIB complex to ALK4 and blocks signaling. Unlike activin, nodal binds directly to Cripto and causes the assembly of an active signaling complex containing nodal, Cripto, ActRII/IIB, and ALK4.
Figure 6.
Model of the proposed mechanism by which Cripto antagonizes activin. (A) Activin signals by binding ActRII/IIB and then recruiting ALK4. ActRII/IIB phosphorylates (P) the GS domain of ALK4, thereby activating the ALK4 kinase and initiating downstream signaling. Nodal does not bind activin receptors and therefore does not signal in the absence of Cripto. (B) Cripto antagonizes activin signaling by forming a complex with activin and ActRII/IIB. We propose that this complex precludes the formation of a functional activin–ActRII/IIB–ALK4 complex and therefore blocks signaling. Nodal binds directly to Cripto, leading to the assembly of ActRII/IIB and ALK4 followed by ALK4 phosphorylation and downstream signaling.
Discussion
The results presented here provide evidence that Cripto can block activin signaling, and they support a mechanism in which Cripto binds activin in a complex with activin type II receptors. Our crosslinking data show that Cripto inhibits binding of activin to ALK4 and the association of ALK4 with ActRII/IIB. By disrupting activin binding to ALK4, Cripto likely prevents phosphorylation of ALK4 by ActRII/IIB to block subsequent downstream signaling. We showed that the mCFC mutant, a Cripto construct with disrupted ALK4 binding (27), bound activin in the presence of ActRII/IIB but was unable to block activin binding to ALK4. However, this mutant was capable of blocking activin signaling. This apparent inconsistency may be the result of the fact that the crosslinking experiments involved comparable expression levels of the mCFC mutant and ALK4, whereas the signaling experiments involved overexpression of the mCFC mutant and endogenous levels of ALK4. We therefore propose that the mCFC mutant has reduced affinity for the activin–ActRII/IIB complex relative to wild-type Cripto and ALK4, as illustrated by the crosslinking results but sufficient affinity to block activin function when overexpressed at high levels relative to endogenous levels of ALK4. We have also shown that the EGF-like domain of Cripto is required for the ability of Cripto to bind activin in the presence of type II receptors, to prevent activin binding to ALK4, and to block activin signaling. These data are consistent with previous results demonstrating that the EGF-like domain binds nodal and is required for nodal signaling (21, 27) and suggests that activin and nodal have similar or overlapping binding sites on Cripto. Finally, we have shown that transfected Cripto has opposite effects on activin and nodal signaling. It has been shown that after transfection into 293T cells, cripto can act either in a cell-autonomous manner or as a secreted protein to facilitate nodal signaling (21). Although not addressed in this study, it will be of interest to determine whether Cripto acts in a cell-autonomous fashion, as a secreted protein, or both to exert its effects on activin signaling.
One important area of future research should be to determine how Cripto interacts with its TGF-β superfamily ligands and activin receptors to either facilitate the formation of functional signaling complexes (i.e., in the case of nodal or Vg1/GDF1) or to inhibit signaling (i.e., in the case of activin). Nodal (19, 21, 28) and Vg1/GDF1 (15) each bind EGF-CFC proteins in the absence of activin receptors. However, we were unable to demonstrate binding of activin to Cripto in the absence of type II receptors. With respect to activin, therefore, Cripto behaves like ALK4 in that it requires the presence of type II receptors to bind activin. It has also been demonstrated that, unlike activin, nodal (27, 28) and Vg1/GDF1 (15) can only bind activin receptors and initiate signaling in the presence of Cripto or related EGF-CFC proteins. Structure/function analyses of these ligands, focusing on regions that are divergent between activin and nodal/Vg1/GDF1, will now be required to determine the molecular basis of these differences (15). It will be instructive to map the residues on activin required for type II receptor binding and compare these residues with the corresponding residues on nodal/Vg1/GDF1 with the aim of determining why activin independently binds ActRII/IIB with high affinity, whereas nodal, Vg1, and GDF1 do not. Similarly, identification of the residues on nodal/Vg1/GDF1 required for binding EGF-CFC proteins may shed light on why the corresponding site(s) on activin does not mediate this binding. We recently solved the structure of the activin-related ligand BMP-7 bound to the ActRII extracellular domain (37). In this study we proposed a model in which the dimeric BMP-7 ligand mediates the assembly of two type II receptors and two type I receptors to form a hexameric complex. It will be very interesting to determine how this model generally applies within the TGF-β superfamily and its implications regarding the structure of Cripto bound to its ligands and/or activin receptors.
Cripto was first isolated as a putative oncogene from a human teratocarcinoma cell line (38), and it was subsequently shown to be able to transform mammary epithelial cells (39). Cripto is a stimulator of cell growth and is expressed at high levels in human breast, colon, stomach, pancreas, lung, ovary, endometrium, testis, bladder, and prostate tumors while being absent or expressed at low levels in their normal counterparts (17). The elucidation of the signals and transcriptional events underlying the up-regulation of Cripto expression in these tumors remains an important area of future research. With regard to Cripto's mechanism(s) of action, it has been shown that recombinant, soluble Cripto can activate both the p42 mitogen-activated protein kinase pathway (40) and the phosphatidylinositol 3-kinase pathway (41) in mammary epithelial cells through an unidentified receptor. The ability of Cripto to activate these pathways, which are frequently growth-stimulatory in nature, may at least partially explain its effects on cell growth, differentiation, and oncogenesis (17).
Antagonism of activin signaling provides an additional mechanism of cell growth regulation by Cripto, and the results presented here indicate that the level of Cripto expression will set a threshold for the activin response. Activin, similar to TGF-β (42), is a potent inhibitor of cell growth in multiple cell types, and disruption of activin signaling is associated with tumorigenesis (3, 4). Consistent with a role for activin in inhibiting carcinogenesis, ALK4 mutations were described recently in pancreatic cancer, leading to the designation of ALK4 as a tumor-suppressor gene (43). Activin and TGF-β both signal through Smad2 and Smad3 (1), and this signaling pathway interacts in complex ways with the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3-kinase) pathways (44). In general, decreases in growth-inhibitory Smad2/3 signals and increases in growth-stimulatory MAPK and PI3-kinase signals are associated with increased tumorigenesis (44). Therefore, by activating the MAPK and PI3-kinase pathways and inhibiting the activin pathway, Cripto may play a dual role in promoting the cancer phenotype.
TGF-β superfamily members, including activins, are regulated by multiple diverse mechanisms at every level of their respective signaling pathways (45). For example, the ability of activin to access and assemble its signaling receptors can be inhibited in several distinct ways (5). Inhibins, which share a subunit with activins, are TGF-β superfamily members that act in conjunction with betaglycan to bind ActRII/IIB, thereby preventing these receptors from binding activin and initiating signaling (46). The soluble, extracellular binding protein follistatin binds activin with high affinity and also blocks the ability of activin to bind its cell-surface receptors and initiate signaling (5). The mechanism by which Cripto inhibits activin signaling seems to resemble that of the pseudo (decoy) type I receptor BMP and activin membrane-bound inhibitor (BAMBI), which binds BMPs and activin in nonfunctional complexes with their receptors to block signaling (47). Future studies will be required to further elucidate the mechanisms of Cripto action and to determine the physiological and possible pathophysiological roles Cripto plays as an activin antagonist in normal and neoplastic tissues.
Acknowledgments
We thank Louise Bilezikjian and Ezra Wiater for helpful discussions and Sandra Guerra for assistance in preparing the manuscript. This work was supported by a National Research Service Award (to P.C.G.), a C. J. Martin Fellowship (to C.A.H.), and the Foundation for Medical Research, California Division, National Institutes of Health Grant HD-13527. W.V. is a Foundation for Medical Research Senior Investigator.
Abbreviations
- TGF-β
transforming growth factor β
- BMP
bone morphogenetic protein
- GDF
growth and differentiation factor
- ActRII
activin receptor type II
- ALK4
activin-like kinase 4
- EGF
epidermal growth factor
- CFC
Cripto-FRL1-Cryptic
References
- 1.Massagué J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Piek E, Heldin C H, Ten Dijke P. FASEB J. 1999;13:2105–2124. [PubMed] [Google Scholar]
- 3.Matzuk M M, Kumar T R, Shou W, Coerver K A, Lau A L, Behringer R R, Finegold M J. Recent Prog Horm Res. 1996;51:123–547. [PubMed] [Google Scholar]
- 4.Chen Y G, Lui H M, Lin S L, Lee J M, Ying S Y. Exp Biol Med (Maywood) 2002;227:75–87. doi: 10.1177/153537020222700201. [DOI] [PubMed] [Google Scholar]
- 5.Phillips D J. BioEssays. 2000;22:689–696. doi: 10.1002/1521-1878(200008)22:8<689::AID-BIES2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 6.Mathews L S, Vale W W. Cell. 1991;65:973–982. doi: 10.1016/0092-8674(91)90549-e. [DOI] [PubMed] [Google Scholar]
- 7.Mathews L S, Vale W W, Kintner C R. Science. 1992;255:1702–1705. doi: 10.1126/science.1313188. [DOI] [PubMed] [Google Scholar]
- 8.Attisano L, Wrana J L, Cheifetz S, Massagué J. Cell. 1992;68:97–108. doi: 10.1016/0092-8674(92)90209-u. [DOI] [PubMed] [Google Scholar]
- 9.Cárcamo J, Weis F M, Ventura F, Wieser R, Wrana J L, Attisano L, Massagué J. Mol Cell Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchida K, Vaughan J M, Wiater E M, Gaddy-Kurten D, Vale W W. Endocrinology. 1995;136:5493–5503. doi: 10.1210/endo.136.12.7588300. [DOI] [PubMed] [Google Scholar]
- 11.Attisano L, Wrana J L, Montalvo E, Massagué J. Mol Cell Biol. 1996;16:1066–1073. doi: 10.1128/mcb.16.3.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derynck R, Zhang Y, Feng X H. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 13.Wrana J L, Attisano L. Cytokine Growth Factor Rev. 2000;11:5–13. doi: 10.1016/s1359-6101(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 14.Schier A F, Shen M M. Nature. 2000;403:385–389. doi: 10.1038/35000126. [DOI] [PubMed] [Google Scholar]
- 15.Cheng S K, Olale F, Bennett J T, Brivanlou A H, Schier A F. Genes Dev. 2003;17:31–36. doi: 10.1101/gad.1041203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen M M, Schier A F. Trends Genet. 2000;16:303–309. doi: 10.1016/s0168-9525(00)02006-0. [DOI] [PubMed] [Google Scholar]
- 17.Saloman D S, Bianco C, Ebert A D, Khan N I, De Santis M, Normanno N, Wechselberger C, Seno M, Williams K, Sanicola M, et al. Endocr Relat Cancer. 2000;7:199–226. doi: 10.1677/erc.0.0070199. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Talbot W S, Schier A F. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- 19.Bianco C, Adkins H B, Wechselberger C, Seno M, Normanno N, De Luca A, Sun Y, Khan N, Kenney N, Ebert A, et al. Mol Cell Biol. 2002;22:2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhadanov A B, Bertuzzi S, Taira M, Dawid I B, Westphal H. Dev Dyn. 1995;202:354–364. doi: 10.1002/aja.1002020405. [DOI] [PubMed] [Google Scholar]
- 21.Yan Y T, Liu J J, Luo Y, Chaosu E, Haltiwanger R S, Abate-Shen C, Shen M M. Mol Cell Biol. 2002;22:4439–4449. doi: 10.1128/MCB.22.13.4439-4449.2002. .Y. E. C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding J, Yang L, Yan Y T, Chen A, Desai N, Wynshaw-Boris A, Shen M M. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- 23.Song J, Oh S P, Schrewe H, Nomura M, Lei H, Okano M, Gridley T, Li E. Dev Biol. 1999;213:157–169. doi: 10.1006/dbio.1999.9370. [DOI] [PubMed] [Google Scholar]
- 24.Gu Z, Nomura M, Simpson B B, Lei H, Feijen A, van den Eijnden-van Raaij J, Donahoe P K, Li E. Genes Dev. 1998;12:844–857. doi: 10.1101/gad.12.6.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Sasaki H, Lowe L, Hogan B L, Kuehn M R. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]
- 26.Conlon F L, Lyons K M, Takaesu N, Barth K S, Kispert A, Herrmann B, Robertson E J. Development (Cambridge, UK) 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 27.Yeo C, Whitman M. Mol Cell. 2001;7:949–957. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 28.Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico M G, Ibanez C F, Brivanlou A H. Genes Dev. 2001;15:2010–2022. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaughan J M, Vale W. Endocrinology. 1992;132:2038–2050. doi: 10.1210/endo.132.5.7682939. [DOI] [PubMed] [Google Scholar]
- 30.Mathews L S, Vale W W. J Biol Chem. 1993;268:19013–19018. [PubMed] [Google Scholar]
- 31.Gray P C, Greenwald J, Blount A L, Kunitake K S, Donaldson C J, Choe S, Vale W. J Biol Chem. 2000;275:3206–3212. doi: 10.1074/jbc.275.5.3206. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Novoselov V, Celeste A J, Wolfman N M, ten Dijke P, Kuehn M R. J Biol Chem. 2001;276:656–661. doi: 10.1074/jbc.M004649200. [DOI] [PubMed] [Google Scholar]
- 33.Cárcamo J, Zentella A, Massagué J. Mol Cell Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hata A, Seoane J, Lagna G, Montalvo E, Hemmati-Brivanlou A, Massagué J. Cell. 2000;100:229–240. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Dou C L, Prabhu L, Lai E. Mol Cell Biol. 1999;19:424–430. doi: 10.1128/mcb.19.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Pouponnot C, Massagué J. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenwald J, Groppe J, Gray P C, Wiater E, Kwiatkowski W, Vale W, Choe S. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 38.Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, Persico M G. EMBO J. 1989;8:1987–1991. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ciardiello F, Dono R, Kim N, Persico M G, Salomon D S. Cancer Res. 1991;51:1051–1054. [PubMed] [Google Scholar]
- 40.Kannan S, De Santis M, Lohmeyer M, Riese D J, II, Smith G H, Hynes N, Seno M, Brandt R, Bianco C, Persico G, et al. J Biol Chem. 1997;272:3330–3335. doi: 10.1074/jbc.272.6.3330. [DOI] [PubMed] [Google Scholar]
- 41.De Santis M L, Kannan S, Smith G H, Seno M, Bianco C, Kim N, Martinez-Lacaci I, Wallace-Jones B, Salomon D S. Cell Growth Differ. 1997;8:1257–1266. [PubMed] [Google Scholar]
- 42.Massagué J, Blain S W, Lo R S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 43.Su G H, Bansal R, Murphy K M, Montgomery E, Yeo C J, Hruban R H, Kern S E. Proc Natl Acad Sci USA. 2001;98:3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakefield L M, Roberts A B. Curr Opin Genet Dev. 2002;12:22–29. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 45.Massagué J, Chen Y G. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 46.Lewis K A, Gray P C, Blount A L, MacConell L A, Wiater E, Bilezikjian L M, Vale W. Nature. 2000;404:411–414. doi: 10.1038/35006129. [DOI] [PubMed] [Google Scholar]
- 47.Onichtchouk D, Chen Y G, Dosch R, Gawantka V, Delius H, Massagué J, Niehrs C. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]