Abstract
Congenital diaphragmatic hernia (CDH) is a significant cause of pediatric mortality in humans with a heterogeneous and poorly understood etiology. Here we show that mice lacking Slit3 developed a central (septum transversum) CDH. Slit3 encodes a member of the Slit family of guidance molecules and is expressed predominantly in the mesothelium of the diaphragm during embryonic development. In Slit3 null mice, the central tendon region of the diaphragm fails to separate from liver tissue because of abnormalities in morphogenesis. The CDH progresses through continuous growth of the liver into the thoracic cavity. This study establishes the first genetic model for CDH and identifies a previously unsuspected role for Slit3 in regulating the development of the diaphragm.
Congenital diaphragmatic hernia (CDH) was described in 1679 by Riverius (1). CDH occurs in ≈1 in 3,000 newborns and is associated with a 30–60% mortality rate with significant morbidity among survivors (2–4). There are several different types of CDH including Bochdalek, Morgagni, and central (septum transversum) diaphragmatic hernia (5). The Bochdalek CDH accounts for ≈70% of CDH and occurs in a posteriolateral region of the diaphragm. Morgagni CDH is less common and forms in the anterior retrosternal diaphragm. Central (septum transversum) CDH occurs in the midline of the septum transversum and accounts for 1–2% of the total cases of CDH. The pathogenesis of CDH results as abdominal contents enter the thoracic cavity. The primary consequence is hypoplasia of the lung due to decreased thoracic volume, resulting in direct compression of the lung and inadequate fetal breathing movements. The resulting compromised pulmonary capacity often results in neonatal death.
Although CDH has been recognized for nearly 350 years, the pathogenesis and etiology of CDH is not clear. Evidence from teratogen-induced rodent models of CDH suggests that defects in diaphragm muscle arises from malformations of the primordial diaphragm, the pleuroperitoneal fold (6). Other evidence from rodent models demonstrates that there may be concomitant insults to lung development associated with CDH (7, 8).
The diaphragm is a complex structure that is thought to arise primarily from the septum transversum, the pleuroperitoneal membrane, paraxial mesoderm of the body wall, and esophageal mesenchyme (9, 10). Diaphragmatic musculature is thought to originate from condensations within the pleuroperitoneal fold (11). The diaphragm is innervated primarily by the phrenic nerve. Previous embryological studies have focused on the ontogeny of the phrenic nerve and muscle. The contribution made by connective tissue to diaphragm development has not been reported.
Slit proteins are large (>200 kDa) molecules with multiple functional domains (12–14). Slit homologs have been identified in multiple organisms (12–16). In Drosophila, mutations in the slit gene result in collapse of the CNS commissural axon scaffold due to the failure of commissural axons to leave the midline (15–17). In contrast to the single slit gene in Drosophila, there are three homologues in vertebrates that show both overlapping and distinct expression patterns. Slit proteins function as guidance cues for motor axons, olfactory bulb axons, neuronal cells, myoblasts, and leukocytes and as a branching factor for DRG sensory axons (18–23). These activities have been established for Slit1 and Slit2 in vitro. The Slit1/Slit2 double-null mouse shows an axon-guidance defect in retinal ganglion cells and in several major axonal pathways in the forebrain (24, 25).
The roundabout (robo) gene encodes the receptor for Slit in both Drosophila and vertebrates (17–19). Four Robo genes, including Robo1, Robo2, Rig-1 (Robo3), and magic Robo, have been identified in vertebrates (26–28). Robo genes encode large transmembrane proteins in which the extracellular Ig-like domains directly interact with Slit (29, 30). Dutt1/Robo1 mutant mice die at birth with defects in lung development (31).
Slit and Robo genes are expressed in both neuronal and nonneuronal tissues. In contrast to Slit1 and Slit2, which are expressed prominently in the CNS, Slit3 is expressed weakly in the CNS but strongly in some peripheral tissues (14). No specific function for Slit3 has been identified either in vivo or in vitro. To investigate in vivo functions for Slit3, a null mutation was introduced into Slit3 in mice by targeted mutagenesis in embryonic stem (ES) cells. Interestingly, Slit3 null mice develop a CDH similar to a central (septum transversum) CDH in humans.
Methods
Construction of Slit3 Null Mice.
The Slit3 targeting vector was made by introducing a 1-kb deletion encompassing 194 bp of the 3′ half of exon 1, which contains the initiation codon and signal peptide and ≈800 bp of intron 1 (Fig. 1a). This region was replaced by the β-galactosidase (β-gal) gene and a loxp-PGK-neo selection cassette. ES cell colonies were screened by DNA-blot analysis using both 5′ and 3′ probes. Positive clones were injected into C57BL/6J blastocysts to obtain chimeric mice, which then were bred with C57BL/6J mice to obtain germ-line transmission. In some sublines, the Neo cassette was removed by mating Slit3−/+ mice with β-actin-cre mice. The Slit3−/− mice with the neo cassette removed showed the same phenotype as mice that contained a PGK-neo gene. In this study we primarily analyzed mice containing the neo insert to avoid further mixing of the genetic background. The progeny were genotyped by either DNA-blot analysis or PCR. For DNA-blot analysis, an internal probe was used to detect the changes in size of an EcoRI fragment (from 6.0 to 2.0 kb). For the PCR analysis, three primers were used. The wild-type allele amplified a 250-bp fragment (primers a and b), whereas the null allele amplified a 410-bp fragment (primers a and c) (Fig. 1). PCR conditions were 32 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min.
Figure 1.
Generation of Slit3−/− mice. (a) Structure of the Slit3 genomic locus (Upper) and targeted allele (Lower). Exon 1 is represented by a black box. The location of β-gal (open bar), loxp-neo (gray bar), DNA-blot probes [5′ probe, internal probe (IP), and 3′ probe], and the lengths of expected restriction fragments are shown. X, XbaI; RV, EcoRV; RI, EcoRI; K, KpnI; B, BamHI. (b) DNA blot of genomic DNA from a targeted ES cell clone (+) and a wild-type ES cell clone (−) hybridized with 5′ and 3′ genomic probes after XbaI digestion. (c) DNA-blot genotyping using an internal probe after EcoRI digestion. (d) PCR genotyping. A 250-bp wild-type band is generated by using primers a and b, and a 410-bp mutant allele is detected by using primers a and c. (e) RNase protection assay of E13.5 wild-type (+/+) and homozygous (−/−) Slit3 embryo RNA. S, Slit3 probe; R, RPL32 probe. Note the 283-nt protected fragment in wild-type but not Slit3−/− mRNA. (f) Detection of β-gal activity in an E11.5 Slit3+/− embryo by whole-mount 5-bromo-4-chloro-3-indolyl β-d-galactoside staining.
Oligonucleotide Sequences.
The oligonucleotide sequences used were primer a (5′-GCG CCT CCT CGG GCT CCT CGT GTC -3′, sense), primer b (5′-TGC GGG GGA TGC CCC GAG GAA-3′, antisense), and primer c (5′-CGG ATT CTC CGT GGG AAC AAA CGG-3′, antisense) (Fig. 1a).
RNase Protection Assay.
A 283-bp NarI fragment from the Slit3 gene was used to synthesize the probe. This fragment included 171 bp of the 197-bp coding region of exon 1, 72 bp of exon 2, and the first 40 bp of exon 3. RPL32, a 197-bp ribosomal protein L32 fragment, was included in the experiment as a control. Assays were done according to manufacturer instructions (RNase protection kit, Roche Diagnostics). Tissue RNAs were prepared from embryonic day (E)13.5 Slit3−/− and wild-type embryos. Fifty micrograms of total RNA was incubated with labeled probe for >8 h at 45°C, digested with RNaseA and T1, and electrophoresed on a 6% denaturing polyacrylamide gel.
Immunohistochemistry and in situ hybridization were performed as described (32–34). MF-20 (anti-myosin) was obtained from Developmental Studies Hybridoma Bank (Iowa City), anti-neurofilament 200 was from Sigma, anti-laminin-γ1 was from Chemicon, anti-desmin was from DAKO, and α-bungarotoxin was from Molecular Probes.
Cell proliferation assays were carried out as described (35). All the BrdUrd-positive nuclei in sections through the anterior medial central tendon were counted. The area of the region counted was measured by using AXIOVISION 3.0 image software (Zeiss). The number of BrdUrd-positive nuclei per unit area was calculated for Slit3−/− and wild-type littermates. At least three sections were counted for each embryo examined. The calculation was based on three Slit3−/− embryos and two control embryos at E13.5.
Results and Discussion
Slit3 gene targeting eliminated 194 bp of the protein-coding region downstream of the initiation ATG in exon 1 and ≈800 bp of the first intron (Fig. 1a). This deletion eliminated the amino terminus of Slit3 up to the first leucine-rich repeat. The amino terminus of Slit3 is hypothesized to contain either a signal peptide for secretion or a signal sequence for mitochondrial localization (14, 36). The deletion of this region is predicted to result in a null mutation for Slit3. To follow the fate of cells that normally express Slit3, the β-gal gene was inserted in frame with the initiation ATG (Fig. 1a). Three independently targeted ES cell lines (Fig. 1b) were used to generate chimeric male mice, and all three passed the targeted allele to offspring. Mice heterozygous for the targeted allele (Slit3+/−) were phenotypically normal and bred to produce homozygous offspring. Genotyping by DNA-blot analysis and PCR identified viable homozygous mice (Fig. 1 c and d). Because the phenotype of mice derived from all three ES cell lines were similar, only one line was selected for further analysis. RNase protection analysis with a probe covering exons 1–3 did not detect alternative splicing around exon 1 in the wild-type allele and demonstrated that no 5′ Slit3 transcript was produced from the targeted allele (Fig. 1e). β-Gal staining of both whole embryos (Fig. 1f) and tissue sections, when compared with Slit3 expression patterns determined by in situ hybridization (14), showed similar expression patterns. Prominent β-gal staining was observed in the floor plate of the spinal cord, hippocampus, blood vessels in the lung and kidney, outer nuclear layer of the eye, exocrine pancreas, hair follicles, and the anterior region of the limb bud (Figs. 1f and 2g and data not shown).
Figure 2.
CDH and lung pathology in Slit3−/− mice. (a) E17.5 wild-type diaphragm viewed from the top showing the liver underneath. (b) E17.5 Slit3−/− diaphragm showing a CDH sac containing liver tissue (arrow). (c) Sagittal section of a CDH in a P14 Slit3−/− mouse. (d–f) Higher-magnification views of the boxed areas indicated in c. (g) Detection of β-gal in the lung of an E18.5 Slit3−/− embryo by 5-bromo-4-chloro-3-indolyl β-d-galactoside staining. (h) Lung section of a P14 Slit3−/− mouse with CDH. (i and j) Lung sections of a 5-week-old wild type (i) and Slit3−/− mouse showing pulmonary congestion in the Slit3−/− tissue (j).
Slit3+/− mice were maintained on a mixed C57B6/J/129/SVJ genetic background. Among 224 genotyped offspring from heterozygous matings, the ratio of wild type to heterozygote to homozygote was 59:116:51 (1.2:2.3:1), which approximated a normal Mendelian ratio. Although Slit3−/− mice were viable, the body weight of homozygous animals was consistently less than that of wild-type and heterozygous animals and became statistically significant (P < 0.05) after postnatal day (P)30 (data not shown). After P30 the morbidity and mortality of homozygous mice increased. Among homozygous animals that died before 9 months of age, >90% presented with a central (septum transversum) CDH in which the liver entered the thoracic cavity (Fig. 2 a–f). The cause of death appeared to be lung compression (Fig. 2j) and intestinal obstruction due to later-stage herniation of the transverse colon. Actuarial analysis showed that 24% of the Slit3−/− mice died by 40 days of age, and 58% died by 150 days of age (data not shown). The overall penetrance of the CDH phenotype was 68% and was higher in males (74%) than females (60%) with a ratio of 1.2:1.
Based on the midline central tendon location (Figs. 2b and 3a), the diaphragmatic hernia in Slit3−/− mice is similar to the central (septum transversum) CDH seen in humans. At early postnatal stages up to P14, the liver was the only contents of the hernia sac and was always adherent to the hernia sac (Fig. 2 c–e). The sac wall was composed of connective tissue (Fig. 2 d–f) and, in some areas, liver tissue (Fig. 2 d and e), indicating failure of the two cell types to separate during development. This pathology was similar to several cases reported in humans (37, 38). The herniated liver tissue often appeared vacuolated and in early stages of necrosis (Fig. 2 c–e). At later stages of herniation, other abdominal organs have been observed in the sac including intestine and gall bladder but never the stomach. The size and degree of herniation was variable and progressive with age.
Figure 3.
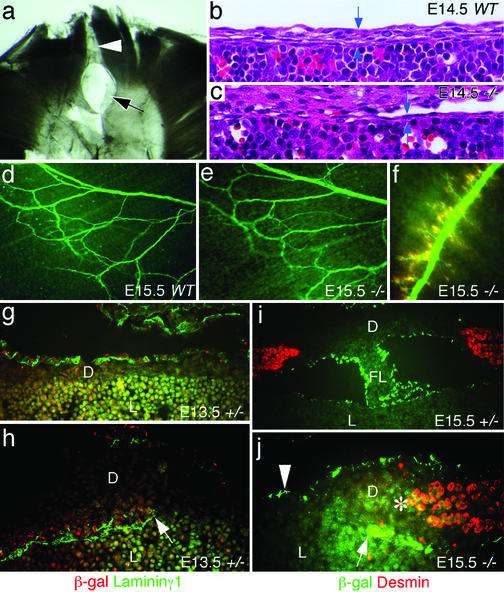
Analysis of the developing diaphragm. (a) Whole-mount immunohistochemistry of an E16.5 Slit3−/− diaphragm using the MF-20 (antimyosin) antibody. The arrow indicates a defect in the central tendon, and the arrowhead indicates a defect in diaphragm muscle. (b and c) Frontal sections of E14.5 wild-type and Slit3−/− embryos showing the thickness of the anterior central tendon (between arrows). (d and e) Whole-mount immunostaining of the phrenic nerve in E15.5 wild-type and Slit3−/− diaphragms using an anti-neurofilament 200 antibody. (f) Merged image showing the phrenic nerve and neuromuscular junction in an E15.5 Slit3−/− diaphragm visualized with anti-neurofilament 200 (green) and rhodamine α-bungarotoxin (red). (g) Pleural mesothelium in E13.5 Slit3+/− mice double-labeled with anti-laminin-γ1 (green) and anti-β-gal (red). (h) Migratory peritoneal mesothelial buds (arrow) in the lateral diaphragm of E13.5 Slit3+/− mice labeled with both anti-laminin-γ1 (for basal lamina, green) and anti-β-gal (red). (i) Developing falciform ligament (FL) in E15.5 Slit3+/− mice labeled with anti-β-gal (green) and anti-desmin (red). (j) Peritoneal mesothelial bud (arrow), defect region in the central tendon (arrow head) and tendon–muscle junction (asterisk) in an E15.5 Slit3−/− diaphragm labeled with anti-β-gal (green) and anti-desmin (red). D, diaphragm; L, liver.
Slit3 was also expressed in the developing lung (Fig. 2g). Because defective lung growth has been considered as a possible etiology of CDH in animals treated with Nitrofen and in humans, and Dutt1/Robo1 mutant mice also have a defect in lung development, the lungs of Slit3−/− mice were examined at different stages of development. No lung pathology was observed in Slit3−/− embryos or in newborn and young mice in the early stages of herniation (Fig. 2 g and h). In the 32% of Slit3−/− mice that do not develop CDH, the lung remained normal throughout life. In contrast, Slit3−/− mice with end-stage CDH were short of breath due to lung atelectasis and hemorrhage (Fig. 2j). These observations suggest that lung pathology is not the primary etiology of the CDH in Slit3−/− mice and that the development of the lung and diaphragm are independent of each other. This conclusion is further supported by the observation that the diaphragm develops normally in other mouse models with primary defects in lung development (39). The different lung phenotypes in Slit3−/− and Dutt1/Robo1−/− mice can be explained by the differential expression patterns of the two genes in the lung. Dutt1/Robo1 was expressed in the bronchial epithelium (31, 40), whereas Slit3 was expressed in blood vessel endothelium (Fig. 2g). Therefore, Dutt1/Robo1 may respond to ligands other than Slit3 for its function in lung development.
The phrenic nerve innervates the diaphragm and divides in specific branching patterns (41). Because Slit proteins function as neuronal guidance molecules we considered that Slit3 might regulate the innervation of the diaphragm. To determine whether defective innervation could contribute to CDH in Slit3−/− mice, the branching patterns and neuromuscular junction of the phrenic nerve were examined from E14.5 to E18.5. No differences were observed in the overall branching patterns, the density and length of subbranches, or the density and pattern of the diaphragmatic neuromuscular junction in Slit3−/− mice and wild-type controls (Fig. 3 d–f).
The observed defects in the anterior muscular part of the diaphragm of some Slit3−/− mice (Fig. 3a) suggested that a defect in myoblast migration or myotube formation could be a possible mechanism. However, frontal sections through E14.5 diaphragm showed that the anterior defect in the central tendon appeared before the formation of muscle bundles in this region (Fig. 3 b and c). Additionally, the region of the “tendon–muscle junction” appeared normal (Fig. 3j), which suggested that defects in diaphragm muscle are likely to be secondary to connective tissue dysgenesis of the central tendon.
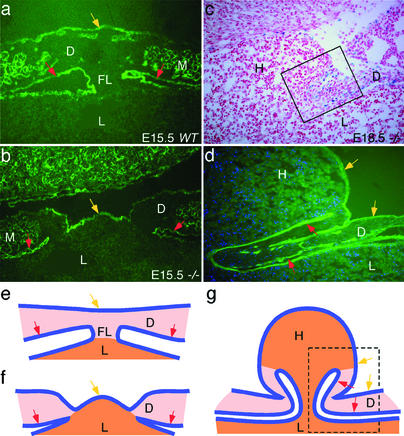
Anti-laminin-γ1 immunohistochemistry showed that the diaphragm at E13.5 consisted of a thin membrane attached directly to the liver, with only pleural mesothelium on top of several layers of mesenchymal cells (Fig. 3g). This part of the liver is the so-called bare area (Fig. 6a). The diaphragm and liver begin to separate due to extension of the peritoneal mesothelial folds (Figs. 3 h and i, 4 a and e, and 6). At E15.5, the mesothelial buds from both sides reach the midline but do not fuse. This results in the formation of a “bridge” of midline connective tissue, called the falciform ligament, attaching the diaphragm and liver (Figs. 3i and 4 a and e).
Figure 6.
Model showing the function of Slit3 in diaphragm development. (a) The peritoneal mesothelium and falciform ligament of the diaphragm arise from the central migration of mesothelial buds. (b) In Slit3−/− mice this process fails to occur, and a CDH is initiated with continuous growth of the liver into the thoracic cavity. D, diaphragm; L, liver; H, diaphragmatic hernia; PlM, pleural mesothelium; PeM, peritoneal mesothelium; FL, falciform ligament.
Figure 4.
Integrity of the pleural mesothelium in Slit3−/− diaphragm. (a and b) Developing central tendon and falciform ligament visualized with an antilaminin-γ1 antibody in an E15.5 wild-type and Slit3−/− embryo. (c and d) Continuity of the pleural mesothelium and the hernia sac. (c) E18.5 Slit3−/− diaphragm stained for β-gal activity. (d) Enlargement of the boxed area in c stained with an anti-laminin-γ1 antibody (green) and 4′,6-diamidino-2-phenylindole (blue). (e) Diagram showing normal development of the diaphragm and falciform ligament at E15.5. (f and g) Diagrams showing herniation in Slit3−/− embryos at E15.5 (f) and E18.5 (g). In g, the boxed area corresponds to the section shown in d. Yellow arrow, pleural mesothelium of the diaphragm or hernia sac; red arrow, peritoneal mesothelium of the diaphragm; orange, liver tissue; pink, connective tissue of the diaphragm; blue line, mesothelium; D, diaphragm; L, liver; H, diaphragmatic hernia; M, muscle; FL, falciform ligament.
Several possible mechanisms could lead to central tendon connective-tissue defects in Slit3−/− mice. First, if central tendon connective tissue fails to differentiate within the septum transversum, a congenital opening could occur, allowing liver tissue to enter the thoracic cavity. A second possibility is rupture due to a thinner or more fragile central tendon. A third possibility is stretching of the central tendon connective tissue. The observation of a hernia sac in all cases of Slit3−/− CDH supports the third possibility.
The pathogenesis of the central tendon first appeared as early as E14.5 where diaphragm mesenchyme was significantly thinner in Slit3−/− mice compared with wild-type control mice (Fig. 3 b and c). At E15.5, the peritoneal mesothelium of the diaphragm was well developed in normal animals such that the diaphragm was only attached to the liver by the developing falciform ligament (Figs. 3i and 4 a and e). In Slit3−/− mice, the central tendon remained adherent to the liver with little mesenchymal tissue and no falciform ligament (Figs. 3j and 4 b and f). The diaphragm also began to distend as the liver continued to grow (Fig. 4 b and f). However, the pleural mesothelium remained continuous, with no congenital opening or rupture (Fig. 4b). At E18.5, the mesothelium of the hernia sac remained continuous with the pleural mesothelium of the diaphragm (Fig. 4 c and d). Therefore, the CDH in Slit3−/− mice formed by stretching of abnormally thin diaphragm tissue, which failed to separate from an expanding liver (Fig. 4 f and g). The origin of the hernia sac appeared to arise from the adherent bare area of the diaphragm and liver (Fig. 4).
To establish a direct link between phenotype and gene expression, in situ hybridization and β-gal immunohistochemistry was used to localize sites of Slit and Robo expression in developing diaphragm. Slit3 was expressed predominantly in the mesothelial cells of the developing diaphragm from E12.5 to E18.5 (Fig. 5 a and b), which suggested a direct relationship between its expression and CDH. Interestingly, Slit2 was expressed in diaphragm mesenchymal cells (Fig. 5c), suggesting possible functional redundancy. Robo1 was also expressed in diaphragm mesenchyme, and Robo2 was expressed at the interface of the liver and diaphragm (Fig. 5 d and e). Slit1 and Robo3 were not detected in developing diaphragm at this stage.
Figure 5.
Expression and cell proliferation in E13 diaphragm tissue. (a) Immunohistochemical detection of β-gal in Slit3−/− diaphragm (frontal section). (b–e) Expression of Slit3, Slit2, Robo1, and Robo2, as indicated, in diaphragm by in situ hybridization. (f and g) Cell proliferation determined by anti-BrdUrd immunohistochemistry in the central tendon of wild-type and Slit3−/− mice. D, diaphragm; L, liver.
Two related processes, proliferation of mesenchymal cells and extension of peritoneal mesothelial buds, occur during the development of the central tendon. To investigate how Slit3 could function in the morphogenesis of the central tendon, cell proliferation was examined during early diaphragm development. At E13.5 there was a 22% decrease in the proliferation rate of mesenchymal cells in the central tendon region of Slit3−/− mice (P < 0.001; Fig. 5 f and g), which may account for the decreased thickness of this region. Cell proliferation can be stimulated by mitogens (e.g., hepatocyte growth factor/scatter factor and fibroblast growth factors) or by mechanical stress (42), and cells can respond to mechanical stress by altering proliferation, protein synthesis, or extracellular matrix structure. At early embryonic stages, the liver is one of the organs that grows rapidly. The mesenchymal and mesothelial cells in the diaphragm are predicted to be under continuous mechanical tension from the expanding liver due to adhesion of the central tendon to the liver. Slit3 is expected to be secreted into the extracellular matrix. Therefore, Slit–Robo interactions in the diaphragm could function to sense this mechanical stress. Interestingly, elastin synthesis was reduced at E13.5 (data not shown), suggesting decreased resiliency of the tissue at this stage of development. Increased apoptosis was not detected in the central tendon at E15.5 (data not shown).
The second critical event for central tendon development is the extension of mesothelial buds that separate liver from diaphragm (Fig. 3 h–j). In organ patterning, branching and bud extension is controlled by local mitogens and/or mechanical tension (42). Slit3 is expressed more intensely at the leading edge and in newly formed mesothelium (Fig. 3 i and j). Because Robo1 and Robo2 are expressed in adjacent cells (Fig. 5e), Slit-Robo interactions could be important for migration of the mesothelial leading edge. If extension of the leading edge is slower than normal, the region of the central tendon that is attached to the liver would remain under increased stress, which could result in the thinning of the central tendon (Fig. 6). Once the central tendon region becomes thinned, further migration of the mesothelial leading edge may be hindered, leading to failure of the falciform ligament to develop and the formation of a hernia defect (Fig. 6).
In vertebrates, molecules that are known to regulate axon guidance have also been found to function in morphogenesis. Class 3 semaphorins were found to inhibit branching morphogenesis in the lung, and KAL-1 was involved in epidermal morphogenesis in addition to olfactory axon guidance and neurite branching (43, 44). Our study indicates that Slit3 may also have functions in regulating cell proliferation, synthesis of extracellular matrix molecules, and/or migration of mesothelial cells. The CDH of Slit3−/− mice could also result from the synergistic outcome of multiple factors. Partial penetrance may result from compensation by other signaling molecules and from redundancy with Slit2.
A small percentage of newborn Dutt1/Robo1 mutant mice developed CDH. The phenotype and the mesenchymal expression of Robo1 in the central tendon (Fig. 5d) suggests a role for Slit-Robo signaling in diaphragm pathogenesis. The low penetrance of CDH in the Dutt1/Robo1−/− mice may arise from redundancy with Robo2 and complex ligand–receptor relationships between Slit and Robo. Double or triple knockouts of Slit and/or Robo genes will be important to address this possibility.
Our current knowledge of the signaling pathways downstream of Robo suggests that Slit and Robo interactions are important for cytoskeletal actin dynamics, which are important for axon guidance and cell migration (45, 46). However, it is still difficult to integrate these pathways with functions in morphogenesis of the developing diaphragm. In Drosophila, Abelson (Abl) is one of the molecules acting downstream of Robo and is critical for morphogenesis of epithelial cells (47). Further studies of signaling pathways downstream of Robo will be required to elucidate functional mechanisms regulating development.
Most cases of CDH are sporadic, but familial cases have been described and include autosomal dominant, autosomal recessive, and X-linked inheritance patterns (48). It has been reported that in familial cases males were more commonly affected, whereas in sporadic CDH females were more commonly affected (49). Interestingly, in the Slit3 genetic model the phenotypic penetrance was higher in males than in females (1.2:1).
The Slit3−/− mouse is the first genetic model with a similar phenotype to human central (septum transversum) CDH. The only other rodent model for CDH resembles the Bochdalek-type hernia and is generated by exposure of the embryos to the herbicide nitrofen. In the nitrofen model, the CDH is preceded by lung hypoplasia, which has led many to view the lung abnormality as the primary etiology of the diaphragm pathology. However, based on the Slit3 CDH model, lung pathology is not a prerequisite for a primary CDH to occur. This is consistent with some human cases in which symptoms appeared late in childhood and lung development was normal (50). The Slit3−/− model therefore should provide insight into the pathogenesis of some forms of CDH and to the devel-opment of the diaphragm. It will also provide a model to study the secondary effects of lung compression on lung development.
Acknowledgments
We thank Jeffrey Miner for advice, Xiang Hua for microinjection, Heather Walker for help with mouse husbandry, Joshua Sanes for anti-β-gal antibody, and Marc Tessier-Lavigne for the Robo2 in situ probe. This work was funded by National Institutes of Health Grant DC04289 and a generous contribution from the Virginia Friedhofer Charitable Trust.
Abbreviations
- CDH
congenital diaphragmatic hernia
- ES
embryonic stem
- β-gal
β-galactosidase
- En
embryonic day n
- Pn
postnatal day n
References
- 1.Laberg J-M, Sigalet D L, Guttman F M. In: Nyhus and Condon's Hernia. Fitzgibbons R J Jr, Greenburg A G, editors. Philadelphia: Lippincott; 1995. [Google Scholar]
- 2.Torfs C P, Curry C J, Bateson T F, Honore L H. Teratology. 1992;46:555–565. doi: 10.1002/tera.1420460605. [DOI] [PubMed] [Google Scholar]
- 3.Harrison M R, Adzick N S, Estes J M, Howell L J. J Am Med Assoc. 1994;271:382–384. [PubMed] [Google Scholar]
- 4.Nobuhara K K, Lund D P, Mitchell J, Kharasch V, Wilson J M. Clin Perinatol. 1996;23:873–887. [PubMed] [Google Scholar]
- 5.Stokes K B. Prog Pediatr Surg. 1991;27:127–147. doi: 10.1007/978-3-642-87767-4_8. [DOI] [PubMed] [Google Scholar]
- 6.Greer J J, Allan D W, Babiuk R P, Lemke R P. Pediatr Pulmonol. 2000;29:394–399. doi: 10.1002/(sici)1099-0496(200005)29:5<394::aid-ppul9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Guilbert T W, Gebb S A, Shannon J M. Am J Physiol. 2000;279:L1159–L1171. doi: 10.1152/ajplung.2000.279.6.L1159. [DOI] [PubMed] [Google Scholar]
- 8.Keijzer R, Liu J, Deimling J, Tibboel D, Post M. Am J Pathol. 2000;156:1299–1306. doi: 10.1016/S0002-9440(10)65000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen W J. Human Embryology. 3rd Ed. New York: Churchill Livingstone; 2001. [Google Scholar]
- 10.Kaufman M H, Bard J B L. The Anatomical Basis of Mouse Development. San Diego: Academic; 1999. [Google Scholar]
- 11. Babiuk, R. P., Zhang, W., Clugston, R., Allan, D. W. & Green, J. J. (2003) J. Comp. Neurol., in press. [DOI] [PubMed]
- 12.Itoh A, Miyabayashi T, Ohno M, Sakano S. Brain Res Mol Brain Res. 1998;62:175–186. doi: 10.1016/s0169-328x(98)00224-1. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama M, Nakajima D, Nagase T, Nomura N, Seki N, Ohara O. Genomics. 1998;51:27–34. doi: 10.1006/geno.1998.5341. [DOI] [PubMed] [Google Scholar]
- 14.Yuan W, Zhou L, Chen J, Wu J Y, Rao Y, Ornitz D M. Dev Biol. 1999;212:290–306. doi: 10.1006/dbio.1999.9371. [DOI] [PubMed] [Google Scholar]
- 15.Rothberg J M, Hartley D A, Walther Z, Artavanis-Tsakonas S. Cell. 1988;55:1047–1059. doi: 10.1016/0092-8674(88)90249-8. [DOI] [PubMed] [Google Scholar]
- 16.Rothberg J M, Jacobs J R, Goodman C S, Artavanis-Tsakonas S. Genes Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- 17.Kidd T, Bland K S, Goodman C S. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 18.Brose K, Bland K S, Wang K H, Arnott D, Henzel W, Goodman C S, Tessier-Lavigne M, Kidd T. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 19.Li H S, Chen J H, Wu W, Fagaly T, Zhou L J, Yuan W L, Dupuis S, Jiang Z H, Nash W, Gick C, et al. Cell. 1999;96:807–818. doi: 10.1016/s0092-8674(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 20.Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu J Y, Rao Y. Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer S G, Kidd T, Simpson J H, Goodman C S. Science. 2001;292:737–740. doi: 10.1126/science.1058766. [DOI] [PubMed] [Google Scholar]
- 22.Wu J Y, Feng L, Park H T, Havlioglu N, Wen L, Tang H, Bacon K B, Jiang Z, Zhang X, Rao Y. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K H, Brose K, Arnott D, Kidd T, Goodman C S, Henzel W, Tessier-Lavigne M. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 24.Plump A S, Erskine L, Sabatier C, Brose K, Epstein C J, Goodman C S, Mason C A, Tessier-Lavigne M. Neuron. 2002;33:219–232. doi: 10.1016/s0896-6273(01)00586-4. [DOI] [PubMed] [Google Scholar]
- 25.Bagri A, Marin O, Plump A S, Mak J, Pleasure S J, Rubenstein J L, Tessier-Lavigne M. Neuron. 2002;33:233–248. doi: 10.1016/s0896-6273(02)00561-5. [DOI] [PubMed] [Google Scholar]
- 26.Kidd T, Brose K, Mitchell K J, Fetter R D, Tessier-Lavigne M, Goodman C S, Tear G. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 27.Yuan S S, Cox L A, Dasika G K, Lee E Y. Dev Biol. 1999;207:62–75. doi: 10.1006/dbio.1998.9141. [DOI] [PubMed] [Google Scholar]
- 28.Huminiecki L, Gorn M, Suchting S, Poulsom R, Bicknell R. Genomics. 2002;79:547–552. doi: 10.1006/geno.2002.6745. [DOI] [PubMed] [Google Scholar]
- 29.Chen J H, Wen L, Dupuis S, Wu J Y, Rao Y. J Neurosci. 2001;21:1548–1556. doi: 10.1523/JNEUROSCI.21-05-01548.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battye R, Stevens A, Perry R L, Jacobs J R. J Neurosci. 2001;21:4290–4298. doi: 10.1523/JNEUROSCI.21-12-04290.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xian J, Clark K J, Fordham R, Pannell R, Rabbitts T H, Rabbitts P H. Proc Natl Acad Sci USA. 2001;98:15062–15066. doi: 10.1073/pnas.251407098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulson C L, Li C, Miner J H. Dev Dyn. 2001;222:101–114. doi: 10.1002/dvdy.1169. [DOI] [PubMed] [Google Scholar]
- 33.Burgess R W, Nguyen Q T, Son Y J, Lichtman J W, Sanes J R. Neuron. 1999;23:33–44. doi: 10.1016/s0896-6273(00)80751-5. [DOI] [PubMed] [Google Scholar]
- 34.Xu J S, Liu Z H, Ornitz D M. Development (Cambridge, UK) 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- 35.Naski M C, Colvin J S, Coffin J D, Ornitz D M. Development (Cambridge, UK) 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 36.Little M H, Wilkinson L, Brown D L, Piper M, Yamada T, Stow J L. Am J Physiol. 2001;281:C486–C495. doi: 10.1152/ajpcell.2001.281.2.C486. [DOI] [PubMed] [Google Scholar]
- 37.Wesselhoeft C W, Jr, DeLuca F G. Am J Surg. 1984;147:481–485. doi: 10.1016/0002-9610(84)90009-6. [DOI] [PubMed] [Google Scholar]
- 38.Chu D Y, Olson A L, Mishalany H G. J Pediatr Surg. 1986;21:897–899. doi: 10.1016/s0022-3468(86)80019-7. [DOI] [PubMed] [Google Scholar]
- 39.Babiuk R P, Greer J J. Am J Physiol. 2002;283:L1310–L1314. doi: 10.1152/ajplung.00257.2002. [DOI] [PubMed] [Google Scholar]
- 40.Clark K, Hammond E, Rabbitts P. FEBS Lett. 2002;523:12–16. doi: 10.1016/s0014-5793(02)02904-6. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski M B, Norton A S, Berger P K. Exp Neurol. 1991;113:212–220. doi: 10.1016/0014-4886(91)90177-e. [DOI] [PubMed] [Google Scholar]
- 42.Huang S, Ingber D E. Nat Cell Biol. 1999;1:E131–E138. doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- 43.Kagoshima M, Ito T. Genes Cells. 2001;6:559–571. doi: 10.1046/j.1365-2443.2001.00441.x. [DOI] [PubMed] [Google Scholar]
- 44.Rugarli E I, Di Schiavi E, Hilliard M A, Arbucci S, Ghezzi C, Facciolli A, Coppola G, Ballabio A, Bazzicalupo P. Development (Cambridge, UK) 2002;129:1283–1294. doi: 10.1242/dev.129.5.1283. [DOI] [PubMed] [Google Scholar]
- 45.Bashaw G J, Kidd T, Murray D, Pawson T, Goodman C S. Cell. 2000;101:703–715. doi: 10.1016/s0092-8674(00)80883-1. [DOI] [PubMed] [Google Scholar]
- 46.Wong K, Ren X R, Huang Y Z, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay S M, Mei L, et al. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 47.Grevengoed E E, Loureiro J J, Jesse T L, Peifer M. J Cell Biol. 2001;155:1185–1198. doi: 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enns G M, Cox V A, Goldstein R B, Gibbs D L, Harrison M R, Golabi M. Am J Med Genet. 1998;79:215–225. [PubMed] [Google Scholar]
- 49.Mishalany H, Gordo J. J Pediatr Surg. 1986;21:372–374. doi: 10.1016/s0022-3468(86)80208-1. [DOI] [PubMed] [Google Scholar]
- 50.Freyschuss U, Lannergren K, Frenckner B. Acta Paediatr Scand. 1984;73:589–593. doi: 10.1111/j.1651-2227.1984.tb09979.x. [DOI] [PubMed] [Google Scholar]