Abstract
Chromatin boundaries or insulators modulate enhancer–promoter interactions in complex genetic loci. However, the mechanism underlying insulator activity is not known. Previous studies showed that the activity of the Drosophila suHw insulator is abolished by the tandem arrangement (pairing) of the insulator elements, suggesting that interactions between insulators or like elements may be involved in their enhancer-blocking mechanism. To test whether such phenomenon reflects a general property of chromatin insulators, we tested the effect of pairing on enhancer-blocking activity of 11 homologous and heterologous insulator combinations using suHw, scs, or SF1 insulators. We found that, unlike the homologous pairing of suHw, the heterologous combinations of suHw with other insulators do not reduce their enhancer-blocking activity. Rather, paired insulators exhibit a higher level of enhancer-blocking activity than either single insulator alone, suggesting that they can function independently or additively. Furthermore, the analyses of two additional chromatin boundaries, scs and SF1, in homologous or heterologous pairing with other boundary elements, also showed no reduction but rather enhancement of insulator activity. We propose that diverse mechanisms may underlie insulator activity, and selective interactions among insulators could influence their function as well as the formations of independent chromatin domains.
Keywords: suHw‖scs‖Fab7‖SF1‖boundary element
Molecular genetic evidence suggests that eukaryotic chromatin is organized into regions or domains, which may separate neighboring genes physically and functionally (1–9). Specialized DNA elements called chromatin boundaries or insulators have been identified to delimit genome regions of distinct chromatin structure and gene activity (10–13). The unique features of chromatin insulators are their ability to block regulatory elements from a gene promoter when in an intervening position and to insulate transgenes from chromosomal position effects. Insulator DNAs have been found in vertebrate β-globin loci, Drosophila heat-shock and homeotic loci, and yeast silent subtelomeric and mating-type loci. Recent studies further showed that the regulated assembly of chromatin boundaries results in differential gene regulation (14, 15). Protein components including several zinc-finger DNA-binding proteins and broad-complex, tramtrack, bric à brac (BTB)-domain proteins are involved in insulator activities.
Several models have been proposed to account for the ability of insulators to block enhancers in a position-dependent fashion. One model proposes that insulators might disrupt enhancer function by propagating silent chromatin structures in a unidirectional fashion (16, 17). The second model postulates that insulators can “trap” the upstream enhancer by acting as a decoy promoter. This model is based largely on the observations that certain insulators contain promoter sequence motifs as well as promoter activities (18–21). The third model proposes that insulators can antagonize enhancer–promoter interactions by directly targeting the “facilitator proteins” that mediate such interactions. An example is the facilitator protein Chip, which mediates the interactions between the Drosophila cut gene and its distal enhancer and was shown to interact with insulator components SuHw and Mod(mdg4) proteins (22–26).
Recent studies of the Drosophila suHw insulator suggest that a previously unknown mechanism may underlie its enhancer-blocking activity. The suHw element, a 340-bp DNA from the gypsy retrotransposon, is one the best characterized insulator elements. It is a potent enhancer blocker and protects transgenes from chromosomal position effects (27, 28). However, the activity of suHw is sensitive to the cis arrangement of the insulator DNA; two tandem copies of the suHw element abolish its enhancer-blocking activity (insulator cancellation) (29, 30). Based on these observations, we proposed that insulator might interact with other insulators or anchor sites in the nucleus to isolate the enhancer and the promoter into different chromatin loop domains (29, 31). The paired suHw insulators, however, would preferentially interact with each other, thus neutralizing their ability to interact with other sites and to separate the enhancer from the promoter. Interestingly, MOD(mdg4), a protein component of the suHw insulator complex, contains a BTB domain known to mediate interactions among distantly located DNAs (16, 31–34). This looping model explains both the structural and functional characteristics of chromatin boundaries.
In this study we tested whether different insulators function through similar mechanisms. If so, what are the characteristics of insulators that determine their interactions (35, 36)? We examined the enhancer-blocking activities of other paired Drosophila insulators including suHw, scs, and SF1 in homologous and heterologous pairs. Eleven pairs of insulators including two homologous pairs (scs/scs and SF1b/SF1b) showed no cancellation of insulator activity. Instead, paired insulators exhibited a stronger enhancer-blocking activity than either the single-insulator component, indicating that they could function independently or in an additive fashion. Our results suggest that interactions between insulators or nuclear anchor sites may be selective. The configuration of chromatin domains could be influenced by the cis arrangement of insulators as well as the selective interactions among them.
Materials and Methods
P-Element Transformation, Whole-Mount in Situ Hybridization, and Visual Assessment of Reporter Gene Expression.
P element-mediated germ-line transformations, in situ hybridizations, and semiquantitative assessment of reporter gene expression were done as described (37, 38). Briefly, y1w67c23 and w1118 Drosophila strains were used to generate all transgenic lines reported. P element-mediated germ-line transformation was carried out as described (39). Three or more independent transgenic lines were characterized for each transgene. Two- to 5-h-old embryos were collected and fixed as described (29). Reporter gene expression was detected by using whole-mount in situ hybridization with digoxigenin-UTP-labeled antisense RNA probes and colorimetric reaction with antidigoxigenin antibody conjugated to alkaline phosphotase (Genius kit, Roche Molecular Biochemicals) (29, 40). It is important to note that all in situ stains were done under the same conditions and by using the same amount of reporter probes. For semiquantitative assessment of insulator activity, 30–200 transgenic embryos from multiple lines were inspected visually. The slides for experimental and control transgenes were mixed and scored anonymously in a large group. The extent of enhancer block was determined by the relative levels of reporter expression directed by the twist proximal element (PE) enhancer as compared with that by eve stripe 3 (E3). Each embryo was assigned to one of the three groups: weak (PE/E3 > 70%), moderate (PE/E3 = 70–30%), and strong (PE/E3 < 30%) block. The most frequently observed staining patterns were used to produce the images in the figures.
Construction of Transgenes.
All P-element constructs used in the embryo enhancer-blocking assays were derivatives of the pCaSPeR vector (39). With the exception of E3-matrix attachment activity (MAR)-PE all transgenes were derived from the pCA-EbP3 backbone plasmid, which contains two embryonic enhancers, PE and E3, between the divergently transcribed eve-lacZ and white reporter genes (ref. 41; also see diagrams in figures). Construction of the PL3, PS3, and PSS3 transgenes has been described (41). The 2.3-kb SF1 DNA was subcloned from a λ phage genomic clone that hybridized to probes from the Scr region (38). Its subfragment b (SF1b) was generated by PCR and cloned into pCRII/TOPO vector (Invitrogen). The 0.9-kb core scs, 1.2-kb core Fab7, and 1.2-kb ftz-MAR DNAs were provided by J. Vasquez (University of California, San Francisco), J. Zhou (Wistar Institute, Philadelphia), and L. Pick (Mount Sinai School of Medicine, New York), respectively (28). All single-insulator elements including suHw, scs, Fab7, SF1, SF1b, and MAR elements were cloned into pBluescript SK(+) vector (38, 41). For suHw double-insulator transgenes, the scs, Fab7, SF1, and MAR elements were inserted into an suHw-containing plasmid to generate SC-, SB-, FS-, and SM-containing subclones. For scs double-insulator transgenes, the scs fragment was inserted into scs-, Fab7-, SF1b-, and MAR-containing vectors to produce CC, CB, SF1bC, and MC subclones. For other SF1b double-insulator transgenes, the SF1b fragment was inserted into SF1b-, Fab7-, and MAR-containing vectors to produce SF1bSF1b, SF1bB, and SF1bM subclones. Single and double insulators were PCR-cloned into pCRII/TOPO vector by using a pair of custom T3 and T7 primers containing NotI ends. NotI fragments containing these insulators were inserted into the unique NotI site between the PE and E3 enhancers in pCA-EbP3 vectors (41). To make the E3-MAR-PE transgene, a 1.2-kb EcoRI fragment containing ftz-MAR was inserted in a pCA-Eb3P vector that contains the distal E3 and proximal PE enhancers between the eve-lacZ and the white reporters (see diagram in Fig. 3A). The positions and orientations of enhancers and insulators were determined by restriction digestions, PCR analyses using both vector and insulator-specific primers, and in some cases by DNA sequencing. The relative position of the elements are as indicated in the diagrams under the embryo photographs and in Table 1. The orientations of the elements and the sequence of the oligonucleotides used for the PCRs are available on request.
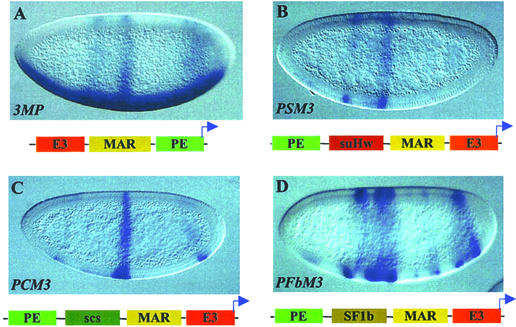
Figure 3.
Enhancer-blocking activity of insulators pair with the MAR element. (A) LacZ expression in a PM3 transgenic embryo, showing a composite pattern directed by three enhancers. (B–D) LacZ expression in transgenic embryos containing double insulators. Enhancement in the block of PE is seen with PSM3 (B) and PCM3 (C) but not in PSF1bM3 (D; also see Table 1).
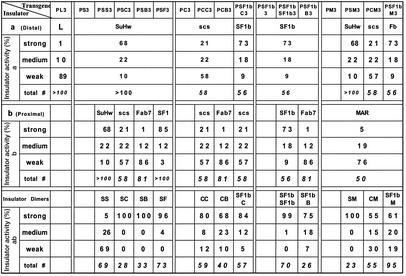
Table 1.
Semiquantitation of enhancer block by various insulator combinations
Results and Discussion
Insulator Activity of Paired SuHw and Heterologous Insulators.
We have shown previously that the enhancer-blocking activity of SuHw is abolished by the tandem arrangement of the insulator element. The observation suggests that the interactions between the two neighboring suHw elements, possibly mediated by the BTB domain of MOD(mdg4), may impede their ability to interact with other sites and abolish their enhancer-blocking activity (insulator cancellation). We wondered whether such interactions would occur between suHw and a heterologous insulator arranged in tandem. To answer this question, we tested enhancer-blocking activity of suHw paired with other insulators in transgenic Drosophila embryos.
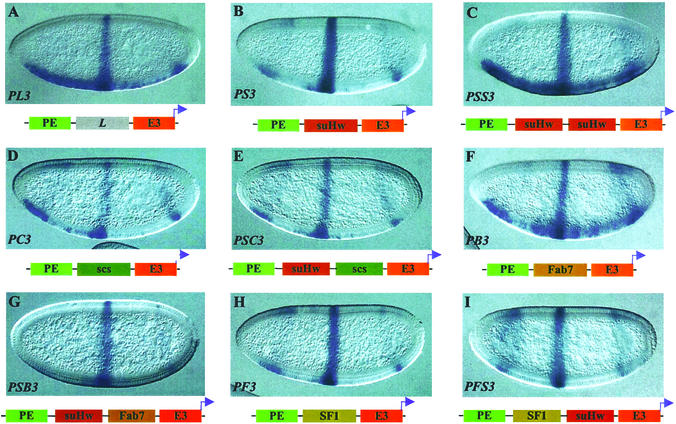
Two embryonic enhancers were included in the transgene constructs to direct tissue-specific expression of the lacZ reporter. At the blastoderm stage the PE enhancer is active in the ventral region of the embryos, and the E3 enhancer is active in a midembryo stripe. The two enhancers separated by a neutral DNA spacer (L) from the λ phage directed the expression of the eve-lacZ fusion gene in a composite pattern (PL3 in Fig. 1A and Table 1; also see Materials and Methods) (41). Insertion of a single suHw insulator reduced the reporter expression directed by the distal PE enhancer but not that by the proximal E3 enhancer, indicating the insulator activity (PS3 in Fig. 1B and Table 1). The embryos shown in all figures represent the most frequently observed type of pattern (see Table 1) and not necessarily the extent of enhancer block of the transgene (see Table 1 for semiquantitative assessment of insulator activity). As shown previously, insertion of a second copy of the suHw insulator in tandem resulted in a dramatic loss of the enhancer-blocking activity, seen in the strong lacZ expression in the ventral region directed by the distal PE enhancer (PSS3 in Fig. 1C and Table 1).
Figure 1.
Enhancer-blocking activity of suHw paired with heterologous insulators. Expression of the eve-lacZ fusion gene in blastoderm-stage transgenic embryos is visualized by whole-mount in situ hybridization (see Materials and Methods). Embryos are shown anterior to the left and dorsal up. Each transgene is diagrammed underneath the embryos. (A) LacZ reporter expression in a PL3 transgenic embryo showing comparable level of expression is directed by the distal PE and proximal E3 enhancers. (B) LacZ expression in PS3 transgenic embryos showing a reduction in the expression directed by the PE enhancer relative to that directed by E3, indicating insulator activity. (C) LacZ expression in PSS3 embryos. The intense ventral stain directed by PE indicates a loss of insulator activity. (D, F, and H) The lacZ expression in transgenic embryos containing single insulators between the PE and E3 enhancers showing reduction of in the PE-directed expression to different extents: PC3 (D), PB3 (F), and PF3 (H). (E, G, and I) Transgenic embryos containing double insulators between the PE and E3 enhancers: PSC3 (E), PSB3 (G), and PSF3 (I). Little or no PE-directed lacZ expression is seen in the ventral region of these embryos.
We first tested pairing between suHw and the Drosophila scs insulator, one of the authentic chromatin boundaries that flank the Drosophila hsp70 loci (42). The scs insulator has been shown to block various enhancers and protect transgenes from chromosomal position effects (28, 42). A zinc-finger protein, Zw5, binds to scs and is required for its insulator activity (43). The 0.9-kb core scs element exhibited enhancer-blocking activity in our assay, shown by the reduced level in PE-directed expression as compared with the spacer control (PC3 in Fig. 1D and Table 1). When suHw and scs were placed in tandem between PE and E3, the distal PE enhancer is blocked completely, showing no reduction in the insulator activity (PSC3, Fig. 1E and Table 1). The enhancer block by the suHw/scs pair was in fact significantly stronger than that of either insulator alone (see Table 1 for quantitative assessment). This result is very different from that observed with the paired suHw that resulted in loss of insulator activity.
The suHw element was next paired with Fab7 and SF1, two chromatin boundary elements from the Drosophila bithorax complex and Antennapedia complex, respectively (38, 44–46). Fab7 and SF1 both contain binding sites for GAGA factor, a BTB-domain protein, and depend on these sites for their enhancer-blocking activity (38). The 1.2-kb Fab7 element showed little enhancer-blocking activity as a single insulator, as seen by the obvious PE activity in the ventral region (PB3 in Fig. 1F and Table 1). However, the enhancer block is augmented greatly by the addition of an suHw element next to Fab7, shown by the complete block of the PE by the suHw/Fab7 insulator pair, as seen above with the suHw/scs transgene (PSB3 in Fig. 1G and Table 1). The 2.1-kb SF1 element, an insulator from the Scr-ftz region of the Antennapedia complex, exhibited potent enhancer-blocking activity when assayed alone (PF3 in Fig. 1H and Table 1) (38), yet its insulator activity is enhanced further by the addition of an suHw element in tandem (PSF3 in Fig. 1I and Table 1). Thus, unlike the homologous paring of suHw, pairing of suHw with Fab7 or SF1 resulted in no loss but an increase of enhancer-blocking activity. This difference is unlikely to be explained by the change in insulator sizes, because PSC3 and PSB3 transgenes contain inserts of comparable or smaller size than PL3.
The results described above suggest that interactions similar to that between two suHw insulators may not occur between suHw and a heterologous insulator or may not dominate and exclude the interaction required for insulator function. Furthermore, our results showed that such noninteracting insulator pairs exhibit augmented activity than either element alone, suggesting that both insulators can function independently in an additive or parallel fashion (see below).
Insulator Activity of Insulator Pairs Containing scs.
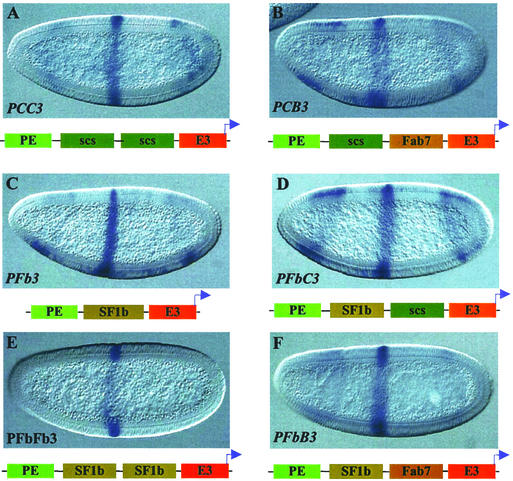
The results discussed above raise the possibility that loss of insulator activity could occur only with homologous pairing of insulator elements. Alternatively, SuHw may represent a unique type of boundary activity that is more prone to insulator cancellation. To distinguish between these possibilities, we examined scs, a native chromatin boundary from the Drosophila heat-shock loci, in both homologous and heterologous pairing combinations. As shown above, the 0.9-kb core element of scs can block the distal PE enhancer (PC3 in Fig. 1D and Table 1). When we inserted two tandem copies of the scs element between the test enhancers, the paired insulators showed a much stronger block of PE (PCC3 in Fig. 2A and Table 1). This result indicates that homologous pairing of boundary elements does not necessarily lead to insulator cancellation.
Figure 2.
Enhancer-blocking activity of scs and SF1b insulators paired with homologous and heterologous insulators. (A, B, and D–F) LacZ expression in transgenic embryos containing double insulators between the PE and E3 enhancers. Enhancement in the block of the PE enhancer is seen with PCC3 (A), PCB3 (B), PSF1bC3 (D), PSF1bSF1b3 (E), and PSF1bB3 (F). (C) LacZ expression in a PSF1b3 transgenic embryo.
We next tested heterologous combinations of scs with Fab7 or SF1 elements. In transgenic embryos the scs/Fab7 dimer blocks the distal PE enhancer more effectively than either scs or Fab7 alone (PCB3 in Fig. 2B and Table 1). For the test of the scs/SF1 combination, we used SF1b, a subfragment SF1, which was shown to contain much of the activity of SF1 (38). SF1b strongly blocks the distal PE enhancer (PSF1b3 in Fig. 2C and Table 1) in our enhancer-blocking assay. When paired with the 0.9-kb scs element, the scs/SF1b insulator dimer again enhanced the block of PE as compared with either single insulator (PCSF1b3 in Fig. 2D and Table 1).
The results discussed above showed that, unlike SuHw, the activity of the 0.9-kb scs element was enhanced when combined in tandem with a second insulator, either homologous or heterologous. Our results suggest that SuHw could possess unique characteristics that render it more prone to insulator cancellation (see below).
Insulator Activity of SF1b Insulator Pairs.
To analyze the characteristics that may contribute to the unique property of the suHw insulator, we also considered the transacting factors. The insulator function of suHw requires MOD(MDG4), a BTB-domain protein shown to mediate protein–protein interactions (32). We wonder whether insulators involving certain transacting factors such as the BTB-domain proteins are particularly sensitive to homologous pairing. To this end, we tested insulator combinations using SF1b, which contains the critical sites for GAGA factor, a BTB-domain protein shown to tether distant DNA together through protein-mediated interactions (33). As shown before, SF1b monomer can block the distal PE enhancer effectively (PSF1b3 in Fig. 2C and Table 1). Transgenic embryos containing the SF1b/SF1b homodimer showed a complete block of the PE enhancer activity (PSF1bSF1b3 in Fig. 2E and Table 1). This result represents a further increase of the insulator activity from that of the strong SF1b monomer. Next we tested a combination of SF1b with Fab7, both from the Drosophila Hox clusters, and both depend on GAGA-binding sites for insulator activity. The SF1b/Fab7 insulator dimer also augmented the enhancer-blocking activity as compared with either insulator alone (PSF1bB3 in Fig. 2F and Table 1). These results indicate that the tendency for insulator cancellation is not simply correlated with the presence of certain protein factors or domains that mediate protein–protein interactions. Our results are also consistent with the previous findings that showed that two copies of the Fab7 core element exhibited a stronger enhancer-blocking activity than the single element (45, 46).
Insulator Activity of Insulator Pairs Containing an MAR/Scaffold Attachment Activity (SAR) Element.
Another characteristic unique to the suHw insulator is the presence of MAR/SAR within the insulator element (36). MAR/SAR elements are regions of DNA proposed to be involved in attachment of chromatin fibers to nuclear matrix. The MAR/SAR activity is defined biochemically: these DNA elements preferentially partition or associate with the insoluble nuclear pellet (matrix) after certain nuclear extractions and nuclease digestions (47, 48). The proposed function of MAR/SAR elements in chromatin attachment is reminiscent of the proposed role of insulators in chromatin looping, and certain MAR/SAR members have been shown to exhibit aspects of boundary function (49). Therefore we tested whether MAR/SAR elements interact with selected insulator elements including suHw. We first tested the combination of suHw with the ftz-MAR, an MAR element near the Drosophila ftz gene (50). The 1.2-kb DNA that harbors the MAR activity also contains an ftz distal enhancer. In transgenic embryos, the 1.2-kb ftz-MAR placed between E3 and PE showed no detectable insulator activity, as seen in the composite lacZ pattern derived from all three elements in the transgene: the ventral expression directed by the distal E3, the midembryo stripe by the proximal PE, and the partial pair-rule expression of the early pattern directed by the ftz-MAR enhancer (E3-MAR-PE in Fig. 3A and Table 1). When the suHw insulator is added in tandem to the MAR element, the distal PE enhancer is blocked completely (PSM3 in Fig. 3B and Table 1). Interestingly, the extent of the block by the suHw-MAR pair is greater than that of suHw or MAR alone despite the little activity shown by the MAR element alone (see Table 1). Similarly, the ftz-MAR element also enhanced the insulator activity of scs when placed with it in tandem (PMC3 in Fig. 3C and Table 1). Curiously, the activity of the ftz enhancer is very weak in most of the PMC3 embryos, possibly because of interaction among the multiple enhancers. Finally, the ftz-MAR element was tested with the SF1b boundary element, both native from the Scr-ftz region in the Antennapedia complex, for possible interactions (PSF1bM3 in Fig. 3C and Table 1). Surprisingly, there appeared be a slight loss of insulator activity in the MAR/SF1b pair when compared with SF1b alone (Fig. 3D and Table 1). However, lacZ expression in the ventral region of these embryos is often limited in bands that coincide with the ftz stripes. It is likely that the proximal ftz-MAR enhancer synergies with the partially blocked PE activity, giving the appearance of a weaker block.
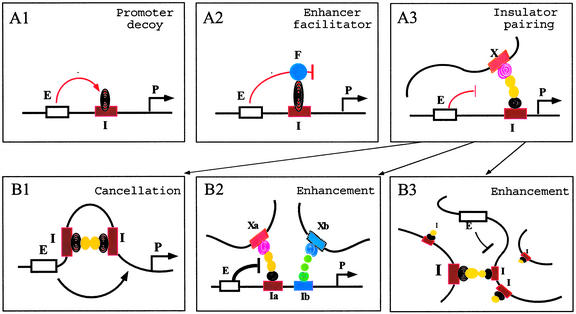
We have investigated whether pairing-mediated cancellation of insulator activity, seen with suHw, reflects a common property of other Drosophila insulators. All combinations of insulator pairs that we tested showed no cancellation but rather an enhancement in insulator activity. Rather, the enhanced insulator activity of the paired insulators suggests that the two insulators could function in parallel or additively. These results raised the possibility that although the suHw insulator may depend on insulator interaction for enhancer-blocking activity, other insulators could function through different mechanisms depending on their unique DNA and protein components. Furthermore, insulator interactions may not be required for enhancer-blocking activity, although such interaction between paired suHw interferes with their normal insulator function. For example, insulator components such as the SuHw, Mod(mdg4), and GAGA proteins contain protein–protein interaction domains that are essential to their insulator activity. The native targets of these proteins could be chromatin-remodeling complexes (directional silencing model), enhancer-binding proteins (Fig. 4A1, promoter decoy model), facilitator proteins (Fig. 4A2, enhancer-facilitator model), or insulator-binding proteins (Fig. 4A3, insulator-pairing model). The SuHw and Mod(mdg4) proteins recruited to the tandem suHw insulators may compete against their native targets and disrupt the insulator function. Alternatively, the insulator cancellation by the suHw pair could be due to the unique ability of the SuHw protein to induce change in DNA topology, which may increase the flexibility of the local chromatin and therefore augment enhancer–promoter interactions (51).
Figure 4.
(A) Models for insulator mechanism. A single intervening insulator between an enhancer and a promoter may block the enhancer by interacting with enhancer-binding proteins as proposed in promoter decoy model (A1), with enhancer facilitator proteins as proposed by enhancer-facilitator model (A2), or with insulators/nuclear sites as proposed by the insulator-pairing model (A3). (B) Selective insulator interaction determines outcome of insulator pairing. According to the insulator-paring model, the block of the distal enhancer may be (i) abolished when two tandem insulators interact with each other, neutralizing their ability to interact with the external sites (B1), or (ii) enhanced when the two neighboring insulators interact independently with the external sites. An insulator may select one among multiple available sites depending on insulator type (B2), or insulator strength, which would preclude interactions between neighboring insulators of even same type in the presence of a stronger competitor (B3). E, enhancers; P, promoters; F, facilitators; I, Ia; Ib, insulators; X, Xa, and Xb, external sites with which insulators interact.
In the context of the insulator-pairing model, our results suggest that paired insulators do not interact obligatorily with each other, even those sharing common characteristics even complete homology. Rather, insulator interaction could be selective. For example, tandem pairing of insulators may lead to cancellation or enhancement of insulator activity depending on selective insulator interactions: paired insulators may interact with each other (Fig. 4B1), or they may each interact with external sites, enhancing their activity (Fig. 4 B2 and B3). The selection of interacting insulators could be influenced by (i) qualitative differences among insulators such as different transacting factors that prevent them from recognizing each other (Fig. 4B2) or (ii) the quantitative differences such as their ability to interact with other insulators or nuclear sites, i.e., their insulator strength (28, 29, 52). Multiple insulators within the local genomic or nuclear environment may compete with each other for the strongest interaction, which may or may not be the nearest (Fig. 4B3). Therefore, the configuration of chromatin domains is influenced by both the cis arrangement of insulators as well as the selective interactions among them.
Acknowledgments
We thank Vladimir Belozerov and Ping Shen for suggestions for the manuscript. We also thank J. Vasquez, J. Zhou, and L. Pick for providing various insulator DNA elements. This work was supported by the National Institutes of Health.
Abbreviations
- BTB
broad-complex, tramtrack, bric à brac
- PE
twist proximal element
- E3
eve stripe 3
- MAR
matrix attachment activity
- SAR
scaffold attachment activity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mihaly J, Hogga I, Barges S, Galloni M, Mishra R K, Hagstrom K, Muller M, Schedl P, Sipos L, Gausz J, et al. Cell Mol Life Sci. 1998;54:60–70. doi: 10.1007/s000180050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillon N, Grosveld F. Curr Opin Genet Dev. 1994;4:260–264. doi: 10.1016/s0959-437x(05)80053-x. [DOI] [PubMed] [Google Scholar]
- 3.Dillon N, Sabbattini P. BioEssays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Grunstein M. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 5.Jackson D A. BioEssays. 1995;17:587–591. doi: 10.1002/bies.950170704. [DOI] [PubMed] [Google Scholar]
- 6.Bode J, Schlake T, Rios-Ramirez M, Mielke C, Stengert M, Kay V, Klehr-Wirth D. Int Rev Cytol. 1995;162A:389–454. doi: 10.1016/s0074-7696(08)61235-8. [DOI] [PubMed] [Google Scholar]
- 7.Pirrotta V, Rastelli L. BioEssays. 1994;16:549–556. doi: 10.1002/bies.950160808. [DOI] [PubMed] [Google Scholar]
- 8.Tartof K D. BioEssays. 1994;16:713–714. doi: 10.1002/bies.950161004. [DOI] [PubMed] [Google Scholar]
- 9.Razin S V, Vassetzky Y S. Cell Biol Int Rep. 1992;16:697–708. doi: 10.1016/s0309-1651(05)80014-1. [DOI] [PubMed] [Google Scholar]
- 10.Labrador M, Corces V G. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 11.Gerasimova T I, Corces V G. Annu Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 12.West A G, Gaszner M, Felsenfeld G. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 13.Kellum R, Elgin S C. Curr Biol. 1998;8:R521–R524. doi: 10.1016/s0960-9822(07)00337-5. [DOI] [PubMed] [Google Scholar]
- 14.Bell A C, Felsenfeld G. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 15.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 16.Gerasimova T I, Gdula D A, Gerasimov D V, Simonova O, Corces V G. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 17.Bi X, Braunstein M, Shei G J, Broach J R. Proc Natl Acad Sci USA. 1999;96:11934–11939. doi: 10.1073/pnas.96.21.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parnell T J, Geyer P K. EMBO J. 2000;19:5864–5874. doi: 10.1093/emboj/19.21.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourel G, Boscheron C, Revardel E, Lebrun E, Hu Y F, Simmen K C, Muller K, Li R, Mermod N, Gilson E. EMBO Rep. 2001;2:124–132. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith P A, Corces V G. Genetics. 1995;139:215–228. doi: 10.1093/genetics/139.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogga I, Karch F. Development (Cambridge, UK) 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- 22.Gause M, Morcillo P, Dorsett D. Mol Cell Biol. 2001;21:4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morcillo P, Rosen C, Baylies M K, Dorsett D. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorsett D. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 25.Rollins R A, Morcillo P, Dorsett D. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torigoi E, Bennani-Baiti I M, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Proc Natl Acad Sci USA. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roseman R R, Pirrotta V, Geyer P K. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai H, Levine M. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 29.Cai H N, Zhang Z, Adams J R, Shen P. Development (Cambridge, UK) 2001;128:4339–4347. doi: 10.1242/dev.128.21.4339. [DOI] [PubMed] [Google Scholar]
- 30.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 31.Gerasimova T I, Byrd K, Corces V G. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh D, Gerasimova T I, Corces V G. EMBO J. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmoudi T, Katsani K R, Verrijzer C P. EMBO J. 2002;21:1775–1781. doi: 10.1093/emboj/21.7.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida C, Tokumasu F, Hohmura K I, Bungert J, Hayashi N, Nagasawa T, Engel J D, Yamamoto M, Takeyasu K, Igarashi K. Genes Cells. 1999;4:643–655. doi: 10.1046/j.1365-2443.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 35.Cremer T, Kreth G, Koester H, Fink R H, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Crit Rev Eukaryotic Gene Expression. 2000;10:179–212. [PubMed] [Google Scholar]
- 36.Nabirochkin S, Ossokina M, Heidmann T. J Biol Chem. 1998;273:2473–2479. doi: 10.1074/jbc.273.4.2473. [DOI] [PubMed] [Google Scholar]
- 37.Cai H N, Shen P. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 38. Belozerov, V. E., Majumder, P., Shen, P. & Cai, H. N. (2003) EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 39.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 40.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 41.Cai H N, Levine M. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kellum R, Schedl P. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 43.Gaszner M, Vazquez J, Schedl P. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galloni M, Gyurkovics H, Schedl P, Karch F. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagstrom K, Muller M, Schedl P. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 46.Zhou J, Barolo S, Szymanski P, Levine M. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- 47.Laemmli U K, Kas E, Poljak L, Adachi Y. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 48.Hart C M, Laemmli U K. Curr Opin Genet Dev. 1998;8:519–525. doi: 10.1016/s0959-437x(98)80005-1. [DOI] [PubMed] [Google Scholar]
- 49.Namciu S J, Blochlinger K B, Fournier R E. Mol Cell Biol. 1998;18:2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasser S M, Laemmli U K. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 51.Shen B, Kim J, Dorsett D. Mol Cell Biol. 1994;14:5645–5652. doi: 10.1128/mcb.14.9.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott K C, Taubman A D, Geyer P K. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]