Abstract
East African cichlid fishes represent one of the most striking examples of rapid and convergent evolutionary radiation among vertebrates. Models of ecological speciation would suggest that functional divergence in feeding morphology has contributed to the origin and maintenance of cichlid species diversity. However, definitive evidence for the action of natural selection has been missing. Here we use quantitative genetics to identify regions of the cichlid genome responsible for functionally important shape differences in the oral jaw apparatus. The consistent direction of effects for individual quantitative trait loci suggest that cichlid jaws and teeth evolved in response to strong, divergent selection. Moreover, several chromosomal regions contain a disproportionate number of quantitative trait loci, indicating a prominent role for pleiotropy or genetic linkage in the divergence of this character complex. Of particular interest are genomic intervals with concerted effects on both the length and height of the lower jaw. Coordinated changes in this area of the oral jaw apparatus are predicted to have direct consequences for the speed and strength of jaw movement. Taken together, our results imply that the rapid and replicative nature of cichlid trophic evolution is the result of directional selection on chromosomal packages that encode functionally linked aspects of the craniofacial skeleton.
Each of the three large lakes in East Africa (Tanganyika, Malawi, and Victoria) houses an endemic assemblage of several hundred cichlid species that have evolved in the last several thousand to a few million years (1, 2). The phylogenetic history of the Lake Malawi flock suggests a pattern of radiation in three stages (3, 4). The first step was the occupation of different macrohabitats (e.g., rocky versus sandy substrates). This was followed by a radiation in feeding morphology that produced most of the currently recognized genera. Finally, certain lineages have diversified in male breeding colors, presumably under the action of sexual selection. We focus on the second stage of the radiation: functional divergence of head and jaw morphology.
Cichlid fishes forage by one of three general modes (biting, sucking, and ram feeding), which can be predicted by the functional design of the feeding apparatus (5, 6). Ram feeders are characterized by a long, streamlined head optimized for pursuit and overtaking of prey. Both sucking and biting species have a short, cone-shaped head, but differ at discrete anatomical points on the upper and lower jaw (6). Further specialization of jaws and teeth is correlated with occupation of specific foraging niches (i.e., biting fins or biting algae).
Because remarkably similar feeding morphologies have evolved convergently in different lakes (7) (Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org), a role for ecological selection in the radiation of jaw shape has been widely assumed, but never documented (8–10). Recent theoretical modeling suggests that disruptive selection on ecological traits can lead to speciation, even in sympatry, if selection can increase the association of ecological traits and markers controlling assortative mating (11, 12). These quantitative genetic models demonstrate that competition for resources is sufficient for sympatric speciation, and predict a history of directional selection on traits associated with ecological divergence. We hypothesized that if natural selection has driven the radiation of cichlid jaw shape, then the genes that underlie the shape of the oral jaw apparatus (OJA) will carry the signature of directional selection.
We used quantitative genetics to characterize the genetic architecture of the cichlid head, and uncovered a pattern of quantitative trait locus (QTL) effects consistent with a history of divergent selection on traits associated with the OJA. Moreover, we found that QTL for different skeletal elements are clustered in the genome, implicating pleiotropy as part of the underlying genetic structure of this character complex. We suggest that consistent selection on genetically correlated traits may partially explain the pattern of rapid and parallel evolution that characterizes East African cichlids.
Materials and Methods
Experimental Cross.
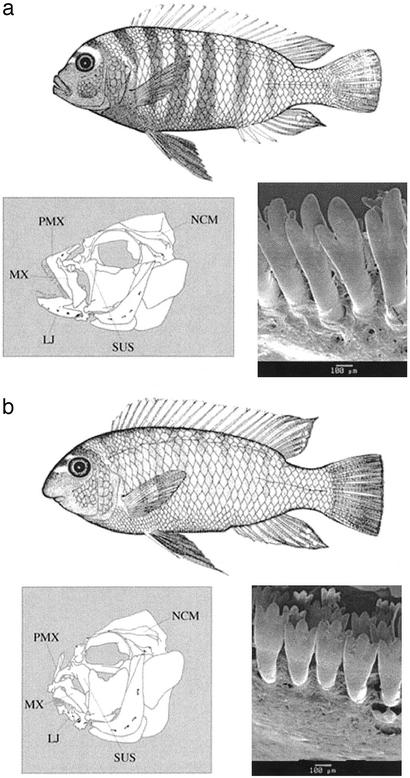
We crossed two closely related Lake Malawi species with distinct feeding morphologies and behaviors (13). Labeotropheus fuelleborni (“LF”) has a robust subterminal mouth that it uses to crop attached algae with a specialized biting mode of feeding. Metriaclima zebra (“MZ”) has a relatively narrow, terminally oriented mouth and is one of a few rock-dwelling species that forages on plankton in the water column with a suction mode of feeding (Fig. 1). LF and MZ coexist at nearly every rocky locality examined in Lake Malawi (14), and partition their environment according to diet, water depth, and other microhabitat characteristics (10, 13, 14).
Figure 1.
Morphological differences between LF and MZ. (a) LF is optimally designed for a biting mode of feeding with a short, robust, U-shaped oral jaw apparatus, an inferior subterminal mouth, and an outer row of closely spaced tricuspid teeth on both the upper and lower jaws. (b) In contrast, MZ is designed for a suction mode of feeding with a long, narrower lower jaw, a terminally oriented mouth, and an outer row of widely spaced bicuspid teeth on both jaws. LJ, lower jaw; MX, maxilla; NCM, neurocranium; PMX, premaxilla; SUS, suspensory apparatus.
Mapping Population.
Male LF were placed in a 500-gallon pool with female MZ. Females of all Lake Malawi rock-dwelling cichlids incubate their clutch in their mouths. Brooding female MZ were removed from the breeding pool and placed in 10-gallon tanks until they released free-swimming fry (≈3 weeks). Two LF sires and two MZ dams were used to produce two F1 families, which were grown to sexual maturity. Fourteen F2 families were generated via brother–sister matings of the F1. All F2 were reared in 40-gallon tanks for 3 months, and in 500-gallon pools for an additional 9–12 months. To minimize the functional demand on the feeding apparatus, F2 animals were fed high-quality spirulina flake food (Aquatic Ecosystems, Apopka, FL). A total of 173 F2 were used in this experiment.
Phenotypic Assay.
F2 families were collected between 12 and 15 months of age, and killed with MS222 in accordance with a protocol approved by the University of New Hampshire ACUC. Specimens were prepared for morphometric analysis with dermestid beetles, which cleaned and disarticulated the skeleton. Craniofacial skeletal elements were digitized by using a SPOT digital camera (Diagnostic Instruments, Burlingame, CA) mounted on a Zeiss SV11 dissecting scope. Images were imported into nih image (version 2.1), and landmark coordinates were captured in two-dimensional (x, y) space.
Shape was quantified via geometric morphometrics and traditional linear measures. Our geometric methods were performed in MORPHOMETRIKA 7.0 (15), and are described in detail elsewhere (16, 17). In brief, landmark coordinates were superimposed by using a generalized least-squares fit algorithm to adjust for size, translation, and rotation. Orthogonal geometric descriptors of shape difference, called partial warps, were generated via thin-plate spline analysis. A principal component analysis was performed on partial warp scores for LF and MZ. F1 and F2 animals were evaluated against the axis that distinguished LF and MZ (PC1 in all instances) by multiplying hybrid partial warp scores by the parental eigenvectors in the space of partial warps. The adjusted PC score for hybrid animals was the phenotype mapped in the QTL analysis. Linear distances between superimposed landmarks were used to assess differences in four functionally significant characters: (i) length of the lower jaw in the lateral view, (ii) width of the lower jaw in the ventral view, (iii) length of the ascending arm of the articular, and (iv) length of the palatinad wing of the maxilla.
Tooth shape was also evaluated in the F2. Morphology of the teeth in the outer row of the upper and lower jaws is a continuum between the fully tricuspid dentition of LF and the fully bicuspid dentition of MZ. Differences in tooth shape were assessed by evaluating the height of the third cusp in 28 teeth per individual (seven teeth on either side of the mandibular and premaxillary symphysis). Tooth shape ranged from bicuspid (a score of 2) to tricuspid (a score of 3). Teeth were assigned a score of 2, 2.25, 2.5, 2.75, or 3, depending on cuspidness. The mean tooth score was calculated separately for the upper and lower jaws but was highly correlated within individuals (17). In every instance upper jaw and lower jaw tooth shape mapped to the same intervals.
Linkage Analysis.
A MZ genomic DNA library enriched for microsatellites was constructed following methods published elsewhere (18). A total of 480 clones were sequenced, and primer pairs flanking 208 microsatellites with 10 or more dinucleotide repeats were designed. We also had >500 microsatellite markers available from a linkage map for the tilapia, Oreochromis niloticus (B.-Y. Lee, W.-J. Lee, J.T.S., K. L. Carleton, G. Hulata, A. Slettan, Y. Terai, and T.D.K., unpublished data). We tested 248 of these in the LF × MZ cross. Ultimately, the F2 were typed for 137 codominant microsatellite or gene-derived restriction fragment length polymorphism markers (91 MZ-derived, 46 tilapia-derived). Microsatellite genotypes were obtained with an ABI 377 (Applied Biosystems) automated sequencer, which determined the size of fluorescently labeled alleles amplified by PCR. We used 20-μl PCRs containing 0.5 μM of each primer, 20 ng of DNA, 0.8 mM dNTPs, 2 mM MgCl2, and 1 unit of Taq polymerase. Primer pairs were multiplexed up to three ways whenever product size and the color of the fluorescent label allowed. When multiplexing, primer concentrations were 0.25 μM. Cycling conditions were 94°C for 2 min; 30–35 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 30 s; 72°C for 5 min. For genotypic analysis, a 1-μl PCR was added to 0.24 μl of GeneScan 500 TAMRA size standard (PE Applied Biosystems) and 2.0 μl of formamide loading buffer. The solution was denatured and loaded on a 4% acrylamide gel. ABI genescan software (version 3.1.2) was used to analyze genotypes.
A linkage map was constructed by using JOINMAP 3.0 (19). The locus file consisted of genotypes for 173 F2 hybrid progeny at 137 microsatellites. One hundred twenty-nine microsatellites (95%) were informative in both F2 families. The grouping module of joinmap assigned 127 of 137 marker loci to 24 linkage groups by using a logarithm of odds (LOD) score threshold of 4.0. The mapping module of joinmap built the genetic map for each linkage group by using the Kosambi mapping function, a LOD threshold of 1.0, a recombination threshold of 0.450, and a jump threshold of 5.0. A ripple function was performed after each locus was added to ensure optimal marker order.
Cloning and Mapping of bmp4.
A pair of primers (F primer: 5′-CCTGGTAATCGAATGCTGATG-3′, R primer: 5′-CCACAATCCAGTCRTTCCAGC-3′) was used to amplify a ≈900-bp fragment from cDNAs derived from LF embryos (14 days after fertilization). Resulting amplicons were assayed for expected fragment size by agarose gel electrophoresis, cloned, sequenced, and verified by blast. Gene-specific primers were designed, and SNPs distinguishing Lf and Mz were identified by comparative sequence analysis and mapped by restriction fragment length polymorphism. Sequences have been deposited in GenBank as accession numbers AF534534 and AF534535.
QTL Analysis.
The genome was scanned for markers segregating with putative QTL by using interval mapping in MAPQTL 4.0 (20) and linear regression (spss version 10.0 for Macintosh). Marker cofactors that accounted for a significant amount of the phenotypic variance were verified by using backward elimination multiple regression of phenotype on cofactors. Markers remaining after backward elimination were used as cofactors in multiple QTL mapping (MQM) to eliminate the variation induced by unlinked QTL and more accurately assess the effects of individual QTL (21).
Significant LOD thresholds were determined for each trait by a permutation test. Here, phenotypic data were shuffled over individuals and marker data remained fixed. The maximum LOD score over the genome was calculated for each iteration, and the LOD frequency distribution was generated under the null hypothesis of no QTL. Genome-wide significance levels of α = 0.05 and 0.01 were identified for each trait from 1,000 permutations of the data and corresponded to LOD scores ranging from 2.60 to 10.54 (average = 4.05).
QTL Sign Test.
The selective history of the OJA was investigated by examining the direction of QTL effects (22, 23). The neutral expectation is that populations diverging via drift will accumulate alleles of both positive and negative effect, whereas populations evolving under directional selection will accumulate alleles with consistent effects. Formalized as the QTL sign test, this approach compares the number of antagonistic QTL within a species with those expected under neutral conditions (22). Because at least six QTL need to be detected for a given trait before the null model of neutral divergence can be rejected, we grouped traits according to whether they were components of the OJA (23). QTL effects for the OJA, head, and suspensorium were fit to a gamma distribution. We measured the phenotypic difference between species in environmental standard deviation units (SD of the F1), and we used the heterozygous effect of the smallest QTL detected as our QTL effect threshold. Trait categories with QTL effects that significantly deviate from neutrality implicate directional selection as a major contributor to phenotypic divergence.
Results and Discussion
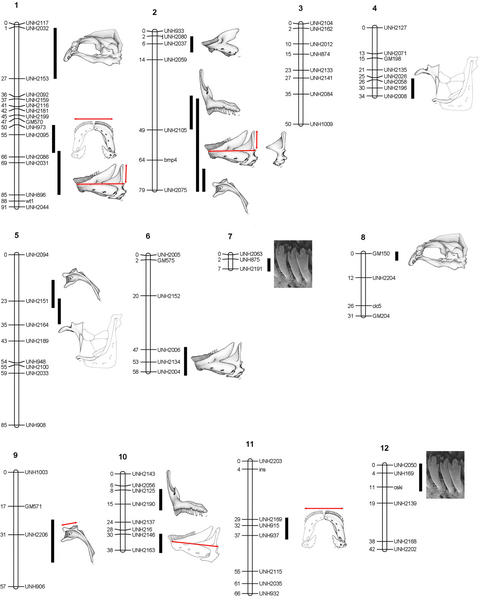
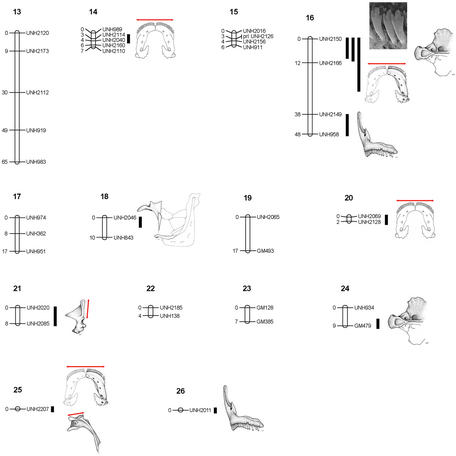
We scored 137 genetic markers in 173 F2 progeny from the LF × MZ cross and constructed a linkage map spanning 845 cM over 24 linkage groups. The average spacing between markers was 6.6 centimorgans, and >92% of the markers were linked. We identified 46 significant QTL that explained between 6.8 and 52.2% (average = 31.4%) of the phenotypic variance in each trait. QTL were detected on 17 of 24 linkage groups (Fig. 2 and Table 2, which is published as supporting information on the PNAS web site). In addition, five traits segregated with unlinked markers UNH2207 and UNH2011.
Figure 2.
QTL position on the genetic linkage map. Marker names are to the right of linkage groups, map positions (in centimorgans) are to the left. The map locations of QTL affecting feeding morphology are identified by illustrations of the corresponding structure. Individual bony elements indicate QTL affecting geometric variables. Red bars indicate QTL affecting linear measures. Where geometric and linear variables map to the same interval, red bars overlay stippled illustrations. When only linear measurements map to an interval, bars are shown with line drawings. Black bars indicate the regions exceeding the 95% genome-wide significance threshold for the corresponding QTL.
Our data offer definitive evidence that directional selection has shaped the cichlid OJA. The ratio of positive and negative QTL effects for the OJA significantly deviated from the neutral expectation (P < 1 × 10−6), consistent with the hypothesis that natural selection has facilitated trophic specialization (22, 23). In contrast, the ratio of QTL that affect the suspensorium and skull (e.g., non-OJA QTL) did not deviate from neutrality (P = 0.498) (Table 1 and Fig. 5, which is published as supporting information on the PNAS web site). Although additional research may show these traits are also under directional selection, our results are consistent with the effects of other forces, such as drift or stabilizing selection.
Table 1.
Tests for directional selection
| n | ncon | QTL ratio | P(ncon) | |
|---|---|---|---|---|
| Total | 46 | 42 | 0.913 | <1 × 10−6* |
| OJA | 39 | 37 | 0.949 | <1 × 10−6* |
| Head/suspensorium | 7 | 5 | 0.714 | 0.498N/S |
The QTL ratio describes the proportion of QTL acting in the expected direction for different categories of traits. n, total number of QTL. ncon, number of QTL with consistent effects. P(ncon), probability of finding the number of consistent QTL by chance alone. N/S, no significant deviation from neutral conditions.
Significant deviation from neutrality at P < 0.0001, corrected for multiple tests by sequential Bonferroni.
QTL were significantly more clustered in the genome than expected under a Poisson distribution (χ2 = 50, df = 5, P < 0.0001), implicating pleiotropy, or the cumulative action of several closely linked genes, as a prominent feature in the genetic architecture of the cichlid OJA. We identified eight intervals that contained two or more overlapping QTL (Fig. 2). For example, QTL for tooth, jaw, and skull shape mapped to the same interval on linkage group 16, and shape differences for both the upper and lower jaws mapped to the same interval on linkage group 2. Selection on these genomic “hot-spots” will propagate change to several morphological characters, leading to rapid evolution of trophic types. This clustering of QTL in the genome likely imposes constraints on evolution, leading to the covariation of traits and the recurrence of particular phenotypes (7, 24). Strong divergent selection on these chromosomal intervals may help explain why rapid, parallel bursts of evolution have occurred in these fishes.
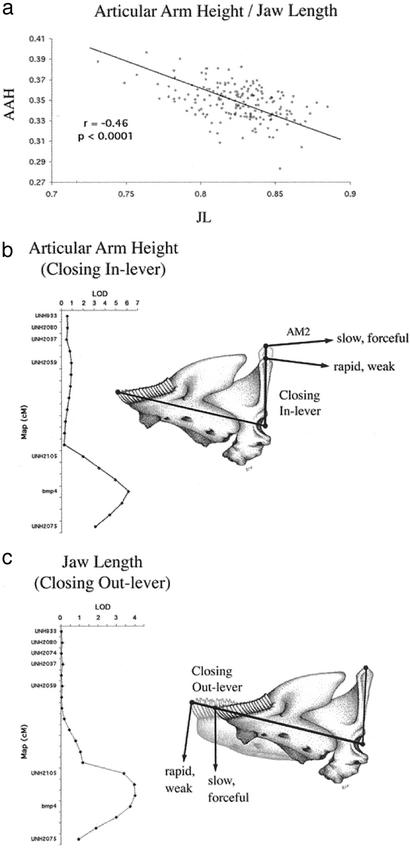
We also identified intervals with a concerted effect on the function of the lower jaw. In the action of biting the articular process (on which the primary jaw closing muscle, the adductor mandibulae, inserts) acts as the in-lever, whereas the length of the lower jaw acts as the out-lever. The ratio between the height of the ascending arm of the articular and the length of the lower jaw will affect the speed and force of jaw closing (25–27). A long articular process will increase force transmission of the adductor mandibulae resulting in a stronger bite. In contrast, a short process will increase the speed of jaw rotation and is more typical in species that employ suction feeding (6). We observed a negative correlation between jaw length and articular height in the F2 (Fig. 3a), which is largely explained by a few QTL with pleiotropic effects on the two structures. QTL for articular arm and lower jaw length are coincident on LG2 (Fig. 3 b and c). F2 individuals that are homozygous for LF alleles in this region have a relatively long ascending arm of the articular and a short lower jaw, whereas those that are homozygous for MZ alleles have a short articular process and a long lower jaw (Table 2, which is published as supporting information on the PNAS web site). A similar pair of coincident QTL is found on LG1. Together, these two intervals explain ≈28% of the phenotypic variance in the length of the articular arm, and ≈21% of the variance in the length of the lower jaw. Selection on these regions will simultaneously affect both the in- and out-levers, altering the mechanical advantage of the lower jaw for the classically described biting, sucking, and ram-feeding behaviors (5, 6).
Figure 3.
The intersection of genetic architecture and functional biology in the lower jaw. (a) Jaw length (LJ) is negatively correlated with articular arm height (AAH) in the F2. As the length of the jaw increases, the height of the articular arm decreases, and vice versa. The major muscle involved in jaw closing, the adductor mandibulae (AM2), inserts onto the articular arm. Thus, in the lexicon of functional morphology, the articular arm is the closing in-lever, and the length of the lower jaw is the closing out-lever. Altering the in- and out-lever ratio will affect the mechanical advantage of the lower jaw for the alternate food-gathering strategies of biting or sucking. QTL for the articular arm (b) and jaw length (c) map to the same interval on LG2. This interval also contains bmp4, a gene known to be involved in embryonic cartilage formation.
It is notable that QTL affecting the shape of the lower jaw on LG2 segregate with variation at a gene known to regulate morphogenesis of embryonic cartilage (28). Bone morphogenic protein 4 (bmp4) exhibits restricted patterns of mRNA expression in the vertebrate mandible (29), has the capacity to induce ectopic cartilage formation (30), and interestingly, is characterized by unusually high rates of evolution among East African cichlids (31). These observations suggest that bmp4 may play an important role in coordinating shape differences in the cichlid OJA.
The modest size of our F2 mapping population limits our power to detect QTL of minor effect, and leads to overestimates of the variance explained by individual QTL (32, 33). Although our relative QTL magnitudes are most likely accurate (34), the absolute values will be upwardly biased because of sampling error (33). Although these sample size effects prevent us from fully quantifying the genetic architecture of the cichlid head, they do not affect our main conclusions: that consistent directional selection has shaped the cichlid OJA, and that QTL for different skeletal elements are clustered in the genome.
Our results bolster the idea that adaptive evolution is facilitated by divergent natural selection acting on genomic regions that control multiple functionally related phenotypes (34–37). They are also consistent with theoretical and empirical evidence that genetic correlations will evolve between functionally and developmentally related traits (37–40). Pleiotropy among functionally linked characters may partly explain the rapid and parallel diversification of East African cichlids. Studies to identify the genes underlying these traits, and their relationship to genes controlling mate preference, will further elucidate the mechanisms generating biological diversity.
Supplementary Material
Acknowledgments
We thank J. Bolker, K. Carleton, J. Hanken, I. Kornfield, J. Webb, M. Zelditch, and two anonymous reviewers for critical reading of earlier versions of this manuscript. Access to dermestid beetles was provided by M. Scott. We thank Janet Conroy, Woo-Jai Lee, and Karen Carleton, who isolated microsatellite loci (UNH169–UNH1009) from tilapia DNA. Woo-Jai Lee kindly provided primers for Genomar markers. This work was supported by grants from the National Science Foundation, the U.S. Department of Agriculture (to T.D.K.), the Alfred P. Sloan Foundation, and the National Institutes of Health (to J.T.S).
Abbreviations
- OJA
oral jaw apparatus
- LF
Labeotropheus fuelleborni
- MZ
Metriaclima zebra
- QTL
quantitative trait locus
Footnotes
References
- 1.Fryer G, Iles T D. The Cichlid Fishes of the Great Lakes of Africa: Their Biology and Evolution. Edinburgh: Oliver and Boyd; 1972. [Google Scholar]
- 2.Meyer A, Kocher T D, Basasibwaki P, Wilson A C. Nature. 1990;347:550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 3.Albertson R C, Markert J A, Danley P D, Kocher T D. Proc Natl Acad Sci USA. 1999;96:5107–5110. doi: 10.1073/pnas.96.9.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danley P D, Kocher T D. Mol Ecol. 2001;10:1075–1086. doi: 10.1046/j.1365-294x.2001.01283.x. [DOI] [PubMed] [Google Scholar]
- 5.Liem K F. In: Cichlid Fishes: Behavior, Ecology and Evolution. Keenleyside M H A, editor. London: Chapman and Hall; 1991. pp. 129–150. [Google Scholar]
- 6.Otten E. Neth J Zool. 1983;33:55–98. [Google Scholar]
- 7.Kocher T D, Conroy J A, McKaye K R, Stauffer J R. Mol Phys Evol. 1993;2:158–165. doi: 10.1006/mpev.1993.1016. [DOI] [PubMed] [Google Scholar]
- 8.Liem K F. Am Zool. 1980;20:25–31. [Google Scholar]
- 9.Bouton N, Seehausen O, van Alpen J J M. Ecol Fresh Fish. 1997;6:225–240. [Google Scholar]
- 10.Genner M J, Turner G F, Hawkins S J. Oecologia. 1999;121:283–292. doi: 10.1007/s004420050930. [DOI] [PubMed] [Google Scholar]
- 11.Kondrashov A S, Kondrashov F A. Nature. 1999;400:351–354. doi: 10.1038/22514. [DOI] [PubMed] [Google Scholar]
- 12.Dieckmann U, Doebeli M. Nature. 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- 13.Ribbink A J, Marsh A C, Ribbink C C, Sharp B J. S Afr J Zool. 1983;18:149–310. [Google Scholar]
- 14.Reinthal P N. Environ Biol Fish. 1990;2:215–233. [Google Scholar]
- 15. Walker J. (2000) MORPHOMETRIKA: Geometric Morphometric Software for the Power Macintosh, www.usm.maine.edu/∼walker/software.html.
- 16.Albertson R C, Kocher T D. J Exp Zool. 2001;289:385–403. doi: 10.1002/jez.1020. [DOI] [PubMed] [Google Scholar]
- 17.Albertson R C. Ph.D. thesis. Durham: Univ. of New Hampshire; 2002. [Google Scholar]
- 18.Carleton K L, Streelman J T, Lee B-Y, Garnhart N G, Kidd M, Kocher T D. Anim Genet. 2002;33:140–144. doi: 10.1046/j.1365-2052.2002.00817.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Ooijen J W, Voorrips R E. JOINMAP 3.0: Software for the Calculation of Genetic Linkage Maps. Wageningen, The Netherlands: Plant Research International; 2001. [Google Scholar]
- 20.Van Ooijen J W, Boer M P, Jansen R C, Maliepaard C. QTLMAP 4.0: Software for the Calculation of QTL Positions on Genetic Maps. Wageningen, The Netherlands: Plant Research International; 2002. [Google Scholar]
- 21.Jansen R C. Genetics. 1994;138:871–881. doi: 10.1093/genetics/138.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr H A. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieseberg L H, Widmer A, Arntz A M, Burke J M. Proc Natl Acad Sci USA. 2002;99:12242–12245. doi: 10.1073/pnas.192360899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruber L, Adams D C. J Evol Biol. 2001;14:325–332. [Google Scholar]
- 25.Barel C D N. Neth J Zool. 1983;33:357–424. [Google Scholar]
- 26.Westneat M W. Zoomorphology. 1994;114:103–118. [Google Scholar]
- 27.Ferry-Graham L A, Lauder G V. J Morph. 2001;248:99–119. doi: 10.1002/jmor.1023. [DOI] [PubMed] [Google Scholar]
- 28.Monsoro-Burq A H, Duprez D, Watanabe Y, Bontoux M, Vincent C, Brickell P, Le Douarin N. Development (Cambridge, UK) 1996;122:3607–3616. doi: 10.1242/dev.122.11.3607. [DOI] [PubMed] [Google Scholar]
- 29.Semba I, Nonaka K, Takahashi I, Takahashi K, Dashner R, Shum L, Nuckolls G H, Slavkin H C. Dev Dyn. 2001;217:401–414. doi: 10.1002/(SICI)1097-0177(200004)217:4<401::AID-DVDY7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka K, Shum L, Takahashi I, Takahashi K, Ikura T, R, Dashner R, Nuckolls G H, Slavkin H C. Int J Dev Biol. 1999;43:795–807. [PubMed] [Google Scholar]
- 31.Terai Y, Morikawa N, Okada N. Mol Biol Evol. 2002;19:1628–1632. doi: 10.1093/oxfordjournals.molbev.a004225. [DOI] [PubMed] [Google Scholar]
- 32.Beavis W D. In: Proceedings of the Corn and Sorghum Industry Research Conference. Wilkinson D B, editor. Washington, DC: American Seed Trade Association; 1994. pp. 250–266. [Google Scholar]
- 33.Beavis W D. In: Molecular Dissection of Complex Traits. Paterson A H, editor. Boca Raton, FL: CRC Press; 1998. pp. 145–162. [Google Scholar]
- 34.Bradshaw H D, Otto K G, Frewen B E, McKay J K, Schemske D W. Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peichel C L, Nereng K S, Ohgi K A, Cole B L E, Colosimo P F, Buerkley C A, Schluter D, Kingsley D M. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- 36.Hawthorne D J, Via S. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- 37.Cheverud J M, Routman E J, Irschik D J. Evolution (Lawrence, Kans) 1997;51:2006–2016. doi: 10.1111/j.1558-5646.1997.tb05122.x. [DOI] [PubMed] [Google Scholar]
- 38.Riedl R. Order in Living Organisms. New York: Wiley; 1979. [Google Scholar]
- 39.Lande R. Evolution (Lawrence, Kans) 1979;33:402–416. doi: 10.1111/j.1558-5646.1979.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 40.Cheverud J M. Am Nat. 1995;145:63–89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.