Abstract
The mammalian amelogenin (AMEL) genes are found on both the X and Y chromosomes (gametologous). Comparison of the genomic AMEL sequences in five primates and three other mammals reveals that the 5′ portion of the gametologous AMEL loci began to differentiate in the common ancestor of extant mammals, whereas the 3′ portion differentiated independently within species of different mammals. The boundary is marked by a transposon insertion in intron 2 and is shared by all species examined. In addition, 540-kb DNA sequences from the short arm of the human X chromosome are aligned with their Y gametologous sequences. The pattern and extent of sequence differences in the 5′ portion of the AMEL loci extend to a proximal region that contains the ZFX locus, and those in the 3′ portion extend all the way down to the pseudoautosomal boundary (PAB)1. We concluded that the AMEL locus spans an ancient PAB, and that both the ancient and present PABs were determined by chance events during the evolution of mammals and primates. Sex chromosome differentiation likely took place in a region that contains the male-determining loci by suppressing homologous recombination.
Keywords: chromosomal rearrangement‖evolutionary strata‖recombination suppression
Lahn and Page (1) have proposed that there are four distinct evolutionary strata on the human X chromosome, and that differentiation of the X from the Y chromosome was initiated one stratum at a time. This hypothesis is based on the observation that the average extent of the sequence divergences at synonymous sites between X and Y homologous, or more precisely gametologous, loci is ≈10% in stratum 4 in contrast to 30% in stratum 3, 50% in stratum 2, and 100% in stratum 1. Stratum 4 spans ≈20 megabases on the short arm region of the X chromosome and is bounded by the amelogenin (AMEL) locus and pseudoautosomal boundary (PAB)1. Among seven loci examined in stratum 4 (1), AMEL has been more extensively studied in animals other than humans (2–7). Notably, primate intron 3 sequences suggest that AMEL on the X chromosome (AMELX) began to differentiate from that on the Y chromosome (AMELY) before the split of Old World and New World monkeys (4). On the other hand, cDNA or amino acid sequences analysis of gametologous AMELs shows greater relatedness within a species than among different mammalian species (5).
Recently, Iwase et al. (8) compared human BAC clones that encompass the AMELX and AMELY loci. They found that, although the region downstream from intron 2 exhibits ≈10% sequence differences per site, the upstream region exhibits a high level that is similar to stratum 3 (> 20%; ≈30% if multiple-hit substitutions are taken into account). This finding does not contradict previous results (4, 5), because AMEL exons 1 and 2 almost exclusively encode the 5′ untranslated region and are excluded from comparisons of intron 3 or amino acid sequences. Therefore, Iwase et al. (8) pointed out that the boundary between strata 3 and 4 on the human X chromosome lies in AMEL intron 2. Their preliminary study of genomic sequences of cattle AMELs also suggested the same boundary position, but the initiation timing of stratum 4 formation may differ greatly between humans and cattle (8).
On the other hand, Ellis et al. (9) investigated 440-bp sex chromosomal regions that span the PAB1 in humans, three great apes, and two Old World monkeys. They found an Alu insertion in the middle of the hominoid Y chromosomal region as well as an abrupt change in sequence differences between the distal and proximal parts of the Alu. Depending on species, the extent of sequence differences is 21.2–26.7% per site in the proximal 240 bp of Alu and 0.8–5.6% in the distal 200 bp of Alu. In humans, sequence differences in these regions are considerably higher compared with 10% in stratum 4 (1, 8) and 0.1% in the pseudoautosomal region (PAR)1. The high sequence similarity in PAR1 is thought to be due to the high rate of obligatory recombination (9).
The purpose of this paper is 2-fold. First, we examine whether the boundary between strata 3 and 4 occurs in AMEL intron 2 of non-human primates and other mammals. To this end, we determine the genomic sequences of AMELs in chimpanzees, squirrel monkeys, greater bushbabies, and ring-tailed lemurs in addition to those of cattle, pigs, and horses. To confirm the location of AMEL loci on sex chromosomes of non-human primates, we map the locations by fluorescence in situ hybridization (FISH). Second, we examine the pattern and extent of sequence differences between the human X and Y chromosomes and ask whether any characteristic sequence motifs are shared by the PAB1 and the boundary between strata 3 and 4. To this end, we compare 540-kb X and Y sequences from the Human Genome Project.
Materials and Methods
Sources and Isolation of DNA Samples.
Genomic DNA from males of humans (Homo sapiens), chimpanzees (Pan troglodytes), squirrel monkeys (Saimiri sciureus), greater bushbabies (Otolemurs crassicaudatus), and ring-tailed lemurs (Lemur catta) were provided by the sources in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Genomic DNA of cattle (Bos taurus), pigs (Sus scrofa), and horses (Equus caballus) were isolated from tissue or blood (Table 1) with the genomic DNA Purification Kit (Qiagen, Chatsworth, CA).
PCR Amplification and Sequencing.
Genomic PCR was performed in 20-μl reactions containing 20 pmol of each PCR primer, 100 ng of genomic DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 200 μM dNTPs, and 2.5 units of ExTaq DNA polymerase (Takara Shuzo, Kyoto). Amplifications were carried out in a RoboCycler Gradient 96 (Stratagene) under the following standard conditions: denaturation at 95°C for 3 min; 30 cycles of 95°C for 30 sec, 56–62°C for 30 sec, and 72°C for 6 min; and an additional extension at 72°C for 10 min. These conditions were slightly modified for some PCR primer sets. In greater bushbabies, ring-tailed lemurs, and pigs, the genome-walking method and “step-down” PCR-based techniques (10) were used. Genome-walking libraries were constructed with Universal Genome Walker Kits according to the manufacturer's instructions (CLONTECH). The PCR primer sequences are given in Tables 2–4, which are published as supporting information on the PNAS web site.
All PCR products were purified through QIAquick PCR Purification Kits (Qiagen). Although some products were directly sequenced, others were cloned by using TOPO TA Cloning Kits (Invitrogen). Sequencing reactions were performed with ABI BigDye Terminator Kits and analyzed on an ABI 377 DNA sequencer (Applied Biosystems). To minimize sequencing errors, PCR products or plasmid DNA were read twice in both directions. These sequences were further confirmed by obtaining templates from independent PCR reactions. These sequence segments were assembled by dnasis (Hitachi, Tokyo) and deposited in DNA Data Base in Japan (see Table 1 for accession nos.).
Data Analysis.
Sequence alignments were made by dotter (11) and clustalw (12) and then manually adjusted. For phylogenetic analysis, we used the neighbor-joining (NJ) (13) and maximum parsimony (MP) methods (14, 15) in MEGA2 (16). The NJ tree was based on p distance (the number of nucleotide differences per site), and reliability was assessed by bootstrap values with 1,000 replications.
Chromosomal Locations of AMELs.
To map the AMEL loci in chimpanzees, squirrel monkeys, and ring-tailed lemurs, we carried out FISH by using the species-specific AMELX and AMELY sequences. Slides of human (as a control) and chimpanzee chromosomes were prepared with phytohemagglutinin-stimulated peripheral white blood cells, and those of squirrel monkeys and ring-tailed lemurs were made with fibroblast cells (17). After aging the slides for a few days at 37°C, FISH was conducted by using 200-ng plasmid clones of AMELs as probes. These probes were labeled with a BioNick Labeling Kit containing biotin-14-dATP (GIBCO/BRL) (17). Fluorescent signals were amplified by using the “sandwich” technique (18). The FISH images were saved on a computer (Mac 8500/120) by using iplab imaging software (Signal Analytics) through a charge-coupled device camera (Photometrics, Fairfax, VA) attached to an epifluorescent microscope (Zeiss).
Results
Primate AMELs Mapped by FISH.
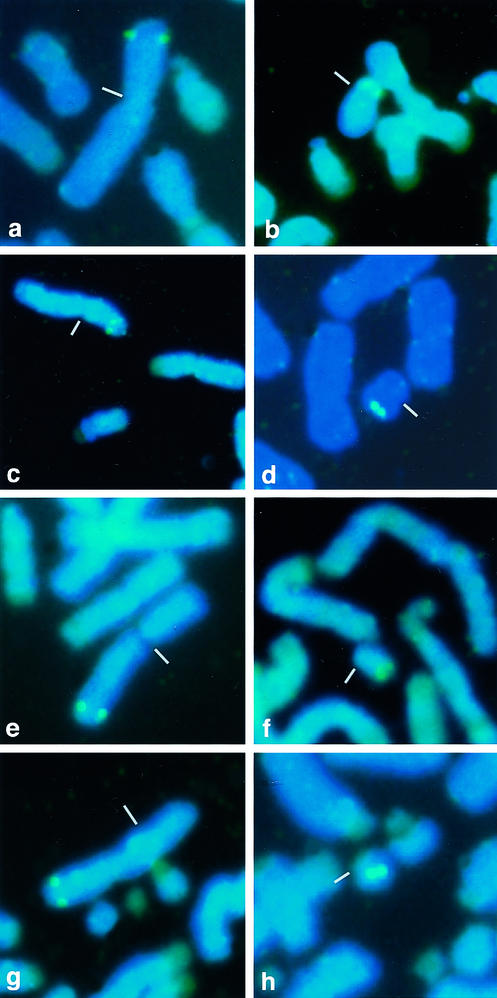
Together with 4,6-diaminido-2-phenylidole (DAPI) staining for chromosome identification, FISH demonstrated that the AMELX and AMELY genes in all chimpanzees, squirrel monkeys, and ring-tailed lemurs tested are single-copy genes located on the X and Y chromosomes, respectively (Fig. 1). The chromosomal location is not the same in all primates; AMELX is located on the distal part of the X short arm in humans and chimpanzees but on the distal part of the X long arm in squirrel monkeys and ring-tailed lemurs. Likewise, AMELY is located on the Y short arm in humans but on the Y long arm in all non-human primates. These results indicate that rearrangements have occurred not only on the Y but also on the X chromosome, even within primate lineages, and that the evolutionary strata observed in the human X chromosome (1) might be shuffled in non-human primates.
Figure 1.
FISH by species-specific AMELX and AMELY probes. FISH was carried out on metaphase chromosomes of humans (a and b), chimpanzees (c and d), squirrel monkeys (e and f), and ring-tailed lemurs (g and h). The X (Left) and Y (Right) chromosomes are marked by small bars. The AMELX probes attached on the X short arm in humans and chimpanzees and on the X long arm in squirrel monkeys and ring-tailed lemurs. The AMELY probes attached on the Y short arm in humans and on the Y long arm in chimpanzees, squirrel monkeys, and ring-tailed lemurs. Green fluorescence indicates a positive signal. A small white bar marks the centromeric region. (Bar = 10 μm.)
Phylogenetic Trees of Mammalian AMELs.
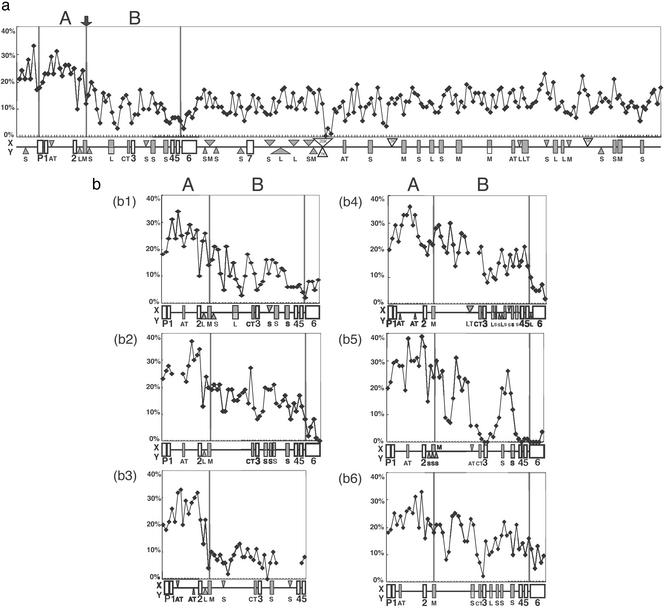
We obtained 5.5-kb genomic sequences of the AMELX and AMELY ranging from the promoter region to exon 6 at all but one AMELY locus. For greater bushbabies, we obtained a slightly shorter sequence of AMELY that ranged from intron 1 to exon 5. A number of regions containing small insertions and deletions (indels) or chunks of DNA with no sequence similarity between the gametologous AMELs were excluded when we computed the p distances between the gametologous AMELs in nonoverlapping windows of 100 bp each (Fig. 2). Relatively small p distances are noticeable in the promoter and exons, which are presumably subjected to stronger functional constraints than the flanking regions and introns. Yet it is clear that the overall p distances are significantly greater in the 5′ portion from intron 2 (>20%) than in the 3′ portion (8–15%). Although this transition looks rather gradual in most species, it is very sharp in ring-tailed lemurs, where the 3′ portion exhibits the smallest p distances. In any case, the transition boundary in all species is marked by a medium reiterated frequency repeat (MER) or short interspersed repetitive element (SINE) transposon. For convenience, the 5′ portion from the transposon was designated region A; it is 844 bp long and ranges from the 130-bp upstream site of exon 1 to the transposon. The 3′ portion was designated region B; it is 1,479 bp long and ranges from the transposon to exon 5.
Figure 2.
The p distances between the AMELX and AMELY in each species. The p distances were computed in nonoverlapping windows of 100 bp each. The p distances between two human BAC clone sequences (a) are designated as region IX in Fig. 4. The p distances between gametologous AMELs are shown for chimpanzees (b1), squirrel monkeys (b2), ring-tailed lemurs (b3), cattle (b4), pigs (b5), and horses (b6). The promoter region (P), exons (open boxes), and introns (horizontal lines) are indicated below each diagram. Shaded rectangles indicate various insertion or repetitive elements shared by chromosomes X and Y (S, SINE; L, LINE; M, MER; AT, AT-rich; CT, CT-rich; LT, long terminal repeats), whereas triangles are either X (above)- or Y (below)-specific. Open triangles indicate regions with no sequence similarity by DOTTER (11). Regions A and B used for separate phylogenetic analyses are marked by vertical bars.
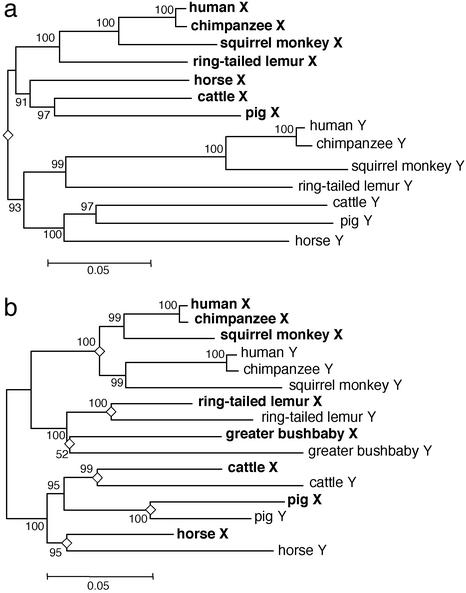
For region A, the average p distances between the gametologous AMELs are greater than those among orthologous AMELs. As a consequence, all AMELX and all AMELY cluster into two distinct clades in the NJ tree. This reciprocal monophyly is supported by a 93% bootstrap value (Fig. 3a) and is also supported by the MP method. To date the divergence in region A, we converted the p distances to sequence divergences (d) with multiple-hit corrections (19) and constructed a NJ tree. The average height (the average d distances from a common node to tips) of the primate AMELX and AMELY clade is 0.073 ± 0.009 and 0.146 ± 0.010, respectively. Thus, the latter evolved twice as fast as the former, supporting the male-driven hypothesis of mutations (20–22). If simians and prosimians diverged 60–80 million years ago (mya) (23), we can calibrate the nucleotide substitution rate at 0.91–1.21 × 10−9 for the AMELX and 1.83–2.43 × 10−9 for the AMELY. Assuming rate constancy, we determined the root of the NJ tree based on the calibrated substitution rates of AMELX and AMELY. The root was dated at 88–117 mya by the AMELX and 76–102 mya by the AMELY. Therefore, region A began to differentiate before the mammalian radiation.
Figure 3.
NJ trees in AMEL regions A and B. The trees for region A (a) and region B (b) are based on p distances. An open diamond at a node indicates AMELX and AMELY differentiation, and the number beside a node is the bootstrap value after 1,000 replications.
The phylogenetic relationships among the AMELs in region B are robust. All but one node in the NJ tree of region B is supported by a >95% bootstrap value, and the NJ tree is topologically identical to the MP tree (Fig. 3b). The tree for region B is entirely different from that for region A in that the mammalian gametologous AMELs diverged from each other during the evolution of individual lineages. Such relatively recent divergences are also observed in ring-tailed lemurs and greater bushbabies, suggesting that differentiation in region B was initiated after these prosimians diverged from the common ancestor. On the other hand, the orthologous AMELX or AMELY in simian primates reflect species relatedness, indicating that differentiation of the gametologous AMELs took place in the stem lineage (4, 5). Accordingly, there are two distinct AMELX and AMELY clades among the simian primates. The average height of the simian AMELX clade is 0.045 ± 0.006, and that of the simian AMELY clade is 0.083 ± 0.007, again in good agreement with the male-driven hypothesis of mutations. The length of the terminal branch leading to the mammalian AMELX or AMELY is estimated as 0.61 ± 0.006 or 0.115 ± 0.009 in cattle, 0.072 ± 0.007 or 0.050 ± 0.006 in pigs, and 0.052 ± 0.006 or 0.114 ± 0.009 in horses. In pigs, the AMELX terminal branch is significantly longer than the AMELY and the biological cause remains obscure. In any event, differentiation in region B is species-specific and was initiated 27–70 mya in different mammals.
Sequence Differences in Human Stratum 4.
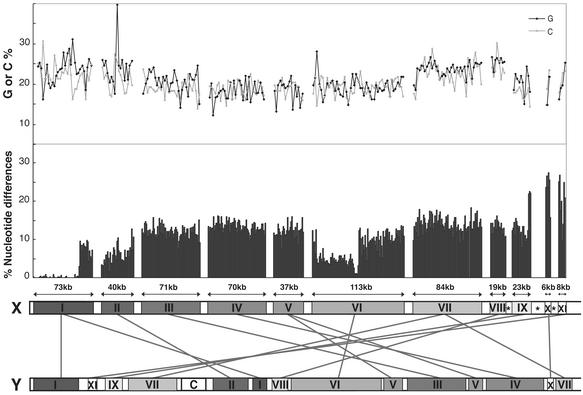
We retrieved 11 sequences (designated regions I–XI) along the short arm of the human X chromosome that contain the PAR1 and the ZFX locus in stratum 3. Although the total length of the X chromosome sequences was more than 2.5 megabases, only one-fifth (544 kb) could be aligned with Y gametologous sequences. We identified one or two Y regions for each X region (Table 5, which is published as supporting information on the PNAS web site); two separate Y regions were found for X regions I, V, and VII, giving a total of 14 Y regions. Except for region IX, the chromosomal positions and orientations of the X and Y regions correspond to the map produced by National Center for Biotechnology Information (NCBI) as of December 2002. In Fig. 4, the 11 X regions are ordered from the leftmost PAR1 in region I to the rightmost ZFX in region XI. We have also indicated the relative positions of the 14 Y regions, the p distances, and the pattern of nucleotide frequencies.
Figure 4.
Sequence profiles of the short arm of the human X chromosome compared with the Y gametologous regions. The p distances were calculated for the 11 separate regions on the X chromosome (designated by roman numerals) in nonoverlapping windows of 1 kb each. The orientation of region IX sequence follows the 2001 NCBI map version. (Upper) The G and C content in 2.5-kb windows. (Lower) The relative positions of 14 gametologous Y chromosomal regions (connected by lines to the X chromosome). The gap size between adjacent regions is arbitrarily drawn, but it is generally much larger than the size of each region. On the X chromosome, open boxes indicate that the corresponding chromosomal regions do not build “contigs” in NCBI MAP VIEWER, and open boxes with asterisks indicate that Y gametologs were not detected by Genomic blast Human Genome Search (NCBI). The “C” on the Y chromosome marks the centromere.
The order of the 14 Y regions is substantially different from that of the corresponding X regions. Supposing that gene order has been conserved for the X chromosome (24), it would be necessary to invoke eight paracentric and two pericentric inversions in the Y chromosome to achieve the same gene order as in the X chromosome (data available on request). Also, it should be noted that a large gap exists between adjacent X regions due to the lack of sequence data in GenBank or the severe lack of sequence similarity. One gap is found at the proximal part of region IX containing the AMELX. We noted that the gene orientation of the AMELX is reversed in the present version of the NCBI map placing the 5′ portion of the locus away from the ZFX. There are two possible explanations. It may be difficult to assign the proper orientation due to the lack of sequence overlap with region IX. Alternatively, region IX may have undergone a paracentric inversion in humans. Because pericentric inversions have been identified in squirrel monkeys and ring-tailed lemurs (Fig. 1), it is possible that some other X chromosomes have undergone rearrangements shuffling the evolutionary strata. However, both the gene content and the gene order among human and other mammalian X chromosomes, with the exception of the mouse X chromosome, which shows considerable rearrangements within it, are well conserved (24–29). For this reason, we believe that the 2001 NCBI version of the AMELX gene orientation was correct.
In region I, the p distances (1-kb window size) are generally 0.1% in the PAR1 and abruptly increase to 10% within the XG locus. The distances stay ≈10% in region II through the distal part of region IX. The distances again abruptly increase to >20% in the proximal part of region IX and stay at the same high level in regions X and XI in stratum 3. If multiple-hit corrections are made, these p distances are in excellent agreement with the synonymous divergences reported by Lahn and Page (1). However, there are noticeable fluctuations in the p distances within stratum 4. For instance, the distal part of region VI, which contains the KAL locus, exhibits fairly small p distances (≈5%). By contrast, region VII, which contains the TBL1X and OA1 loci, exhibits rather large p distances (15%).
Sex chromosomal differentiation can occur as a result of suppressing homologous recombination in male meiosis (1, 30, 31). It is known that the human genome has long-range GC mosaic structures or isochores that are correlated with events such as DNA replication timing, recombination, and chromosomal condensation (32–34). It is also known that the GC or AT skew is useful for identifying the replication origin and strand asymmetry in the prokaryote genome (35), although its application to human genomic sequences is suggestive at best (36). We therefore examined the GC content, the GC and AT skews, and the relative abundance of the consensus sequence 5′-(A/T)TT(G/C) or 5′-(C/G)AA(T/A) at sites where Holliday intermediates in Escherichia coli are resolved (37).
The human X short arm is generally AT-rich, but the GC content is relatively high in regions I, II, VII, and VIII, regions that may replicate in the early S phase (32–34). In terms of the GC and AT skews, T (C) is more abundant than A (G) in the distal half of region I, whereas the reverse is true in the proximal half. The transition occurs near the PAB1. In regions VIII and IX, A is always more abundant than T, but the relative abundance of G and C is reversed in the middle of each region. One may therefore speculate that recombination suppression is somehow related to long-range mosaic structures of the genome in terms of the GC content and the GC or AT skew. However, because these mosaic structures of the genome are not restricted to the PAB1 and the boundary between strata 3 and 4, it seems difficult to evaluate their significance for recombination suppression.
The frequency of the resolution sequences of Holliday intermediates is fairly uniform throughout the X short arm, although some resolution sequences tend to be significantly overrepresented. A resolution sequence exists within the MER transposon in intron 2 of mammalian AMEL loci and may act as a signal of recombination suppression. Despite this possibility, MERs are relatively abundant in the eukaryote genome, and there is no evidence that both prokaryotes and eukaryotes use the same sequences to resolve Holliday intermediates. It therefore seems that neither MERs nor resolution sequences had played any important roles in suppressing homologous recombination.
Discussion
The mammalian sex chromosomes arose from an ordinary pair of autosomes ≈300 mya (38). After this event, the sex-determining region Y (SRY) evolved from its X-linked progenitor gene (SOX3), and the need to avoid exchange of SRY led to the suppression homologous recombination (1, 39, 40). The Y long arm had already diverged from the X and had begun to degenerate. After the mammalian lineage diverged from monotremes and marsupials, an autosomal region containing the AMEL locus appears to have been added to form the X and Y short arms ≈80–130 mya (38, 41). As the nonrecombining portion of sex chromosomes expanded, this newly added region too became differentiated into strata 3 and 4. Our analysis suggests that the expansion of the nonrecombining portion in the short arm of mammalian sex chromosomes occurred almost immediately after the addition.
Through direct comparison of the nucleotide sequences of mammalian AMEL genes, we have demonstrated that the 5′ and 3′ portions of the locus from a transposon inserted in intron 2 belong to evolutionary strata 3 and 4, respectively. There must, therefore, have been an evolutionary stage at which the 5′ portion had already accumulated ≈10% sequence difference but the 3′ portion was still allelic and had been almost identical between the gametologous AMELs. Because this pattern of sequence differences is consistent over the short arm of the human X chromosome, we have postulated that AMEL intron 2 was once a distal boundary of stratum 3. The entire region of stratum 4 and PAR1 must have been included in the pseudoautosomal precursor region that was bounded by the ancient PAB in AMEL intron 2.
Our finding that the ancient PAB located between strata 3 and 4 is commonly found in different mammals supports the hypothesis that it formed in the common ancestor of eutherian mammals. It is therefore likely that the ubiquity of PAB is simply a result of the common ancestry rather than due to molecular or cytological mechanisms. It is also plausible that the ancient PAB may have been determined largely by chance. One indirect way of examining this plausibility is to ask how the present PAB1 was determined and whether it is the same among different mammals. The gene content in the PAR1 was studied in various mammals (42, 43). Recent genome projects have characterized PAR1 in several mammals (25–27, 44, 45; see also Table 6, which is published as supporting information on the PNAS web site). These studies show that the gene content in the PAR1 differs greatly among mammals and reflects the stochastic processes of differential addition, rearrangement, and Y degradation (42). If the present mammalian PAR1 was determined in this way, there is no reason to reject the hypothesis that similar stochastic processes played an important role in forming the ancient PAB as well.
A possible mechanism that had generated the ancient PAB is chromosomal inversion. Although any inversion that truncates the AMEL locus is irrelevant (8), the present human PAB1 resides in the gametologous XG locus, and the Y locus is truncated by a pericentric inversion. In addition, the distal boundary of the Y pericentric inversion differs from the PAB1 by only 88 bp. One may therefore postulate that the inversion is responsible for the formation of stratum 4. However, it turns out that the pericentric inversion corresponds to a small region in stratum 4 (Fig. 4), and the extent of sequence differences between the gametologous XG loci is ≈10% within the inverted Y region. Clearly, the pericentric inversion occurred relatively recently and could not have caused the formation of stratum 4.
Alternatively, recombination suppression in a chromosomal region can be accounted for if the sex determination process requires at least two linked genes (40). For example, if the mammalian male-determination requires not only the SRY but also additional genes, their tight linkage should be preferred and should evolve by natural selection. These genes should be as old as the SRY and have gametologs on the X chromosome. On the basis of these criteria, we surveyed the functional genes on the human Y chromosome. One candidate gene is RBMY1 on the Y long arm. Its ortholog has been found in mammals, indicating that the gene originated before the mammalian radiation. Although the gametolog on the X long arm is ubiquitously expressed and encodes a nuclear protein that binds nascent pre-mRNA (46), the RBMY1 is a spermatogenesis gene, and its expression is limited to spermatocyte nuclei in the meiosis phase. Thus, in addition to the SRY, RBMY1 is critical for determining maleness in mammals. Initially, these loci may have been linked on the long arm of the sex chromosomes, and recombination suppression between them may have been favored by natural selection. However, whenever subsequent Y chromosomal rearrangements changed their locations, the nonrecombining portion between the sex chromosomes must have expanded.
The above model explains the observations found in this study. However, the model does not specify an upper boundary of the nonrecombining region, only requiring that it include male-determining genes. An attractive argument for this aspect can be made based on the fact that larger nonrecombining regions are likely to contain more genes and mutations. If these genes were dispensable, the mutations would be harmless and eventually deteriorate the genes. On the other hand, if these genes were indispensable, most mutations would be detrimental. To reduce the genetic load, there is an obvious advantage to minimizing the size of a nonrecombining region. In an extreme case, a nonrecombining region should be strictly bounded by the male-determining gene loci. If this is the case, the boundary between adjacent evolutionary strata (1) is the fossilized chromosomal location of a set of male-determining loci.
Supplementary Material
Acknowledgments
This research was supported in part by Japan Society for Promotion of Science Grants 13557021 (to H.H.) and 12304046 (to N.T.).
Abbreviations
- mya
million years ago
- AMEL
amelogenin
- FISH
fluorescence in situ hybridization
- MER
medium reiterated frequency repeat
- SINE
short interspersed repetitive element
- PAR
pseudoautosomal region
- PAB
pseudoautosomal boundary
- NCBI
National Center for Biotechnology Information
Footnotes
References
- 1.Lahn B T, Page D C. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 2.Snead M L, Lau E C, Zeichner-David M, Fincham A G, Woo S L, Slavkin H C. Biophys Res Commun. 1985;129:812–818. doi: 10.1016/0006-291x(85)91964-3. [DOI] [PubMed] [Google Scholar]
- 3.Nakahori Y, Takenaka O, Nakagome Y. Genomics. 1991;9:264–269. doi: 10.1016/0888-7543(91)90251-9. [DOI] [PubMed] [Google Scholar]
- 4.Huang W, Chang B H-J, Gu X, Hewett-Emmett D, Li W-H. J Mol Evol. 1997;44:463–465. doi: 10.1007/pl00006166. [DOI] [PubMed] [Google Scholar]
- 5.Toyosawa S, O'hUigin C, Figueroa F, Tichy H, Klein J. Proc Natl Acad Sci USA. 1998;95:13056–13061. doi: 10.1073/pnas.95.22.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen E, Yuan Z A, Collier P M, Greene S R, Abrams W R, Gibson C W. Gene. 1998;216:131–137. doi: 10.1016/s0378-1119(98)00300-x. [DOI] [PubMed] [Google Scholar]
- 7.Hasegawa T, Sato F, Ishida N, Fukushima Y, Mukoyama H. J Vet Med Sci. 2000;62:1109–1110. doi: 10.1292/jvms.62.1109. [DOI] [PubMed] [Google Scholar]
- 8.Iwase M, Satta Y, Takahata N. Mol Biol Evol. 2001;18:1601–1603. doi: 10.1093/oxfordjournals.molbev.a003948. [DOI] [PubMed] [Google Scholar]
- 9.Ellis N, Yen P, Neiswanger K, Shapiro L J, Goodfellow P N. Cell. 1990;63:977–986. doi: 10.1016/0092-8674(90)90501-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Gurr S J. Gene. 2000;253:145–150. doi: 10.1016/s0378-1119(00)00289-4. [DOI] [PubMed] [Google Scholar]
- 11.Sonnhammer E L, Durbin R. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 14.Fitch W M. Syst Zool. 1971;20:406–416. [Google Scholar]
- 15.Hartigan J A. Biometrics. 1973;29:53–65. [Google Scholar]
- 16.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 17.Hirai H, Hirai Y, Kawamoto Y, Endo H, Kimura J, Rerkamnuaychoke W. Chromosome Res. 2002;10:313–327. doi: 10.1023/a:1016523909096. [DOI] [PubMed] [Google Scholar]
- 18.Hirai H, LoVerde P T. Parasitol Today. 1995;11:310–314. doi: 10.1016/0169-4758(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 19.Jukes T H, Cantor C R. In: Mammalian Protein Metabolism III. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 20.Miyata T, Hayashida H, Kuma K, Mitsuyasu K, Yasunaga T. Cold Spring Harbor Symp Quant Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 21.Makova K D, Li W-H. Nature. 2002;416:624–625. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 22.Bohossian H B, Skaletsky H, Page D C. Nature. 2000;406:622–625. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- 23.Martin R D. Nature. 1993;363:223–234. doi: 10.1038/363223a0. [DOI] [PubMed] [Google Scholar]
- 24.Murphy W J, Sun S, Chen Z-Q, Pecon-Slattery J, O'Brien S J. Genome Res. 1999;9:1223–1230. doi: 10.1101/gr.9.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raudsepp T, Kata S R, Piumi F, Swinburne J, Womack J E, Skow L C, Chowdhary B P. Genomics. 2002;79:451–457. doi: 10.1006/geno.2002.6723. [DOI] [PubMed] [Google Scholar]
- 26.McCoard S A, Fahrenkrug S C, Alexander L J, Freking B A, Rohrer G A, Wise T H, Ford J J. Anim Genet. 2002;33:178–185. doi: 10.1046/j.1365-2052.2002.00878.x. [DOI] [PubMed] [Google Scholar]
- 27.Quilter C R, Blott S C, Mileham A J, Affara N A, Sargent C A, Griffin D K. Mamm Genome. 2002;13:588–594. doi: 10.1007/s00335-002-3026-1. [DOI] [PubMed] [Google Scholar]
- 28.Stone R T, Grosse W M, Casas E, Smith T P, Keele J W, Bennett G L. Mamm Genome. 2002;13:211–215. doi: 10.1007/s00335-001-2124-9. [DOI] [PubMed] [Google Scholar]
- 29.Band M R, Larson J H, Rebeiz M, Green C A, Heyen D W, Donovan J, Windish R, Steining C, Mahyuddin P, Womack J E, Lewin H A. Genome Res. 2000;10:1359–1368. doi: 10.1101/gr.145900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bengtsson B O, Goodfellow P N. Ann Hum Genet. 1987;51:57–64. doi: 10.1111/j.1469-1809.1987.tb00865.x. [DOI] [PubMed] [Google Scholar]
- 31.Navarro A, Bretrán E, Barbadilla A, Ruiz A. Genetics. 1997;146:695–709. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernardi G. Gene. 1993;135:57–66. doi: 10.1016/0378-1119(93)90049-9. [DOI] [PubMed] [Google Scholar]
- 33.Ikemura T, Wada K, Aota S. Genomics. 1990;8:207–216. doi: 10.1016/0888-7543(90)90273-w. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe Y, Fujiyama A, Ichiba Y, Hattori M, Yada T, Sakaki Y, Ikemura T. Hum Mol Genet. 2002;11:13–21. doi: 10.1093/hmg/11.1.13. [DOI] [PubMed] [Google Scholar]
- 35.Lobry J R. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 36.Shioiri C, Takahata N. J Mol Evol. 2001;53:364–376. doi: 10.1007/s002390010226. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa M, Iwasaki H, Shinagawa H. J Biol Chem. 2001;276:10432–10436. doi: 10.1074/jbc.M010138200. [DOI] [PubMed] [Google Scholar]
- 38.Graves J A M. Trends Genet. 2002;18:259–264. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- 39.Lahn B T, Pearson N M, Jegalian K. Nat Rev Genet. 2001;2:207–216. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- 40.Nei M. Genetics. 1969;63:681–699. doi: 10.1093/genetics/63.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcox S A, Watson J M, Spencer J A, Graves J A M. Genomics. 1996;35:66–70. doi: 10.1006/geno.1996.0323. [DOI] [PubMed] [Google Scholar]
- 42.Graves J A M, Wakefield M J, Toder R. Hum Mol Genet. 1998;7:1991–1996. doi: 10.1093/hmg/7.13.1991. [DOI] [PubMed] [Google Scholar]
- 43.Gläser B, Myrtek D, Rumpler Y, Schiebel K, Hauwy M, Rappold G A, Schempp W. Hum Mol Genet. 1999;8:2071–2078. doi: 10.1093/hmg/8.11.2071. [DOI] [PubMed] [Google Scholar]
- 44.Ferretti L, Urquhart B G, Eggen A, Olsaker I, Harlizius B, Castiglioni B, Mezzelani A, Solinas-Toldo S, Thieven U, Zhang Y, et al. Mamm Genome. 1997;8:29–36. doi: 10.1007/s003359900341. [DOI] [PubMed] [Google Scholar]
- 45.Moore S S, Byrne K, Johnson S E, Kata S, Womack J E. Anim Genet. 2001;32:102–104. doi: 10.1046/j.1365-2052.2001.00740.x. [DOI] [PubMed] [Google Scholar]
- 46.Bennett M, Pinol-Roma S, Staknis D, Dreyfuss G, Reed R. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.