Abstract
Wrinkle-free (wrfr) is a previously uncharacterized, spontaneous, autosomal recessive mouse mutation resulting in very tight, thick skin. wrfr mutant mice exhibit severe breathing difficulties secondary to their tight skin and die shortly after birth. This phenotype is strikingly similar to a very rare human genetic disorder, restrictive dermopathy. wrfr mutant mice display a defective skin barrier, which is normally imparted by the cornified envelope, a composite of protein and lipid that prevents loss of water from within and entry of potentially harmful substances from without. In addition, hair growth from grafted wrfr skin is impaired. Positional cloning of the wrfr mutation revealed a retrotransposon insertion into a coding exon of Slc27a4, the gene encoding fatty acid transport protein (FATP)4. FATP4 is the primary intestinal FATP and is thought to play a major role in dietary fatty acid uptake; it therefore is viewed as a target to prevent or reverse obesity. However, its function in vivo had not been determined. Our results demonstrate an unexpected yet critical role for FATP4 in skin and hair development and suggest Slc27a4 to be a candidate gene for restrictive dermopathy.
The skin of mammals is composed of a dermis and an epidermis separated by a basement membrane. The epidermis is stratified, consisting of basal keratinocyte, spinous/prickle, granular, and squamous/cornified layers. Keratinocytes move upward from the basement membrane and progress through a scheduled program of differentiation, with cell death marking their final differentiation step. During this differentiation program, keratinocytes flatten and accumulate lipids that are discharged into the intercellular space (1). In the stratum corneum the lipids are crosslinked with a number of proteins including loricrin and involucrin to form the cornified envelope (2). The insoluble cornified envelope is a composite of protein and lipid that serves as a barrier to loss of water from within and to entry of potentially harmful substances from without. Without such a barrier, life on land could not exist (3).
We discovered a spontaneous, autosomal recessive mutation in our mouse colony causing extremely tight, thick skin. We named the mutation wrinkle-free (wrfr). The wrfr phenotype is similar to a rare human disease called restrictive dermopathy (4, 5). Newborn wrfr −/− mice have difficulty breathing because of the tight skin, do not suckle, exhibit a defective skin barrier, and die several hours after birth. There are also defects in hair follicle morphogenesis and hair growth. Because of the presumed fundamental importance of the mutated gene to skin development and its potential relevance to human disease, we sought to identify the affected gene. Here we present a characterization of the wrfr phenotype and the positional cloning of the wrfr mutation.
Materials and Methods
Microsatellite Marker Analysis.
Genomic DNA was prepared from tissues by standard proteinase K digestion, phenol/chloroform extraction, and ethanol precipitation (6) or by proteinase K digestion of tail pieces followed by boiling (7). Microsatellite primer pairs for the genome scan were purchased from Research Genetics (Huntsville, AL). For fine mapping, previously uncharacterized markers were identified by finding microsatellites polymorphic between 129X1/SvJ and C57BL/6J mouse strain DNAs by using sequences obtained from the Celera Mouse Genome Assembly (129X1/SvJ) and the Ensembl Mouse Genome Server (C57BL/6J). PCR conditions were as described (8). PCR products were analyzed by separation through 3% super-fine resolution agarose gels (Amresco, Solon, OH). Primer pairs for markers at the margins of the 701-kb wrfr critical region were 5′-AACCTCTCTCCGTGTGGTGAT-3′ and 5′-CCTTTGGTGGTCCTTCTCCT-3′ (for D2Jhm125) and 5′-CTTGAAAGCCTGGGTTCACT-3′ and 5′-AGGTCCCCTTAATGCCTCAT-3′ (for D2Jhm132).
RT-PCR and Sequencing.
Total RNA was extracted from control and wrfr −/− skin by using Tri-Reagent (Molecular Research Center, Cincinnati). RT-PCR was performed with 1 μg of total RNA by using a GeneAmp kit (Applied Biosystems). Products were separated on 1–1.5% agarose gels, cut from the gel, purified on Wizard PCR Preps columns (Promega), and directly sequenced with BigDye Terminator v3.0 (Applied Biosystems). Purified RT-PCR products were also used as probes for Northern blots.
Northern Blotting.
Total RNA was extracted as described above from embryonic day (E)15.5 skin, and poly(A) RNA was isolated by using oligo(dT) magnetic beads (Ambion, Austin, TX). Poly(A) RNA (1.5 μg) was separated in formaldehyde-containing gels and blotted as described (9). Blots were prehybridized at 42°C overnight in a solution containing 5× Denhardt's solution, 50% formamide, 50 mM Tris (pH 7.4), 0.8 M NaCl, 0.1% sodium pyrophosphate, 10% dextran sulfate, 1% SDS, 0.2 mg/ml heparin, and 100 μg/ml denatured salmon sperm DNA and hybridized overnight in the same solution after the addition of 1 × 106 cpm/ml of a denatured 32P random primer-labeled probe. Blots were washed twice for 5 min in 1× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0) and 0.2% SDS at room temperature and once for 15 min in 0.1× SSC and 0.2% SDS at 50°C. Hybridized blots were exposed to a phosphor screen and scanned with a PhosphorImager 445 SI (Molecular Dynamics).
Southern Blotting.
Ten micrograms of genomic DNA purified from lung were digested with ScaI and separated through 1% agarose. Capillary transfer of DNA was performed as described (6). Blots were hybridized with a random primer-labeled Slc27a4/fatty acid transport protein (FATP)4 cDNA probe using QuikHyb (Stratagene) according to manufacturer specifications. Hybridized blots were exposed to BioMax film (Kodak) at −80°C.
Histological Analysis.
Tissues were placed into 10–20 volumes of 10% buffered formalin (DiRuscio and Associates, Fenton, MO) and fixed overnight with gentle rocking at room temperature. Fixed tissues were dehydrated to 70% ethanol and processed into paraffin blocks. Paraffin sections were stained with hematoxylin/eosin. For oil red O staining, fresh tissues or whole embryos were snap-frozen in OCT compound (VWR Scientific), and 7 μm of fresh frozen sections were cut and stained on positively charged slides as described (10).
Antibodies and Immunofluorescence.
Loricrin, keratin 6, and keratin 14 antibodies were purchased from Covance (Princeton). Tissues were frozen and sectioned as described above and blocked in 1% BSA and 10% normal goat serum. Secondary antibodies were conjugated to Cy3 (Chemicon). All washes were performed in PBS and antibody incubations in PBS with 1% BSA. After staining, sections were mounted in 0.1× PBS and 1 mg/ml p-phenylenediamine in 90% glycerol.
Skin-Permeability Assay and Skin Grafts.
The permeability assay was performed exactly as described (11). For the grafts, circular dorsal skin pieces were removed from five control and four wrfr −/− E18.5 embryos and placed in sterile PBS. Recipient nu/nu male mice (Harlan Breeders, Indianapolis) were anesthetized, and circular skin pieces were removed from their backs to make graft sites for the donor skin. Wound edges were sutured and covered with Vaseline gauze. Drinking water was supplemented with penicillin and gentamycin for 5 days after grafting.
Results
Identification of wrfr Mice.
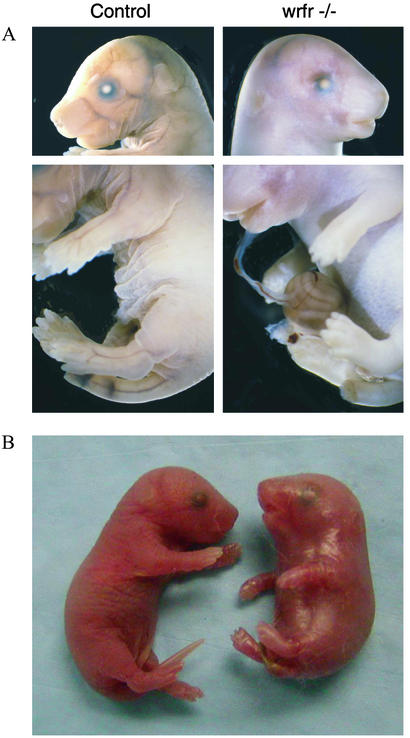
We discovered a spontaneous, neonatally lethal, autosomal recessive mutation in our mouse colony that we named wrinkle-free (wrfr) because the affected mice exhibited abnormally tight, noncompliant skin and lacked the wrinkles typical of perinatal mice (Fig. 1). After birth the skin was abnormally shiny, dry, and peeling, and the mouth was fixed in an open position. Body movements were extremely limited, and we never observed milk in the stomachs of mutants, demonstrating that they did not suckle. No significant abnormalities in other tissues were found, although mutants had poorly expanded alveolar spaces (data not shown). These results suggest that mutants died because of their restricted ability to inspire, secondary to their tight skin. This overall phenotype is remarkably similar to that observed in an extremely rare human disease, restrictive dermopathy (4, 5). These patients exhibit tight, noncompliant skin that prevents the movement needed for proper breathing, leading to death within several hours or days after birth. In addition, restrictive dermopathy patients have several characteristic abnormalities secondary to the rigid skin including a small mouth and nose, low-set and poorly formed ears, a fixed open mouth, and flexion contractures in all limbs. These features are all present to some degree in wrfr mice (Fig. 1).
Figure 1.
The wrfr phenotype. E17.5 (A) and newborn (B) wrfr −/− mice exhibit tight, shiny, noncompliant skin, with an absence of wrinkles typical of the normal mice. Herniated intestines (A) were present in ≈10% of mutants.
Mapping and Positional Cloning of the wrfr Mutation.
The wrfr mutant mice first appeared in the F2 offspring of a chimeric male, the germ line of which was derived from embryonic stem cells carrying a targeted mutation. We initially assumed that the wrfr phenotype resulted from the targeted mutation, but subsequent genotyping revealed that the wrfr mutation was independent. We hypothesized that the mutation occurred in the embryonic stem cells either spontaneously or by nonhomologous insertion of our targeting vector. The R1 embryonic stem cells used to make the chimeric mouse were derived from a 129X1/SvJ × 129S1/SvImJ F1 hybrid embryo (12), and the chimera had been subsequently bred to C57BL/6J females. Therefore, the wrfr mutation should be linked to genetic markers from one of the two parental 129 strains, whereas the wild-type allele of the affected gene should be linked to C57BL/6J genetic markers.
To map the chromosomal location of the wrfr mutation, we began a genome scan of wrfr −/−, +/−, and +/+ mice by using polymorphic microsatellite markers previously identified in the lab (8). This demonstrated a highly significant association of the wrfr mutation with the proximal region of chromosome 2. Additional markers were developed from DNA sequence in the Celera Mouse Genome Assembly and used to screen 886 mice representing 1,606 informative meioses. This localized the mutation to a 701-kb critical region between D2Jhm125 and D2Jhm132, 18–19 centimorgans from the centromere. In human, this region is homologous to chromosome 9q34.
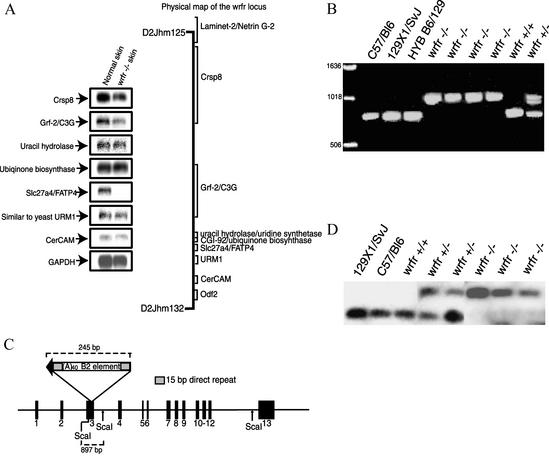
According to Celera's annotations, there were 14 known or predicted genes in the wrfr critical region (Fig. 2A). Of the 14 candidate genes, 5 were only predicted to be genes, there being no ESTs or cDNAs with significant homology. We began to analyze the nine remaining genes by Northern blotting using control and mutant skin mRNA. In contrast to other genes in the region, there was a striking absence of Slc27a4/FATP4 mRNA in mutant skin, although expression was easily detected in control skin (Fig. 2A). This indicated that the wrfr mutation affected expression of Slc27a4 either directly or indirectly.
Figure 2.
Mapping and positional cloning of wrfr. (A Right) Physical map of the wrfr critical region, with the position of known genes indicated. (A Left) Northern blot with mRNA from normal and wrfr −/− skin was probed with RT-PCR products from known genes and GAPDH. No Slc27a4/FATP4 mRNA was detected in wrfr −/− skin, although it was readily detectable in normal skin. (B) PCR amplification of Slc27a4 exon 3. A PCR product ≈225 bp larger than that observed in wild type was amplified from all wrfr +/− and −/− DNAs. (C) Sequencing of Slc27a4 exon 3 from wrfr −/− mice revealed a 230-bp B2-type retrotransposon insertion with a 40-bp poly(A) tail. Fifteen base pairs of Slc27a4 were duplicated after insertion, generating a direct repeat. (D) Southern blot of ScaI-digested genomic DNA from wrfr +/+, +/−, and −/− mice was probed for Slc27a4. An ≈900-bp ScaI fragment was detected in normal mice, and an ≈1,150-bp fragment was detected in wrfr −/− mice. Both fragments were present in heterozygous mice.
In parallel to Northern blotting, we amplified and sequenced cDNAs derived from genes in the critical region. For those cDNAs that could be amplified by RT-PCR, no mutations were found after extensive sequencing. However, we were unable to amplify the 5′ third of the Slc27a4 cDNA from mutant RNA, even though the same region could be amplified from control RNAs. Therefore, the three exons that comprise the 5′ third of the Slc27a4 gene were amplified from wrfr −/− genomic DNA and wild-type 129X1/SvJ genomic DNA. The amplified product from exon 3 was ≈225 bp larger in mutants (Fig. 2B). Sequencing of the mutant exon 3 revealed the insertion of a 230-bp element (Fig. 2C) with both significant homology to and all features typical of a B2-type murine retrotransposon (13, 14). Retrotransposons make up ≈37.5% of the mouse genome and are responsible for 10–20% of spontaneous mouse mutations (15). They use a “copy-and-paste” mechanism of expansion in which the transposable element is copied into RNA and reverse-transcribed into DNA, which then is inserted into a new location in the genome (16). Southern blot analysis confirmed that the insertion was present only in the mutant Slc27a4 gene (Fig. 2D).
The absence of detectable Slc27a4 mRNA in the mutant (Fig. 2A) suggests that the retrotransposon insertion destabilizes the transcript, and thus little if any FATP4 protein should be produced. However, even if low levels of mRNA are present and translated, the insertion into a coding exon of Slc27a4 should result in a truncated, inactive FATP4 protein. From the deduced amino acid sequence, the mutant protein should contain 183 aa; of these, only 133 would be from FATP4 compared with the 644-aa full-length protein. Fifty amino acids would be translated from the retrotransposon sequence. We conclude that the retrotransposon insertion into Slc27a4 represents the wrfr mutation and results in the absence of FATP4. The allele hereafter is to be referred to as Slc27a4wrfr.
FATP4.
Although cells can synthesize a subset of the fatty acids they require, efficient uptake of extracellular fatty acids is an important cellular function and is thought to be mediated in part by FATPs (17). Expression of FATPs in transfected cells increases the rate of uptake of long-chain fatty acids (18). FATP4 is one of five known mouse FATPs and one of six known human FATPs; it is the major FATP of the small intestine (18, 19). FATP4 has been shown to mediate fatty acid uptake by intestinal epithelial cells and therefore is viewed as a potential drug target to prevent or reverse obesity (20), although it is widely expressed (19, 21). Our positional cloning of the Slc27a4wrfr mutation demonstrates that an FATP plays an essential role in skin development.
Slc27a4wrfr and Skin Development.
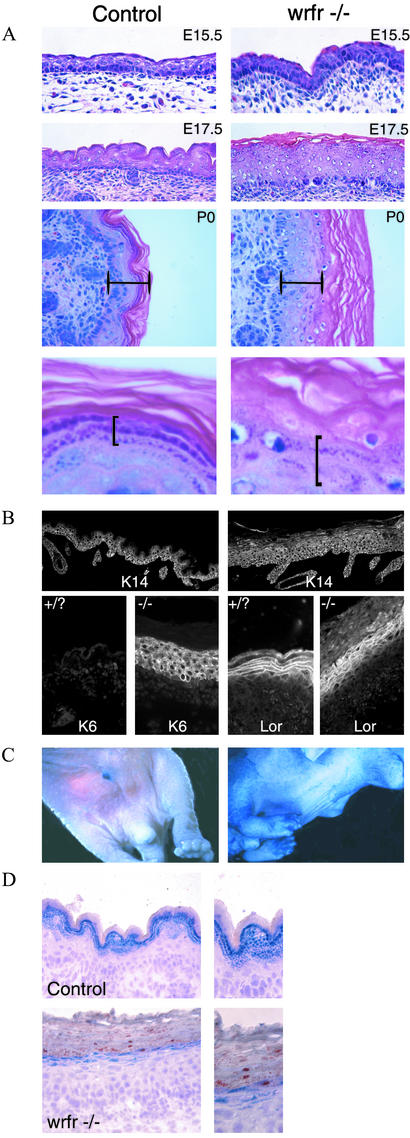
To investigate further how the absence of FATP4 affected skin development, we performed histological analyses. No epidermal abnormalities were present at E15.5, although the dermis appeared more cellular in the mutant (Fig. 3A). By E17.5, the mutant epidermis was significantly thicker than the control, and the skin surface was notably flattened compared with control (Fig. 3A). In newborn mice, the mutant epidermis was on average three times as thick as that of normal littermates, and it displayed a flattened dermal–epidermal junction (Fig. 3A). The spinous/prickle and cornified layers each were thicker than normal, and cells in the granular layer were not flattened as they are in normal skin. In addition, the keratohyalin granules were more sparse and mislocalized in the mutant, and we observed hyperkeratosis, or a thickened squamous/keratinized outer layer (Fig. 3A and data not shown). The flattened dermal–epidermal junction was particularly evident after immunohistochemical staining with an antibody to keratin 14, an epidermal cell marker (Fig. 3B). Histological abnormalities in restrictive dermopathy patients are similar to those we observed in wrfr: a thickened epidermis with hyperkeratosis, abnormal shape of keratohyalin granules, and a flat dermal–epidermal junction (4, 5). Thus, mutations in SLC27A4 may cause restrictive dermopathy.
Figure 3.
Histological analysis of Slc27a4wrfr −/− mice. (A) At E15.5, control and mutant epidermis appeared similar, but more cells were present in the mutant dermis just beneath the basement membrane. At E17.5, the mutant epidermis was obviously thicker than the control, and a flattened skin surface was already apparent. At birth (P0), the mutant epidermis was on average three times thicker than control. (Bars are identical lengths.) At high power (Bottom), fewer and abnormally arranged keratohyalin granules (bracketed) were observed in the mutant. (B) At E17.5, staining for keratin 14 (K14) demonstrated the increased thickness and flattening of the Slc27a4wrfr −/− epidermis. Keratin 6 (K6) was expressed only in Slc27a4wrfr −/− epidermis, demonstrating a differentiation defect. Loricrin (Lor), a protein component of the cornified envelope, was not highly concentrated at the plasma membrane of granular cells in mutant skin. (C) Skin-permeability assay. E17.5 embryos were incubated overnight in 5-bromo-4-chloro-3-indolyl β-d-galactoside. Penetration into mutant but not control skin demonstrated a barrier defect. (D) Oil red O staining revealed numerous fat droplets in the squamous layer of mutant but not control epidermis.
To determine whether keratinocyte differentiation in the Slc27a4wrfr −/− epidermis was abnormal, we examined the expression of keratin 6, a marker of aberrant keratinocyte differentiation or hyperproliferation (22). Keratin 6 was detected in the mutant but was absent from the normal skin (Fig. 3B). BrdUrd assays for keratinocyte proliferation did not reveal a significant difference between mutant and control (data not shown), and thus expression of keratin 6 in the mutant epidermis most likely indicates an aberrant keratinocyte-differentiation program.
Formation of the skin barrier requires a precisely orchestrated program of keratinocyte differentiation, which seemed to be disrupted in the mutants. To examine whether Slc27a4wrfr −/− mutant mice had an intact and functional skin barrier, we performed a skin-permeability assay by incubation in a solution of 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) at low pH. If X-Gal can penetrate the barrier, a blue precipitate forms (11). After overnight incubation, E17.5 Slc27a4wrfr −/− mutant skin turned blue, whereas the skin of the normal littermate did not (Fig. 3C). This result indicates a defect in the barrier of the mutant, which is normally imparted by the cornified envelope. Consistent with this defect, loricrin, a protein component of the cornified envelope initially expressed in granular cells (23), was not well concentrated at the plasma membrane in the mutant, and its staining pattern revealed a failure of granular cells to flatten (Fig. 3B).
The massive hyperkeratosis associated with a thickened stratum corneum observed here was also described in transglutaminase 1 knockout mice, which have a defective skin barrier (24). Indeed, many diseases that affect skin permeability result in a thickening of the stratum corneum and scaling as a protective mechanism (25). Hence, the massively thickened stratum corneum observed in wrfr mice may be an attempt to compensate for the defective skin barrier.
Slc27a4wrfr and Lipid Homeostasis.
Lipids are major components of the cornified envelope and are crucial for barrier formation (1, 26); this provides a plausible reason why the fatty acid transport properties of FATP4 could play a role in establishment or maintenance of the barrier. We therefore hypothesized that lipid homeostasis was affected in Slc27a4wrfr −/− skin. To test this hypothesis, we incubated sections of E17.5 Slc27a4wrfr −/− and control fetuses in oil red O, which stains neutral lipids and cholesterol esters (Fig. 3D). Numerous lipid droplets were present in the lower squamous layers of the epidermis of Slc27a4wrfr −/− mice, but few if any were detected in the epidermis of normal littermates. The presence of lipid droplets in the mutant epidermis suggests defects in lipid transport and/or processing, which could prevent proper formation of the skin barrier.
Slc27a4wrfr and Hair Development.
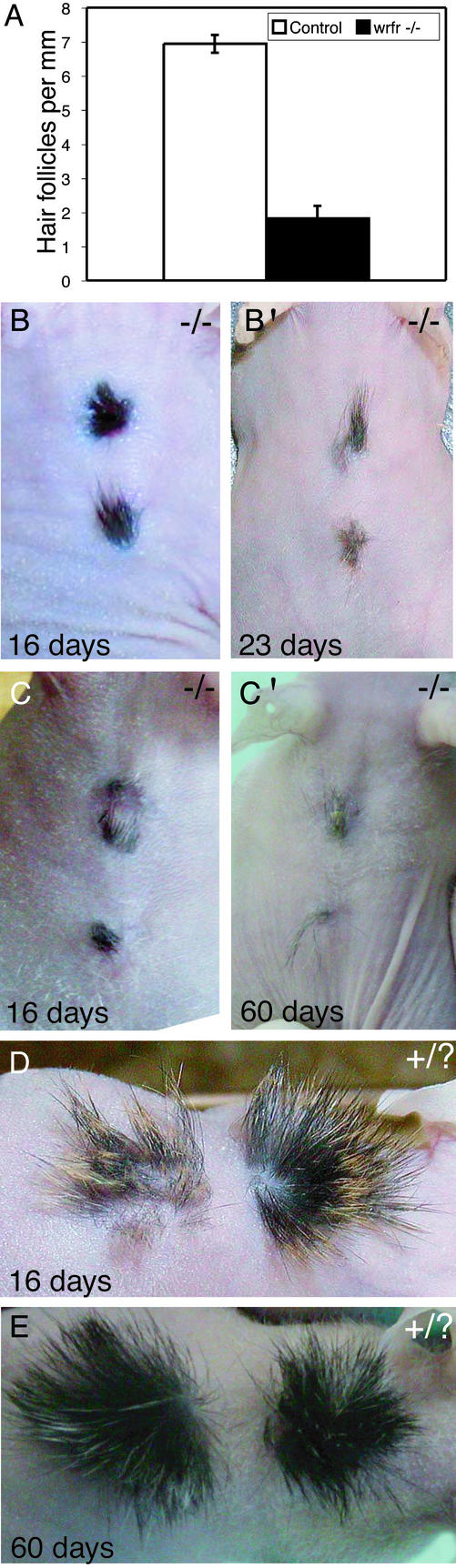
Our initial histological studies suggested that there were fewer developing hair follicles in the mutant skin. Quantitation of hair germs per unit length in multiple sections from late-gestation fetuses confirmed this suggestion (Fig. 4A). To examine how the Slc27a4wrfr mutation affected postnatal development of hair and skin, full-thickness skin from E18.5 mutant and control fetuses was grafted onto the backs of nude mice and left for 23 and 60 days. Thick tufts of hair formed on the control skin grafts (Fig. 4D). Mutant skin grafts also developed hair, but hair growth was not nearly as robust as in the control (Fig. 4 B and C), and hair growth did not recover with time (Fig. 4 B′ and C′). Thus, results from these experiments indicate that (i) there are fewer developing hair follicles present in the mutant, and (ii) although the mutant skin is capable of forming hair, hair growth is clearly impaired.
Figure 4.
Defective hair growth in Slc27a4wrfr −/− mice. (A) Quantitation of developing hair follicles in multiple sections from E17.5 embryos showed that Slc27a4wrfr −/− mice had less than one-third the number present in controls. (B and B′) Skin from an E18.5 Slc27a4wrfr −/− embryo was grafted onto a nude mouse and photographed after 16 (Left) and 23 (Right) days. (C and C′) Skin from a separate E18.5 mutant embryo was grafted and photographed after 16 (Left) and 60 (Right) days. (D) Skin grafts from an E18.5 control littermate; large tufts of hair were present after 16 days. (E) Large tufts of hair were also present in a separate control graft after 60 days.
Discussion
The phenotype of Slc27a4wrfr −/− mice demonstrates a critical and unexpected role for FATP4 in development and function of skin and growth of hair. Why does the lack of FATP4 result in such a severe perinatal skin phenotype? Although additional studies will be necessary to reveal the mechanism leading to the skin defects, there are several possibilities to consider. The observed defects may result from essential fatty acid deficiency, because many of these defects have also been described as symptoms of essential fatty acid deficiency (27, 28). In support of this notion, (i) linoleic acid, one of the two essential fatty acids, is a competitive substrate for FATP4 (20), and (ii) FATP4 is expressed strongly in the placenta (data not shown), where it may have an important role in transport of both essential and nonessential fatty acids from mother to fetus. If true, FATP4 may not be required in the skin, although it is expressed there (Fig. 2; ref. 21). However, the fact that both skin and hair defects were observed when the skin was grafted to a nude mouse, an environment where fatty acids should be readily available, seems to demonstrate a requirement for FATP4 in skin. On the other hand, FATP4 deficiency during fetal development may alter the dermis and/or epidermis irreversibly such that the mutant graft tissue becomes incapable of normal maturation. Alternatively, FATP4 may be important in the vasculature for fatty acid uptake from the bloodstream, and the defective vasculature could be maintained in the graft.
To understand the mechanism of the wrfr defects, it will be important to determine which cells express FATP4. Our attempts to localize the mRNA by in situ hybridization failed, but Northern blotting with mRNA prepared from dermis vs. dermis + epidermis suggested that expression is higher in the epidermis (data not shown). Together with the oil red O results, this result suggests that FATP4 may have an autonomous role in the epidermis.
Lipid droplets in the perinatal mutant epidermis (Fig. 3) suggest defects in fatty acid and lipid homeostasis and are likely associated with the defective skin barrier. However, even at E15.5, before the barrier forms, dermal hypercellularity was evident, and a very subtle lack of wrinkling could be detected (Fig. 3 and data not shown). Thus, FATP4 might have a role in early developmental events, perhaps in signaling. In particular, signaling by the peroxisome proliferator-activated receptor (PPAR) subfamily of nuclear hormone receptors has been demonstrated to play a role in skin development and barrier formation (29, 30), and PPARs are activated by fatty acids shown to be competitive substrates for FATP4 such as oleic and linoleic acids (20, 31). Finally, several other mouse models exhibit skin phenotypes with similarities to wrfr. These include knockout mice lacking follistatin (32) and mice overexpressing transforming growth factor β or Whn, the product of the nude gene, in keratinocytes (33, 34). It will be important to determine whether FATP4 activity and/or fatty acid trafficking play a role in any of these important developmental pathways.
Acknowledgments
We thank Cong Li for technical assistance; Miriam Meisler for advice; the Mouse Genetics Core for care of mice; the Pulmonary Morphology Core for paraffin histology; Chris Stander and Andrey Shaw for DNA sequencing support; and Joshua Sanes, Raphe Kopan, and Bill Parks for comments on the manuscript. This work was supported in part by National Institutes of Health Grant R01 GM060432 (to J.H.M.). Institutional funds supported access to the Celera Mouse Genome Assembly.
Abbreviations
- wrfr
wrinkle-free
- En
embryonic day n
- FATP
fatty acid transport protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY195604).
References
- 1.Wertz P W. Acta Derm Venereol Suppl (Stockh) 2000;208:7–11. doi: 10.1080/000155500750042790. [DOI] [PubMed] [Google Scholar]
- 2.Kalinin A E, Kajava A V, Steinert P M. BioEssays. 2002;24:789–800. doi: 10.1002/bies.10144. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs E. Curr Opin Cell Biol. 1990;2:1028–1035. doi: 10.1016/0955-0674(90)90152-5. [DOI] [PubMed] [Google Scholar]
- 4.Wesche W A, Cutlan R T, Khare V, Chesney T, Shanklin D. J Cutan Pathol. 2001;28:211–218. doi: 10.1034/j.1600-0560.2001.028004211.x. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann R, Lohner M, Bohm N, Leititis J, Helwig H. Eur J Pediatr. 1993;152:95–98. doi: 10.1007/BF02072481. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook J, Fritsch E F, Maniatis F T. In: Molecular Cloning: A Laboratory Manual. Nolan C, editor. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 9.42–9.44. [Google Scholar]
- 7.Hanley T, Merlie J P. BioTechniques. 1991;10:56. [PubMed] [Google Scholar]
- 8.Andrews K L, Mudd J L, Li C, Miner J H. Am J Pathol. 2002;160:721–730. doi: 10.1016/S0002-9440(10)64892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis F T. In: Molecular Cloning: A Laboratory Manual. Nolan C, editor. Vol. 1. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 7.43–7.50. [Google Scholar]
- 10.Stotz E, Schenk E A, Churukian C, Willis C. Stain Technol. 1986;61:187–190. doi: 10.3109/10520298609110730. [DOI] [PubMed] [Google Scholar]
- 11.Hardman M J, Sisi P, Banbury D N, Byrne C. Development (Cambridge, UK) 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- 12.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krayev A S, Markusheva T V, Kramerov D A, Ryskov A P, Skryabin K G, Bayev A A, Georgiev G P. Nucleic Acids Res. 1982;10:7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers J H. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- 15.Dennis C. Nature. 2002;420:458–459. doi: 10.1038/420458a. [DOI] [PubMed] [Google Scholar]
- 16.Kazazian H H, Jr, Goodier J L. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 17.Schaffer J E. Am J Physiol. 2002;282:E239–E246. doi: 10.1152/ajpendo.00462.2001. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch D, Stahl A, Lodish H F. Proc Natl Acad Sci USA. 1998;95:8625–8629. doi: 10.1073/pnas.95.15.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahl A, Gimeno R E, Tartaglia L A, Lodish H F. Trends Endocrinol Metab. 2001;12:266–273. doi: 10.1016/s1043-2760(01)00427-1. [DOI] [PubMed] [Google Scholar]
- 20.Stahl A, Hirsch D J, Gimeno R E, Punreddy S, Ge P, Watson N, Patel S, Kotler M, Raimondi A, Tartaglia L A, et al. Mol Cell. 1999;4:299–308. doi: 10.1016/s1097-2765(00)80332-9. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann T, Buchkremer F, Gosch I, Hall A M, Bernlohr D A, Stremmel W. Gene. 2001;270:31–40. doi: 10.1016/s0378-1119(01)00489-9. [DOI] [PubMed] [Google Scholar]
- 22.O'Guin W M, Schermer A, Lynch M, Sun T-T. In: Cellular and Molecular Biology of Intermediate Filaments. Godman R D, editor. New York: Plenum; 1990. pp. 301–334. [Google Scholar]
- 23.Mehrel T, Hohl D, Rothnagel J A, Longley M A, Bundman D, Cheng C, Lichti U, Bisher M E, Steven A C, Steinert P M, et al. Cell. 1990;61:1103–1112. doi: 10.1016/0092-8674(90)90073-n. [DOI] [PubMed] [Google Scholar]
- 24.Kuramoto N, Takizawa T, Matsuki M, Morioka H, Robinson J M, Yamanishi K. J Clin Invest. 2002;109:243–250. doi: 10.1172/JCI13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams M L. Pediatr Dermatol. 1992;9:365–368. doi: 10.1111/j.1525-1470.1992.tb00632.x. [DOI] [PubMed] [Google Scholar]
- 26.Mao-Qiang M, Elias P M, Feingold K R. J Clin Invest. 1993;92:791–798. doi: 10.1172/JCI116652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anonymous. Nutr Rev. 1969;27:85–88. [Google Scholar]
- 28.Horrobin D F. J Am Acad Dermatol. 1989;20:1045–1053. doi: 10.1016/s0190-9622(89)70130-4. [DOI] [PubMed] [Google Scholar]
- 29.Hanley K, Jiang Y, He S S, Friedman M, Elias P M, Bikle D D, Williams M L, Feingold K R. J Invest Dermatol. 1998;110:368–375. doi: 10.1046/j.1523-1747.1998.00139.x. [DOI] [PubMed] [Google Scholar]
- 30.Thuillier P, Brash A R, Kehrer J P, Stimmel J B, Leesnitzer L M, Yang P, Newman R A, Fischer S M. Biochem J. 2002;366:901–910. doi: 10.1042/BJ20020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Willson T M, Lenhard J M, et al. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matzuk M M, Lu N, Vogel H, Sellheyer K, Roop D R, Bradley A. Nature. 1995;374:360–363. doi: 10.1038/374360a0. [DOI] [PubMed] [Google Scholar]
- 33.Sellheyer K, Bickenbach J R, Rothnagel J A, Bundman D, Longley M A, Krieg T, Roche N S, Roberts A B, Roop D R. Proc Natl Acad Sci USA. 1993;90:5237–5241. doi: 10.1073/pnas.90.11.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nehls M, Pfeifer D, Schorpp M, Hedrich H, Boehm T. Nature. 1994;372:103–107. doi: 10.1038/372103a0. [DOI] [PubMed] [Google Scholar]