Abstract
Although LINE-1 (long interspersed nucleotide element-1, L1) retrotransposons comprise 17% of the human genome, an exhaustive search of the December 2001 “freeze” of the haploid human genome working draft sequence (95% complete) yielded only 90 L1s with intact ORFs. We demonstrate that 38 of 86 (44%) L1s are polymorphic as to their presence in human populations. We cloned 82 (91%) of the 90 L1s and found that 40 of the 82 (49%) are active in a cultured cell retrotransposition assay. From these data, we predict that there are 80–100 retrotransposition-competent L1s in an average human being. Remarkably, 84% of assayed retrotransposition capability was present in six highly active L1s (hot L1s). By comparison, four of five full-length L1s involved in recent human insertions had retrotransposition activity comparable to the six hot L1s in the human genome working draft sequence. Thus, our data indicate that most L1 retrotransposition in the human population stems from hot L1s, with the remaining elements playing a lesser role in genome plasticity.
Preliminary analysis of the human genome has shown that retrotransposons comprise at least 42% of its mass. L1 is one of the most successful retrotransposons and occupies 17% of DNA (1). The overwhelming majority of L1s (>99.8%) are inactive because of 5′ truncations, internal rearrangements, and mutations. However, some L1s remain retrotransposition-competent (i.e., active), and we previously estimated that an average diploid human genome contains ≈30–60 active L1s (2).
An active L1 is 6 kb in length. Transcription is initiated from an internal promoter located within its 5′ UTR (3), and the RNA is transported to the cytoplasm. The L1-encoded proteins, ORF1p and ORF2p, then act on the mRNA that encoded them, a phenomenon known as cis preference (4–6). The resultant ribonucleoprotein particle then reenters the nucleus where L1 integration is thought to occur by target-primed reverse transcription (7, 8). During this process, the L1 endonuclease generates a single-stranded nick in genomic DNA at the loose consensus sequence 5′-TTTTT/A-3′ (9–13), exposing a 3′ OH, which is used as a primer for reverse transcription of L1 RNA by the L1 RT.
All 14 known de novo human L1 insertions are members of the two youngest L1 subsets (14); 13 are derived from the transcribed group a (Ta) subset (15), and one is derived from the pre-Ta subset (1, 16). Five intact L1s are associated with these 14 insertions. Two (L1RP and L1β-Thal) are full-length disease-producing insertions (17, 18), and three (L1.2, LRE2, and LRE3) are the likely progenitors of disease-producing insertions (19–21). When assayed for retrotransposition, L1RP, L1β-Thal, and LRE3, all of which were isolated from affected individuals or family members, are hot L1s (19, 22–24). Hot L1s are defined as showing at least one-third of the activity of L1RP. By contrast, L1.2A and LRE2, which were isolated from commercial libraries, are weakly active (2, 25).
Here we estimate that the average human genome has 80–100 retrotransposition-competent L1s and show that only a small proportion are hot L1s. In contrast, we find that most in vivo human retrotranspositions involve hot L1s. Thus, we conclude that although rare in an individual genome, hot L1s are responsible for the majority of retrotransposition in the human population.
Materials and Methods
Database Searches.
blast (26) searches of nonredundant human genomic databases with a full-length retrotransposition-competent L1 [L1.3, GenBank accession no. L19092 (2, 27)] were performed to identify sequence contigs that contain full-length L1s as described (28). Search details are provided in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Phylogenetic Tree and Consensus Sequences.
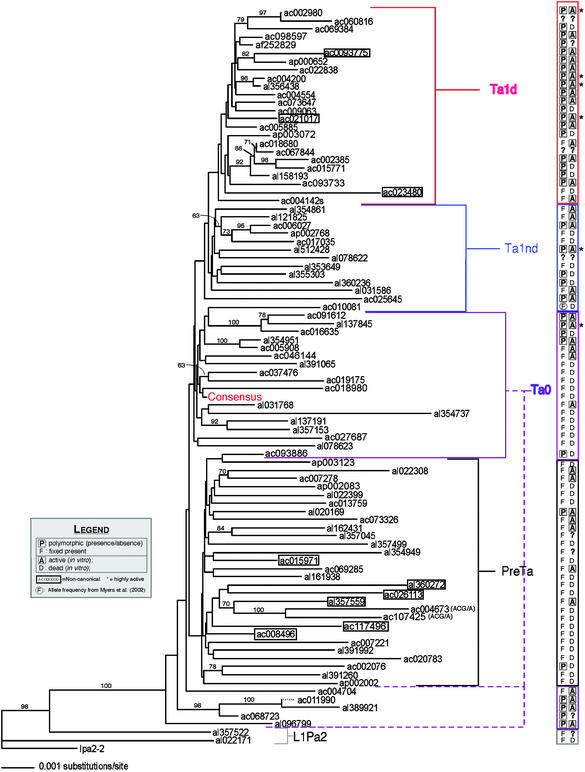
We used the latest version of paup (29) to construct neighbor-joining trees from full-length L1 sequences excluding positions corresponding to consensus CpGs and the G-rich polypurine tract in the 3′ UTR. Distances were corrected by using the 2ρ method of Kimura (30). Element Ac009269 had an ≈400-bp deletion in the 5′ UTR and was not included. Fourteen elements overlap with data from a smaller tree constructed by Boissinot et al. (31). As reported, neighbor joining, with few exceptions, correctly clustered the pre-Ta, Ta-0, Ta-1nd, and Ta-1d elements. These clusters can be readily distinguished by a number of diagnostic characters (31). Not surprisingly, given their short branches, the four clusters are not recovered in the 50% majority neighbor-joining tree of 1,000 bootstrap replicates, although some subgroups within them are (indicated in Fig. 3), as was also found with the smaller data set (31). The consensus sequences were built by using a simple majority rule at each base.
Figure 3.
Neighbor-joining tree with 89 intact L1 elements. The tree was constructed by using the full L1 sequences as described in Materials and Methods. The nodes recovered >60% of the time in 1,000 bootstrap replicates of the data are indicated, excluding CpG dinucleotides and the polypurine tract in the 3′ UTR. Ac009269 with the ≈400-bp deletion in the 5′ UTR was not included. Polymorphism and activity data are appended in the column on the right. A question mark signifies that the experiment was not performed. Hot elements are followed by asterisks, and noncanonical elements are boxed. The consensus sequence of the 89 elements is indicated. The Ta-0 subgroup that clusters between the ancestral L1Pa2 elements and the pre-Ta group had been identified (31). Based on short branch lengths, polymorphism data, and the in vivo activity of one member (LRE2/al389921) (21), the subgroup was predicted to be relatively young (31). We verify that prediction by showing four of five tested members to be polymorphic and all tested members to be active. This group is canonically Ta-0 based on seven defining nucleotides (Table 1). However, the group appears similar to L1Pa2 ancestral L1s based on its position in the tree. Preliminary sequence analysis comparing these L1s to the two L1Pa2 elements shows ancestral nucleotides spread throughout each element in a mosaic pattern and no obvious region where an element is clearly ancestral or young. AC004673 and AC107425 are members of the ACG/A group, distinct from the pre-Ta subgroup. They are marked as such.
Cloning Genomic L1s.
Intact L1s found in the human genome working draft sequence (HGWD) were amplified from 50–200 ng of genomic DNA or ≈200 ng of bacterial artificial chromosome (BAC) DNA by using Expand Long Template PCR (Roche Applied Science) in a 20-μl reaction volume under the following conditions: 10-min denaturation at 94°C, 30 cycles of 30-s denaturation at 94°C, 40-s annealing step at 55–70°C, 6-min elongation at 68°C, and 10-min final elongation at 68°C. The forward primer, containing a NotI site, was placed in a unique sequence 5′ of the L1 and the reverse primer was placed 3′ of the L1. Primer sequences are in Table 2, which is published as supporting information on the PNAS web site. If PCR on genomic DNA did not produce satisfactory yields, the BAC containing the L1 (Invitrogen) was used as a template. BACs were purified by using a Large-Construct Kit (Qiagen, Chatsworth, CA). We cloned 67 L1s from genomic DNA and 15 from BAC DNA. When we tested clones of hot L1s, 4/15 (27%) from BAC template and 6/13 (46%) from genomic DNA displayed high activity; 10/15 (67%) from BAC and 7/13 (54%) from genomic DNA displayed weak activity. One BAC-derived clone was inactive. A similar analysis with weakly active elements showed that ≈50% of clones were inactive regardless of whether the clones came from BAC template or genomic template. This analysis suggests that the template used in PCR does not dramatically affect measured activity.
L1-containing bands from at least two separate PCRs were pooled, band-purified by using a GeneClean Spin Kit (Bio 101), and digested with NotI and BstZ17I (New England Biolabs). Fragments were swapped into pL1RP-enhanced GFP (EGFP) (BstZ17I) (19) [full-length L1RP tagged with the EGFP retrotransposition cassette; pL1RP-EGFP (23), except modified to contain a BstZ17I site] by using T4 Ligase (New England Biolabs). At least four clones of each L1 except al356438 (two clones) were assayed for activity.
EGFP Assay for Retrotransposition Activity.
All L1s were compared with pL1RP-EGFP (BstZ17I) and pL1ac002980-EGFP (BstZ17I), an internal control whose retrotransposition activity was consistently ≈130% of pL1RP-EGFP (BstZ17I). pL1RP(JM111)-EGFP (23) was used as a negative control. Each genomic L1 was cloned and assayed in human 143B TK− osteosarcoma cells. Cell culture, antibiotic selection, and flow cytometry were performed as described (23). Briefly, cells were transfected (22), grown overnight, selected with puromycin (10 μg/ml), and grown for 6 more days. EGFP expression was quantified in a FacsCalibur flow cytometer (Becton Dickinson). A gated sample of 10,000 live cells was analyzed for each transfection. Cells were positive when they showed greater fluorescence intensity than the most fluorescent cell transfected with pL1RP(JM111)-EGFP. All clones for each L1 were assayed in three separate transfections. When at least one clone had measured activity more than one-third that of L1RP, the element was called hot.
One hundred sixty-eight clones were assayed in determining the activity of the 34 weakly active elements. Of those, 56% were inactive and the rest showed weak activity. In determining the activity of the six hot L1s, 28 clones were tested. Clones of each of the six showed widely varying activities. For four hot L1s, two clones per L1 showed high activity. For the remaining two hot L1s only one clone was highly active per L1. Notably, only one of the 28 clones from hot L1s was inactive. As seen with five of six hot L1s, three of 34 weak L1s showed all tested clones to be weakly active. Thus, it is formally possible that these three L1s are unidentified hot L1s. Raw activity data for all clones are available in Table 3, which is published as supporting information on the PNAS web site. Under the assumption that a mutation is rarely activating, the in vitro activity recorded for each element was the average across three transfections for the single clone with the highest activity.
Allele Frequency.
A two-reaction, three-primer technique (19, 31) was used to determine the allele frequency of each L1 in 46 genomes (23 individuals) from five ethnic groups (European descent, Chinese, Indo-Pakistani, Pacific, and Sub-Saharan African). Allele frequency details are in the Supporting Text.
Repairing L1.2A.
We corrected L1.2A (LRE1), an allele of the precursor of a 3,784-bp insertion (20), at nucleotides 5649 and 5765 so that its amino acid sequence was identical to the disease insertion. This was done by swapping a 530-bp SpeI/BstZ17i fragment from L1RP into L1.2A. The two restriction fragments were identical except for the changes at nucleotides 5649 and 5765. DNA sequence analysis confirmed the corrected L1.
Results
Retrotransposition Activity of Intact L1s From the HGWD.
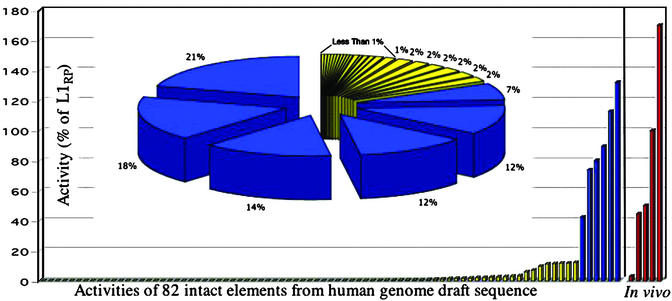
A database search of the December 2001 HGWD (95% complete) revealed 89 intact L1s and one with a ≈400-bp deletion in the 5′ UTR. Using Expand Long Template PCR, we cloned 82 of these L1s and assayed each for its ability to retrotranspose in cultured cells. Forty of the 82 L1s showed activity greater than the negative control, JM111, an ORF1 missense mutant of L1RP (25). We used L1RP as a standard for comparison and found that L1 activity ranged from 0.1% to 130% of L1RP activity (Fig. 1, Table 2). When the measured activities of the L1s were summed, six hot L1s comprised 84% of the total activity of the 82 L1s tested (Fig. 2, Table 4, which is published as supporting information on the PNAS web site).
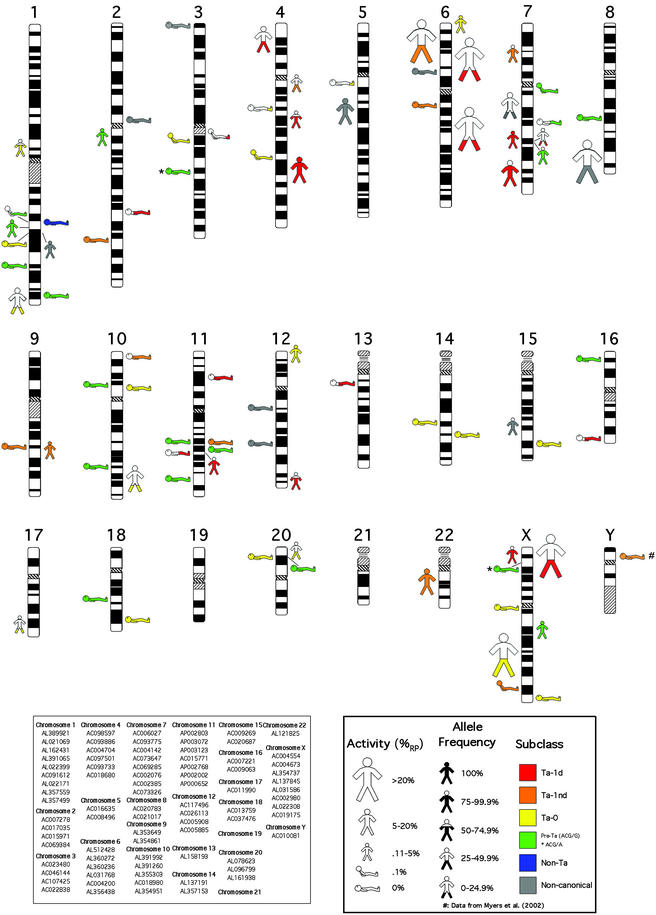
Figure 1.
Chromosomal location, activity, allele frequency, and subclass of 82 full-length L1 elements with two intact ORFs.
Figure 2.
L1 activity distribution. The measured potential activity of L1s from both the HGWD and de novo human insertions is shown. The histogram depicts the activities of 82 intact L1s from the HGWD and five human L1s involved in recent disease-causing insertions. The entire pie in the pie chart represents the total of all of the activity of the 82 L1s from the HGWD. Each slice of the pie represents the activity of a single element. The six hot elements (blue slices) represent 84% of the total measured potential activity in the HGWD.
Relationship Between L1 Age and Activity.
To examine the relationship between L1 age and activity, we used sequence divergence and allele frequency of each L1 as surrogate markers for age (32–34). To assess sequence divergence, we included 89 elements in a neighbor-joining tree (35). Because such a tree depicts nucleotide variation as branch length between L1s, younger, more similar, L1s are nearer one another on the tree (Fig. 3).
We next estimated the allele frequency of 86 of the 90 L1s by using a PCR-based assay with 23 individuals from five different ethnic groups (Fig. 4, which is published as supporting information on the PNAS web site). Although one cannot formally determine the age of a polymorphic insertion based on its allele frequency alone, fixed insertions are likely older than polymorphic insertions. When activity and allele frequency data were added to the tree, we saw a trend similar to that noted by others in which putative young L1s with little sequence divergence (separated by short branch lengths) were generally polymorphic in the population (16, 31) and were active in cultured cells (Fig. 3). Conversely, highly diverged L1 sequences were most frequently fixed and inactive.
To facilitate quantitative analysis of activities of L1s of different ages, we placed L1s first into three groups (0.0–0.49, 0.5–0.99, and 1.0) based on allele frequency and then into six groups [from youngest to oldest: Ta-1d, Ta-1nd, Ta-0, pre-Ta (ACG/G), pre-Ta (ACG/A), and L1Pa2] based on nine nucleotide positions (Table 1) previously associated with L1s in different age groups (15, 31, 36). Ten L1s (11%) were noncanonical, not falling into one of the six groups (see below). Percentage of active L1s, median activity, mean activity, and standard deviations were calculated for each group, either with these 10 L1s excluded or included based on tree position. Regardless of the method used to age the L1s, we found that younger groups generally had higher percentages of active L1s and higher mean and median activities. Age vs. activity data are available in Table 5, which is published as supporting information on the PNAS web site.
Table 1.
Subclass-defining nucleotides, numbering as in Boissinot et al. (34)
| Canonical position
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 74 | 711 | 1820 | 5557 | 5560 | 5954 | 5955 | 5956 | 6040 | |
| Ta-1d | – | t | t | t | g | a | c | a | g |
| Ta-1nd | g | c | c | t | g/c | a | c | a | g |
| Ta-0 | g | c | c | g | c | a | c | a | g |
| Pre-Ta (ACG/G) | g | c | c | g | c | a | c | g | g |
| (ACG/A) | g | c | c | g | c | a | c | g | a |
| L1-Pa2 | g | c | c | g | c | g | a | g | a |
Some Noncanonical L1s Remain Retrotransposition-Competent.
Preliminary sequence analysis of the 10 noncanonical L1s revealed that retrotransposition competence is not affected by the hybrid nature of these elements: four are weakly active and one is a hot L1. Furthermore, sequence analysis did not suggest an obvious mechanism for the generation of these L1s. In contrast to the old/young chimeric L1s (13, 36) and the L1/PAI1b cDNA chimeras (6), we did not find a clear switching point between multiple diagnostic nucleotides of one subclass and those of another. Sequence alignments are in Fig. 5, which is published as supporting information on the PNAS web site, and a detailed description of the noncanonical L1s is in Supporting Text.
Similarity to Hot L1 Consensus Is a Good Predictor of Retrotransposition Activity.
To analyze the relationship between L1 activity and nucleotide sequence, we constructed a consensus sequence with eight of the hot L1s (LRE3, L1RP, ac004200, ac002980, al356438, al512428, ac021017, and al137845). This sequence is identical to the Ta-1d consensus except for a silent ORF1 change at position 1033 and is identical to a consensus of the 90 intact L1s except for 12 polymorphic sites. Consensus sequences are in Fig. 6, which is published as supporting information on the PNAS web site.
We compared the L1s, first as active and inactive groups, then pairwise to the consensus of the hot elements. We analyzed the L1s in their entirety, then by region, and finally by whether differences resulted in amino acid changes. We found no nucleotide changes uniquely associated with active or inactive L1s. As expected, with some exceptions, the closer an L1 was to the hot L1 consensus, the more likely it was to be active. Taken with the above result, our data indicate that a decrease in retrotransposition activity occurs as a function of time. The further an L1 is from the “hot” consensus sequence, the less likely it is to be active. A description of this mutation analysis is in Supporting Text.
Active and Inactive Intact L1s Show Similar Genomic Distributions.
The assayed L1 sequences were divided into “active” (40 L1s that showed some retrotranspositional activity in cell culture) and “inactive” sequences (42 L1s that showed none). Analyses determined that neither class was overrepresented with respect to (i) proximity to or location within known genes, (ii) the GC content of the empty sites, (iii) the recombinagenicity of the empty sites, and (iv) the composition of the target site. A statistical analysis of the genomic distributions of these L1s is in Supporting Text.
Disease-Causing L1s Are as Active as the Hot L1s Isolated from the HGWD.
There are only five known full-length L1s involved in de novo human retrotransposition (17–21). Although all have been assayed for retrotransposition activity (2, 19, 22–25), we repaired L1.2A (20) and compared the activity of all five in the context of our 82 L1s. We found that four of these five disease-causing insertions had activities of 150%, 100%, 50%, and 39% of L1RP, making them among the most active elements ever tested in cell culture (Fig. 2). Thus, hot L1s are responsible for most of the retrotransposition in the human population at the present time.
Discussion
We use two independent methods to estimate that the average human being has 80–100 retrotransposition-competent L1s or about twice the previous estimate (2). One method assumes that the HGWD is representative of a 95% complete haploid genome. Because 49% of 82 tested L1s showed retrotransposition activity, we expect that 44 (0.49 × 90) of 90 full-length L1s in the HGWD are retrotransposition-competent. This number extrapolates to 93 L1s (44 × 2/0.95) in the complete diploid genome.
A second method necessitates knowing the allele frequency of every active L1 present or absent from the HGWD. Our data from the HGWD and earlier L1 studies can be used to begin this estimation. The average allele frequency of the 40 retrotransposition-competent L1s from the HGWD is 0.66 (the average allele frequency of the inactive L1s was 0.86). If we assume that 44 retrotransposition-competent L1s are present in the completed haploid HGWD, the expected number of these L1s in an average human being is 58 (44 × 2 × 0.66). The following seven retrotransposition-competent L1s are absent from HGWD: L1.2 (2, 20), L1.3, L1.4 (2, 27), L1.19, L1.20, L1.39 (2), and LRE3 (19). The average allele frequency of these L1s is 0.60 (2, 19). Thus, an average human carries eight copies of these L1s in their diploid genome for a total of 66 (58 + 8). Furthermore, there are likely many undiscovered L1 insertions that make contributions to the sum proportional to their allele frequencies. Therefore, using 93 L1s from method 1 and 66 + (undiscovered L1s) from method 2, we estimate that the number of retrotransposition-competent L1s in an average human being is between 80 and 100.
There are caveats to this estimation. Without the sequence of the heterochromatic portion of the human genome (1), it is difficult to quantify the degree to which we have underestimated the number of retrotransposition-competent L1s. In addition, the HGWD is a draft. It may be systematically biased for or against repeat sequence and may contain errors such as premature stop codons that disqualify actual active L1s from analysis. Also, as noted (25), a true measurement of in vivo retrotransposition activity would need to account for other variables such as cell type, timing, chromatin effects/genome position, and host defenses. Because the 40 retrotransposition-competent L1 elements are randomly distributed throughout the genome, it is likely that some are inactive in vivo. However, if we group intact elements by activity (hot, weakly active, or inactive), it is unlikely that inactivation mechanisms have disproportionately affected one of these activity groups.
The majority of de novo human insertions are in the Ta group and most are polymorphic (14). Our data support this inverse correlation between L1 activity and age (Fig. 3 and Table 4). Results also suggest that a relatively small number of hot L1s comprise the bulk of L1 activity in the average human genome. Whereas 40 of 82 full-length elements showed activity in the cell culture assay, just six accounted for 84% of the total measured retrotransposition activity in HGWD. All hot L1s are polymorphic and three (ac002980, ac004200, and al356438) come from the youngest Ta-1d group (ref. 31, Table 1). In addition, one is in the Ta-1nd group (al512428), and another is a member of a younger Ta-0 subgroups (al137845). The sequences of these five canonical hot L1s are very similar to the consensus sequences of their respective groups or subgroups, indicating that they have retrotransposed relatively recently in human evolution. Finally, one hot L1 (ac021017) is noncanonical and has the potential to play a role in the foundation of a novel subgroup (33).
Interestingly, although our data demonstrate that there are only six hot L1s in HGWD, it is notable that al356438, a hot element, and ac093886, an inactive element, were not present in any of the 46 genomes we used to determine allele frequency. In addition to al356438, ac093886, and the 14 de novo human insertions reviewed in ref. 14, there are numerous other examples of L1 insertions with very low frequencies (16, 28, 37, 38). Although our data show only six hot L1s in HGWD, if a private hot L1 element were to exist in only 1 in 1,000 individuals, there would be millions of such elements in the entire human population. Thus, although hot L1s only comprise a small proportion of the active L1s in any given individual, it is possible that the total number of extant hot L1s in the human population is greater than the number of weakly active and inactive L1s.
If the L1 activity distribution in the HGWD derived from the cultured cell assay can be extrapolated to in vivo activity, we would predict that a high percentage of L1s involved in de novo human insertions would be derived from hot L1s. Indeed, when the five intact L1s from human in vivo retrotransposition events were reassayed with the EGFP assay, four were found to be hot L1s (Fig. 2). Only LRE2 (21), which was isolated from a commercial library and which therefore may be an allele of the authentic progenitor of a mutagenic insertion into the dystrophin gene, did not show high activity. Thus, although hot L1s are relatively rare in an individual genome, they comprise at least four of five known intact L1s involved in human insertions.
Kazazian (39) and Li et al. (40) estimated that 1 in 10 to 1 in 37 individuals harbor a new L1 insertion by multiplying the percentage of nonrecurrent mutations attributable to retroelements by the number of mutations per diploid genome per generation. We now offer another estimate of retrotransposition frequency by summing the total retrotransposon activity in a genome in terms of L1RP and then estimating how often a single L1RP retrotransposes in vivo. The 40 active elements we tested have a summed activity equal to ≈6.5 times the activity of a single L1RP. Using the estimated 93 L1s in a human being, an entire diploid genome would have ≈15 times (6.5 × 93/40) the activity of a single L1RP element in the entire diploid genome. Our best estimate of how well L1RP retrotransposes comes from two sources. In the cell culture assay, L1RP inserts in at least one cell in every 30. In germ cells of male transgenic mice, an L1RP transgene driven by its endogenous promoter retrotransposes in one mouse line of four at about half the rate of an L1RP transgene driven by a PpolII promoter (one germ cell in 68; ref. 41). Therefore, a conservative estimate of L1 retrotransposition rate would be 1 germ cell in 500 (1/68 × 1/2 × 1/4). Thus, we estimate a retrotransposition frequency in human beings ranging from 1 in 2 to 1 in 33 (1 in 30 to 1 in 500 divided by 15), well within the range of the previous estimates.
In summary, by examining 90 intact L1s from the 95% complete HGWD, this work doubles previous estimates for the number of active L1s in an average human being. More importantly, it demonstrates that, although rare in an individual genome, hot L1s account for the bulk of L1 retrotransposition in human populations.
Supplementary Material
Acknowledgments
We thank A. V. Furano for help with the sequence alignments and carrying out the phylogenetic analysis, N. Brake for assistance with figures, L. Leach for help with HTML coding, and N. Luning Prak, E. Ostertag, and M. Seleme for critically evaluating the manuscript. R.M.B. was supported by a Wellcome Trust grant. J.V.M. was supported by grants from the W. M. Keck Foundation and National Institutes of Health Grant GM60518. H.H.K. was supported by National Institutes of Health Grant GM45398.
Abbreviations
- Ta
transcribed group a
- HGWD
human genome working draft sequence
- BAC
bacterial artificial chromosome
- EGFP
enhanced GFP
References
- 1.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M L, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Sassaman D M, Dombroski B A, Moran J V, Kimberland M L, Naas T P, DeBerardinis R J, Gabriel A, Swergold G D, Kazazian H H., Jr Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 3.Swergold G D. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazazian H H, Jr, Moran J V. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 5.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 6.Wei W, Gilbert N, Ooi S L, Lawler J F, Ostertag E M, Kazazian H H, Jr, Boeke J D, Moran J V. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan D D, Eickbush T H. Mol Cell Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 9.Morrish T A, Gilbert N, Myers J S, Vincent B J, Stamato T D, Taccioli G E, Batzer M A, Moran J V. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 10.Jurka J. Proc Natl Acad Sci USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 12.Cost G J, Boeke J D. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert N, Lutz-Prigge S, Moran J V. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 14.Ostertag E M, Kazazian H H., Jr Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 15.Skowronski J, Fanning T G, Singer M F. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers J S, Vincent B J, Udall H, Watkins W S, Morrish T A, Kilroy G E, Swergold G D, Henke J, Henke L, Moran J V, et al. Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Divoky V, Indrak K, Mrug M, Brabec V, Huisman T H J, Prchal J T. Blood. 1996;88:580–580. [Google Scholar]
- 18.Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen A A, Rosenberg T, et al. Nat Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 19.Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian H H., Jr Am J Hum Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dombroski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H., Jr Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 21.Holmes S E, Dombroski B A, Krebs C M, Boehm C D, Kazazian H H., Jr Nat Genet. 1994;7:143–148. doi: 10.1038/ng0694-143. [DOI] [PubMed] [Google Scholar]
- 22.Wei W, Morrish T A, Alisch R S, Moran J V. Anal Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 23.Ostertag E M, Prak E T, DeBerardinis R J, Moran J V, Kazazian H H., Jr Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimberland M L, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian H H., Jr Hum Mol Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 25.Moran J V, Holmes S E, Naas T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Dombroski B A, Scott A F, Kazazian H H., Jr Proc Natl Acad Sci USA. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Badge R M, Alisch R S, Moran J V. Am J Hum Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swofford D L. paup* 4.0: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 30.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 31.Boissinot S, Chevret P, Furano A V. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 32.Adey N B, Schichman S A, Graham D K, Peterson S N, Edgell M H, Hutchison C A. Mol Biol Evol. 1994;11:778–789. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- 33.Casavant N C, Hardies S C. J Mol Biol. 1994;241:390–397. doi: 10.1006/jmbi.1994.1515. [DOI] [PubMed] [Google Scholar]
- 34.Pascale E, Liu C, Valle E, Usdin K, Furano A V. J Mol Evol. 1993;36:9–20. doi: 10.1007/BF02407302. [DOI] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Ovchinnikov I, Rubin A, Swergold G D. Proc Natl Acad Sci USA. 2002;99:10522–10527. doi: 10.1073/pnas.152346799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheen F M, Sherry S T, Risch G M, Robichaux M, Nasidze I, Stoneking M, Batzer M A, Swergold G D. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ovchinnikov I, Troxel A B, Swergold G D. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kazazian H H., Jr Nat Genet. 1999;22:130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Scaringe W A, Hill K A, Roberts S, Mengos A, Careri D, Tezanos Pinto M, Sommer S S. Hum Mutat. 2001;17:511–519. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- 41.Ostertag E, DeBerardinis R J, Goodier J L, Zhang Y, Yang N, Gerton G, Kazazian H H., Jr Nat Genet. 2002;32:655–660. doi: 10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.