Abstract
The Drosophila melanogaster genome has six physically clustered NK-related homeobox genes in just 180 kb. Here we show that the NK homeobox gene cluster was an ancient feature of bilaterian animal genomes, but has been secondarily split in chordate ancestry. The NK homeobox gene clusters of amphioxus and vertebrates are each split and dispersed at two equivalent intergenic positions. From the ancestral NK gene cluster, only the Tlx–Lbx and NK3–NK4 linkages have been retained in chordates. This evolutionary pattern is in marked contrast to the Hox and ParaHox gene clusters, which are compact in amphioxus and vertebrates, but have been disrupted in Drosophila.
The Drosophila melanogaster 93D/E or NK gene cluster contains six homeobox genes: tinman (tin, NK4), bagpipe (bap, NK3), ladybird late (lbl), ladybird early (lbe), C15 (93Bal), and slouch (slou, NK1) in a gene cluster spanning just 180 kb (1, 2). All six genes possess homeobox sequences of the ANTP class (3), forming a distinct clade within this class along with several NK-related genes (4). The similarity of the genes and their clustered arrangement indicates that they arose through a series of tandem gene duplications, in an analogous way to the Hox gene cluster. We deduce that the NK homeobox gene cluster is ancient, dating at least to the base of the Bilateria. The reasoning is based on the phylogenetic distribution of these genes. Animals from very divergent evolutionary lineages have orthologues of each Drosophila NK gene (except for lbl and lbe, which are recent tandem duplicates). For example, human NKX3.1 and NKX3.2 (BAPX1) genes are orthologues of Drosophila bap, LBX1 and LBX2 genes are orthologues of the ancestral ladybird gene, two human genes equivalent to mouse Nkx1.1 and Nkx1.2 are orthologues of slou, and TLX1, TLX2, and TLX3 (the HOX11 family) are orthologues of C15 (4). It is also likely that human NKX2.3, NKX2.5, and NKX2.6 are orthologues of Drosophila tin, although sequence similarity is less clear (4, 5). Existence of these human genes implies that the tandem duplications that produced these various NK-related genes had occurred before the divergence of humans and Drosophila and, by implication, before the separation of the Deuterostomia and Ecdysozoa. Therefore, Drosophila NK (and related) genes have remained a tight gene cluster since the origin of these genes >500 million years (Myr) ago, which implies the existence of a selective reason for the gene clustering in Drosophila.
To assess whether maintenance of homeobox gene clustering is conserved in other taxa, we have examined a cephalochordate, amphioxus (Branchiostoma floridae). This animal is ideal for this comparison because, as a chordate, it belongs to a phylum very distantly related to arthropods, yet it has a genome uncomplicated by the extensive gene duplications that accompanied vertebrate evolution (6). Furthermore, amphioxus has a single canonical Hox gene cluster (7) and a compact ParaHox gene cluster (8), suggesting that the genome has not undergone wholescale rearrangement from the ancestral bilaterian state.
Materials and Methods
Cloning and Genomic Walking.
Three single-animal genomic libraries were used to isolate B. floridae genes and for genomic walking: cosmid libraries MPMGc117 and MPMGc118, distributed by the Resource Center and Primary Database, Berlin (www.rzpd.de) and a genomic phage library in lambda FIXII (9). Contig 1 was initiated by using an AmphiNK3 cDNA probe (G. Panopoulou, Max Planck Institute for Molecular Genetics, Berlin, personal communication); this probe hybridized to one genomic phage clone, which was used to isolate cosmids for genomic walking. AmphiNK4 and AmphiNK3 were identified from this walk. Two cDNA clones of AmphiNK4 were also isolated from an amphioxus embryo library (10). An AmphiLbx fragment was obtained by degenerate PCR (11) on the cDNA library. The cloned PCR fragment was used to probe the cosmid library MPMGc117; Lbx-positive clone MPMGc117A1852 was then used to initiate genomic walking (contig 2). Because we reasoned that AmphiTlx might be within the resultant contig, we designed degenerate Tlx primers from Ciona intestinalis Tlx (gene identified by BLAST to the Joint Genome Institute Ciona genome assembly at www.jgi.doe.gov) and mammalian Tlx sequences. PCR identified AmphiTlx in clone MPMGc117B1601; a partial cDNA from this gene was also identified by PCR. Contig 3 was initiated by screening cosmid library MPMGc117 with a probe from an AmphiNK1 cDNA, provided by G. Panopoulou, yielding positive clone MPMGc117P1753. Genomic walking from this cosmid established contig 3, which was then screened for homeoboxes by direct sequencing with a degenerate helix three primer (7). Sequencing revealed a second NK1 gene, AmphiNK1b, in cosmid MPMGc118G2336. A partial cDNA of the latter gene was also obtained by PCR. Genomic walking and sequencing identified the two AmphVent and one AmphiLcx genes in this contig. All clone overlaps were confirmed by cross-hybridization and sequencing.
Sequencing.
Genomic phage inserts were recloned into a pUC18 vector modified to contain a NotI site. Shotgun sequencing of phage and cosmid clones was performed at the Wellcome Trust Sanger Institute. Sequence was assembled from reads of Phred quality ≥30, attempts were made to resolve all sequencing problems, compressions, and repeats, and the assembly was confirmed by restriction digestion. Cosmid and phage sequences were analyzed by NIX at the U.K. Human Genome Mapping Project Resource Centre to obtain gene and exon predictions, followed by further comparison by using clustalx alignment to homologues.
Fluorescent in Situ Hybridization.
Fluorescent in situ hybridization to amphioxus embryo metaphase chromosomes used probes from cosmids MPMGc118M0380 (contig 1), MPMGc117A1852 (contig 2), and MPMGc117M0861 (contig 3) and was performed as described (12), except that the hybridizations were overnight at 41°C and washes were at 45°C.
Results
We isolated genomic clones for nine NK-related homeobox genes from the amphioxus B. floridae. Comparisons of deduced homeodomain sequences between amphioxus, vertebrates, and Drosophila (Fig. 1) identifies these genes as orthologues of tin (AmphiNK4), bap (AmphiNK3), Lbx (AmphiLbx), C15 (AmphiTlx), and slou (two genes, AmphiNK1a and AmphiNK1b). We also cloned three divergent amphioxus homeobox genes without homologues in the Drosophila NK cluster [AmphiVent1 (13), AmphiVent2, and AmphiLcx]; these are not discussed further.
Figure 1.
Deduced homeodomain amino acid sequences of amphioxus NK-related homeobox genes aligned to orthologues from D. melanogaster and human. Dashes indicate identical residues.
Genomic walking by using cosmid end fragments allowed us to extend the physical map around each amphioxus gene. We assembled three contigs of 200, 161, and 77 kb (Fig. 2). Five of the cosmids encompassing homeobox genes, and one other cosmid, were completely sequenced; the remaining 12 cosmids were partially sequenced to verify clone overlaps and extend coding sequences. We also fully sequenced two lambda clones from the same genomic regions. The physical maps reveal that the amphioxus NK-like genes are dispersed over an extensive genomic region, far larger than the distance encompassed by the Drosophila 93D/E gene cluster or the amphioxus Hox and ParaHox gene clusters. Contig 1 contains AmphiNK4 and AmphiNK3, the orthologues of Drosophila tinman and bagpipe. No intervening coding sequences are found between these two genes. Contig 2 contains AmphiTlx and AmphiLbx, plus at least three nonhomeobox genes mapping outside the pair of homeobox genes. Contig 3 contains AmphiNK1b, AmphiNK1a, and the three divergent genes, plus at least two nonhomeobox genes (one mapping between two of the homeobox genes; Fig. 2). One of these genes, a member of the synaptojanin gene family, has a distant homologue within the Drosophila NK gene cluster, between C15 and slou.
Figure 2.
Contigs containing amphioxus NK-related homeobox genes. Only cosmid clones are shown, showing RZPD clone ID numbers; phage clones are not shown. Red bars are fully sequenced clones; black bars are partially sequenced clones. Green boxes are homeobox gene exons; yellow circles and ovals are nonhomeobox genes identified; arrows denote transcriptional orientation. G, related to germ cell-less genes; P, catalytic subunit of ser/thr protein phosphatase; S, synaptojanin-related; S18, syntaxin 18-related; Tm, gene for putative transmembrane protein.
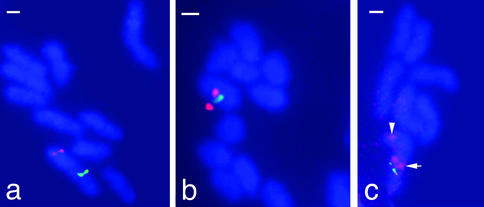
The physical maps alone do not reveal whether the three contigs are adjacent, at distant positions along the same chromosome, or on different chromosomes (B. floridae has 19 chromosome pairs). To resolve between these alternatives, we used two-color fluorescent in situ hybridization to amphioxus metaphase chromosomes with double and triple combinations of cosmid probes. All probe combinations consistently showed that the three contigs map to the same chromosome but are dispersed along it (Fig. 3). Specifically, contig 1 is telomeric, whereas contigs 2 and 3 are closer to the center of a chromosome arm; the physical order of contigs is 1, 3, 2. We conclude that an ancestral tightly linked NK gene cluster has dispersed along a chromosome during evolution. The dispersal is not complete, because tight linkage is retained between AmphiLbx and AmphiTlx (55 kb between coding regions), and especially between AmphiNK3 and AmphiNK4 (10.7 kb between coding regions). Tight linkage also occurs between the two NK1 family genes in amphioxus.
Figure 3.
Fluorescent in situ hybridization to amphioxus metaphase chromosomes. Cosmid probes used were from: (a) contig 1 (red) and contig 2 (green); (b) contig 3 (red) and contig 2 (green); and (c) contig 1 (red, arrowhead), contig 3 (red, arrow), and contig 2 (green). (Bars, 1 μm.)
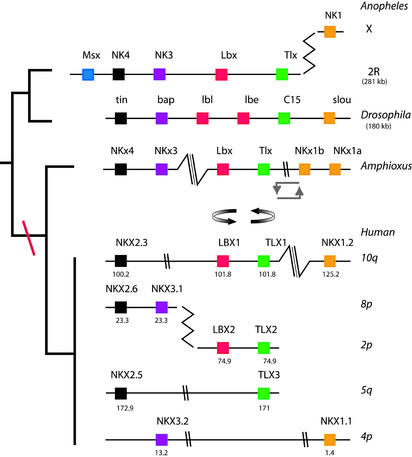
To determine how and when the NK homeobox gene cluster became dispersed, it is necessary to compare with other species. We searched the human and mosquito genome sequences (14, 15) for homologues of the Drosophila and amphioxus NK cluster genes and identified their precise positions and organization (Fig. 4). The genome of the malaria mosquito Anopheles (ensembl release 8.1b.1) was found to include a near intact NK homeobox gene cluster, with tight physical linkage between orthologues of NK4, NK3, a single Lbx and Tlx, plus the Msx gene; the NK1 orthologue was found to be transposed to the X chromosome. We found four dispersed NK homeobox gene clusters in the human genome (ensembl release 7.29.1), as predicted by an earlier study based on band positions (4). These four are clearly descendent from a single set of NK genes on one ancestral chromosome. Duplication from one set to four in the vertebrate lineage mirrors the evolutionary history described for Hox and ParaHox gene clusters in chordates (4, 7, 8).
Figure 4.
Comparison of NK homeobox gene clusters between Anopheles, Drosophila, amphioxus, and human chromosomes. Boxes indicate homeobox genes, color coding denotes orthologous relationships. Cluster breaks are denoted by double-parallel marks (when intergenic distance >1 Mb), triple parallel marks (very large distances), or zigzag (transposition between chromosomes). Angular double arrows denote a large chromosomal inversion in amphioxus; curved double arrows denote a local inversion of a gene pair (relative to the Drosophila gene order). Genomic location is shown for human.
We find that the human NK homeobox gene clusters are broken at the same positions as the amphioxus NK gene cluster. The first split divides homologues of NK3 and NK4 (either single genes or a tightly linked NK3–NK4 gene pair) from TLX and LBX genes (or a TLX–LBX gene pair). This division is seen in at least three of the human clusters (the fourth cluster is lacking TLX and LBX so cannot be assessed for this breakpoint). Two of these splits are manifest as dispersal along a chromosome (HSA10 and HSA5), with the third being transposition to a different chromosome (between HSA8 and HSA2). The second split divides the NK1 genes from the other NK homeobox genes; for example, dispersal of NKX1.2 (10q26.11; RefSeq XM_061241.1) to a large distance from the TLX–LBX genes. The latter genes also reveal a probable inversion in the human lineage. An inversion has also occurred on the amphioxus lineage, involving the Tlx–Lbx pair in relation to the NK1 genes.
It is notable that the intergenic distance between the linked NK4 and NK3 homologues in human (NKX2.6 and NKX3.1) is very small (19.6 kb); this distance is comparable with amphioxus, where an even shorter intergenic spacer separates these genes. Genome data from other species indicate this intergenic distance is consistently short, being <20 kb in all six species examined (3.1 kb in Fugu, 7.2 kb in Drosophila, 8.7 kb in Anopheles, 10.7 kb in amphioxus, 15.4 kb in mouse, and 19.6 kb in human).
Discussion
We have cloned amphioxus orthologues of all of the clustered Drosophila NK-related homeobox genes and mapped their relative positions by genomic walking, DNA sequencing, and chromosomal fluorescent in situ hybridization. The amphioxus data were then compared with the genomic organization of their human orthologues, as deduced from a human genome assembly (14). The most striking finding is that the single amphioxus NK homeobox gene cluster is broken and dispersed at exactly the same intergenic positions as the duplicated human NK homeobox gene clusters, which implies that these two sites of breakage and dispersal arose before the divergence of the vertebrate and cephalochordate lineages; these splits are a chordate or deuterostome character. Neither split has occurred in Drosophila or Anopheles, which retain the ancestral physical linkage of NK4, NK3, Lbx, Tlx, and (in Drosophila) NK1.
The evolutionary history described here for the NK homeobox gene cluster contrasts sharply with that observed for the two other homeobox gene clusters known at present, the Hox and ParaHox clusters. The NK homeobox gene cluster retained an ancestral compact organization in Drosophila, but underwent breakage and dispersal on the chordate lineage. In contrast, both the Hox and ParaHox gene clusters retain the ancestral compact organization in amphioxus and humans, but underwent breakage and dispersal in Drosophila. For example, the Hox cluster is split into two gene complexes (ANT-C and BX-C) in D. melanogaster (16), whereas a split at a different position is seen in Drosophila virilis (17). Furthermore, intergenic distances have expanded in Drosophila; for example, the BX-C spans 279 kb yet contains just three Hox genes. Similarly, a ParaHox gene cluster existed early in metazoan evolution (18), but has been split in Drosophila, with the ind gene now on chromosome 3 and cad on chromosome 2; the third ParaHox gene, Xlox, has been lost.
We suggest that the selective pressures for maintaining homeobox gene clustering are different for the NK class genes and the Hox/ParaHox genes. It has been argued that compact clustering of Hox and ParaHox genes is necessary for an anteroposterior patterning mechanism involving temporal colinearity during and after gastrulation (19). The ancient clustering of Hox or ParaHox genes is prone to disruption; for example, in Drosophila, when evolutionary changes to the developmental program result in less reliance on temporal deployment of these genes during development. We do not know the precise evolutionary reasons underlying maintenance of NK gene clustering in Drosophila, although it is most likely related to sequential functional deployment of these genes during mesoderm development (2). We can deduce, therefore, that this selective pressure was relaxed during the evolution of deuterostome or chordate development.
Total dispersal of the chordate NK homeobox gene clusters has not taken place, because two gene pairs are retained. These linked pairs (NK4 with NK3, and Tlx with Lbx) are maintained in the amphioxus genome; each pair is also evident on at least one chromosome in human, mouse, and Fugu. Indeed, after breakup of the NK gene cluster in chordates, the NK4–NK3 gene pair has remained linked for >1,520 Myr of lineage evolution, comprising at least 515 Myr along each of the cephalochordate and the vertebrate lineages, plus 420 Myr of independent divergence leading to Fugu and at least 70 Myr to mouse (20–23). Whether selection is responsible for preservation of the linkage is difficult to test, because no accurate estimates exist for the rate of local gene order rearrangement in chordate genomes. The rate of intergenic spacer breakage for two nematodes has been estimated as 0.4–1.0 breakages per Mb spacer per Myr (24). Taking the most conservative of these values and extrapolating to chordate evolution, we estimate that an intergenic spacer of 12 kb is expected to be broken 7.3 times in 1,520 Myr. The probability of two randomly chosen genes, separated by such a spacer, staying tightly linked in the absence of selection is approximately P = exp(−7.3) = 0.00067. Because the NK4–NK3 pair was not randomly chosen, but is one of four possible gene pairs from an initial array of five, the corrected probability is ≈1 − (1 − P)4 = 0.0027. Although the extrapolations involved have many caveats, these estimates do accord with the general observation that gene families produced anciently in animal evolution are generally dispersed around genomes. We suggest, therefore, selective reasons exist for maintaining the very tight physical linkage between NK4 and NK3 genes (and possibly between the more loosely linked Tlx and Lbx genes), perhaps because of coregulation.
In summary, a global selective pressure for maintenance of the NK homeobox gene cluster was secondarily relaxed during chordate evolution, allowing dispersal of most genes in the gene cluster. The NK4–NK3 and Tlx–Lbx pairs are evolutionary remnants, possibly retained by a residual selective pressure.
Acknowledgments
We thank Carola Burgtorf, Jim Langeland, and Jordi Garcia-Fernandez for libraries, Pete Currie for Lbx primers; Georgia Panopoulou for kindly providing two cDNAs before publication; and Kerry Ambrose, Hazel Arbery, Joy Davies, Rebecca Deadman, Darren Grafham, Cathy Kidd, Rachel McLean, Suzanna Squares, and Kathy Wright for their contributions to the subcloning and sequencing process. Discussions with Ken Wolfe, Per Ahlberg, Mike Lynch, and Andrew Rambaut were extremely helpful. This work was aided by the efficient work of the Resource Center/Primary Database, Berlin. L.F.C.C. is a Graduate Programme in Applied Basic Biology (GABBA) Ph.D. student funded by Fundação para a Ciência e a Tecnologia, Portugal. This research was funded by a Biotechnology and Biological Sciences Research Council grant (to P.W.H.H.).
Abbreviation
- Myr
million years
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [contig 1 accession nos. are AL513308 (cosmid MPMGc118D2349), AL671994 (cosmid MPMGc118F1011), AL513310 (phage BFP807), and AL513311 (phage BFP809); contig 2 accession nos. are AL672000 (cosmid MPMGc117A1852) and AJ551449 (Tlx sequence); and contig 3 accession nos. are AJ551450 (AmphiNK1b sequence) and AL671989 (cosmid MPMGc117P1753)].
References
- 1.Jagla K, Jagla T, Heitzler P, Dretzen G, Bellard F, Bellard T. Development (Cambridge, UK) 1997;124:91–100. doi: 10.1242/dev.124.1.91. [DOI] [PubMed] [Google Scholar]
- 2.Jagla K, Bellard M, Frasch M. BioEssays. 2001;23:125–133. doi: 10.1002/1521-1878(200102)23:2<125::AID-BIES1019>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Galliot B, de Vargas C, Miller D. Dev Genes Evol. 1999;209:186–197. doi: 10.1007/s004270050243. [DOI] [PubMed] [Google Scholar]
- 4.Pollard S L, Holland P W H. Curr Biol. 2000;10:1059–1062. doi: 10.1016/s0960-9822(00)00676-x. [DOI] [PubMed] [Google Scholar]
- 5.Harvey R P. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 6.Furlong R, Holland P W H. Philos Trans R Soc London B. 2002;357:531–544. doi: 10.1098/rstb.2001.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Fernàndez J, Holland P W H. Nature. 1994;370:563–566. doi: 10.1038/370563a0. [DOI] [PubMed] [Google Scholar]
- 8.Brooke N M, Garcia-Fernàndez J, Holland P W H. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- 9.Ferrier D E K, Minguillon C, Holland P W H, Garcia-Fernandez J. Evol Dev. 2000;2:284–293. doi: 10.1046/j.1525-142x.2000.00070.x. [DOI] [PubMed] [Google Scholar]
- 10.Langeland J A, Tomsa J M, Jackman W R, Kimmel C B. Dev Genes Evol. 1998;208:569–577. doi: 10.1007/s004270050216. [DOI] [PubMed] [Google Scholar]
- 11.Neyt C, Jagla K, Thisse C, Thisse B, Haines L, Currie P D. Nature. 2000;408:82–86. doi: 10.1038/35040549. [DOI] [PubMed] [Google Scholar]
- 12.Castro L F C, Holland P W H. Zool Sci. 2002;19:1349–1353. doi: 10.2108/zsj.19.1349. [DOI] [PubMed] [Google Scholar]
- 13.Kozmik Z, Holland L Z, Schubert M, Lacalli T C, Kreslova J, Vlcek C, Holland N D. Genesis. 2001;29:172–179. doi: 10.1002/gene.1021. [DOI] [PubMed] [Google Scholar]
- 14.International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Holt R A, Subramanian G M, Halpern A, Sutton G G, Charlab R, Nusskern D R, Wincker P, Clark A G, Ribeiro J M, Wides R, et al. Science. 2002;298:129–149. [Google Scholar]
- 16.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 17.Von Allmen G, Hogga I, Spierer A, Karch F, Bender W, Gyurkovics H, Lewis E B. Nature. 1996;380:116. doi: 10.1038/380116a0. [DOI] [PubMed] [Google Scholar]
- 18.Ferrier D E K, Holland P W H. Evol Dev. 2001;3:263–270. doi: 10.1046/j.1525-142x.2001.003004263.x. [DOI] [PubMed] [Google Scholar]
- 19.Ferrier D E K, Holland P W H. Mol Phylogenet Evol. 2002;24:412–417. doi: 10.1016/s1055-7903(02)00204-x. [DOI] [PubMed] [Google Scholar]
- 20.Chen J-Y, Dzik J, Edgecombe G D, Ramskold L, Zhou G Q. Nature. 1995;377:720–722. [Google Scholar]
- 21.Shu D-G, Luo H-L, Conway Morris S, Zhang X-L, Hu S-X, Chen L, Han J, Zhu M, Li Y, Chen L-Z. Nature. 1999;402:42–46. [Google Scholar]
- 22.Zhu M, Yu X, Ahlberg P E. Nature. 2001;410:81–84. doi: 10.1038/35065078. [DOI] [PubMed] [Google Scholar]
- 23.Murphy W J, Eizirik E, Johnson W E, Zhang Y P, Ryder O A, O'Brien S J. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 24.Coghlan A, Wolfe K H. Genome Res. 2002;16:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]