Abstract
The purebred dog population consists of >300 partially inbred genetic isolates or breeds. Restriction of gene flow between breeds, together with strong selection for traits, has led to the establishment of a unique resource for dissecting the genetic basis of simple and complex mammalian traits. Toward this end, we present a comprehensive radiation hybrid map of the canine genome composed of 3,270 markers including 1,596 microsatellite-based markers, 900 cloned gene sequences and ESTs, 668 canine-specific bacterial artificial chromosome (BAC) ends, and 106 sequence-tagged sites. The map was constructed by using the RHDF5000-2 whole-genome radiation hybrid panel and computed by using multimap and tsp/concorde. The 3,270 markers map to 3,021 unique positions and define an average intermarker distance corresponding to 1 Mb. We also define a minimal screening set of 325 highly informative well spaced markers, to be used in the initiation of genome-wide scans. The well defined synteny between the dog and human genomes, established in part as a function of this work by the identification of 85 conserved fragments, will allow follow-up of initial findings of linkage by selection of candidate genes from the human genome sequence. This work continues to define the canine system as the method of choice in the pursuit of the genes causing mammalian variation and disease.
Keywords: dog‖microsatellites‖ESTs‖bacterial artificial chromosome ends
The structure of the canine population offers unparalleled opportunities for understanding the genetic basis of morphology, behavior, and disease susceptibility (1–3). Millions of purebred dogs are newly registered worldwide every year, each of which will be assigned to one of ≈300 well defined “breeds” based on its parentage (4). To maintain physical and behavioral homogeneity, gene flow between breeds is tightly restricted and only a dog whose parents are both registered members of a breed is also eligible for registration.
Global events including world wars and economic depressions have limited the number of founders, and thus restricted the genetic diversity associated with many dog breeds. This, together with the common practice of repeatedly breeding dogs that feature desired physical or behavioral characteristics has resulted in severe population bottlenecks within many breeds, at times reducing the effective breeding stock to only a few individuals (5). The net result is a species characterized by enormous phenotypic diversity, but often at a loss of genome-wide variability (5). As a result, inherited diseases are common in most dog breeds. Researchers concerned with human disease gene mapping are thus afforded a rare opportunity to understand the genetics of diseases that have proven intractable through the study of small, outbred human families (3, 6, 7). In addition, the phenotypic diversity present in modern dog breeds offers developmental biologists an opportunity to decipher the contributions of multiple interacting loci to the seemingly complex phenotypes associated with mammalian development (8). Toward that end, we have developed the resources for mapping and sequencing the dog genome (9–11). Our most recent efforts, summarized herein, encompass a complete mapping resource featuring a 3,270-marker radiation hybrid (RH) map that spans the entire dog genome at 1-Mb resolution, with a well distributed set of microsatellite markers, mapped bacterial artificial chromosome (BAC) ends, and canine-specific genes or ESTs.
Methods
Genotyping.
The panel used in these experiments, RHDF5000-2, comprises 118 cell lines from the original RHDF5000 panel, constructed by fusing dog fibroblasts irradiated at 5,000 rad with TK-HTK3 hamster cells (12). The panel has a retention frequency of 22% with a theoretical resolution limit of 600 kb.
Primers were designed to have an optimal length of 25 nt and a melting temperature of 58–60°C, and result in amplicons of 200–500 bp. PCRs were carried out in 15-μl volumes as described (9–11) by using the following touchdown program: 8 min 95°C, followed by 20 cycles of 30 sec 94°C, 30 sec 63°C decreasing of 0.5°C per cycle, 1 min 72°C and 15 cycles of 30 sec 94°C, 30 sec 53°C, 1 min 72°C, and a final extension of 2 min 72°C. Markers yielding either faint or spurious bands were optimized. Amplification products were resolved and recorded as described (9, 11). Accession numbers, characterization, and PCR conditions for all markers are available at www-recomgen.univ-rennes1.fr/doggy.html and www.fhcrc.org/science/dog_genome/dog.html.
Microsatellite Markers.
New microsatellite markers were isolated and characterized as described (13). The degree of polymorphism was estimated either as a heterozygosity (Het) value or a polymorphic information content (PIC) value after testing a panel of either 5 unrelated mongrel dogs (14) or 10 unrelated purebred dogs representing a subset of the 20 most popular American Kennel Club breeds (13).
BAC-End Sequences.
Plates of BAC clones were randomly selected from the RPC81 canine BAC library (15) for end-sequencing using standard automated approaches (16). Average read lengths were in excess of 700 bp. Primers defining each BAC end were selected from sequence with the highest number of high-quality (HQ) bases. HQ sequence was defined as having 100 continuous sequences with phred scores of 20 or greater. Only one set of primers was used to genotype each BAC; primers designed from the opposite end of the insert were used for genotyping only if the first pair yielded poor-quality data.
Single-Nucleotide Polymorphism (SNP)-Containing Sequence-Tagged Site (STS) Markers.
A genomic library was constructed by cloning 1-kb inserts of mongrel dog genomic DNA into pBluescript KS+II vector. Two hundred clones were sequenced and SNPs were identified after analysis of STS using DNA isolated from 20 dogs representing 20 breeds. Cycle sequencing was performed by using the BigDye Terminator chemistry (Applied Biosystems). Sequencing traces were processed by using phred, phrap, and consed (17–19). SNPs were identified by visual inspection of mismatches detected in the 20 sequencing traces.
Gene and EST-Based Markers.
Primers were designed to amplify known dog gene sequences deposited in public databases by using standard approaches (9, 11). Canine ESTs were isolated from a cDNA library constructed from a Madin–Darby canine kidney cells line by priming with a tailed oligo (dT).
Identification of Orthologous Human Gene Sequences.
Orthologous human sequences were searched by blast analyses (20) against public databases (GenBank “nr” and “HTGS”) by using default criteria and by blat searches (21) against “Human NCBI build 31” sequence. For 95% of the genes, a sequence analogy >80% over 100 nt was observed. The size of 100 bp for sequence comparison was dictated by the size of the available query sequence and not by the absence of analogy. Gene nomenclatures were retrieved from the LocusLink database and human chromosomal locations were confirmed by the University of California Santa Cruz human genome server (Nov. 2002); http://genome.ucsc.edu.
Quality Control.
Approximately 65% of BAC end markers and 30% of gene-based and microsatellite markers were genotyped in duplicate. These correspond to a subset of markers selected at random, as well as to gene markers mapping to regions of synteny breaks. Additional markers were selected from RH groups where ambiguities in ordering were noted and all singletons were also typed in duplicate. Duplicate data were considered consistent when the number of discrepancies between data sets was ≤16%. The percent was calculated as the number of differences over the marker retention value. A threshold limit was determined as corresponding to a distance lower than the resolution limit of the RHDF5000-2 panel. In rare cases, where two independent typings yielded >16% discrepancies, a third typing was done and the resulting vector was either integrated into the map construction, or the marker was discarded if no agreement was observed between two of three genotypes.
Analysis and Map Construction.
Novel markers were incorporated into the previous 1,500-marker RH data set (11) by pairwise calculations using multimap software (22) at a logarithm of odds (lod) threshold ≥8.0. A total of 3,162 markers could be clustered into RH groups. RH groups were ordered by using the traveling salesman problem (TSP) approach as specified by the concorde computer package (23). tsp/concorde computes five independent RH maps; three are variants of the maximum-likelihood estimate approach, and two are constructed by using obligate chromosome breaks. The resulting maps were evaluated to produce a consensus map (24). For markers whose map position was not well supported, genotyping data were reexamined, and genotypes were repeated. When no erroneous genotypes were observed, the problematic linkage group was split into two or more RH groups by using the multimap algorithm and a lod threshold of >9.0.
Inter-marker distances were determined with the rh_tsp_map1.0 version of tsp/concorde, which delivers map positions in arbitrary units. For each chromosome the sum of the arbitrary units was converted into kb by using the known physical size of each chromosome, as determined by cytofluorimetry (25). When more than one RH group was assigned to a chromosome, 350 units were added for each gap, corresponding to the upper limit of our ability to detect linkage between adjacent markers.
Results
General Map Characteristics.
The 1,770 markers added to the canine RH map were typed on the RHDF5000-2 panel described (11, 26). Mapping vectors were added to the previous 1,500-marker map data set (11), and the complete dataset of 3,270 markers was recomputed by using multimap (22) and tsp/concorde (23) software programs. Pairwise linkage analysis at a lod threshold ≥8.0 by using multimap allowed the localization of 3,162 markers to the 38 autosomes and sex chromosomes, leaving only 16 orphan RH groups and 108 unlinked markers. Of the 16 orphan groups, comprising 2–19 markers, 12 could be incorporated into RH groups already assigned to chromosomes by using two-point analyses with lod scores between 5.0 and 8.0. For eight groups the resulting map position is in full agreement with predictions from syntenic human data, and for one group a synteny break is introduced. The four remaining orphan RH groups contain only 14 markers.
Ordering of markers within each RH group was performed by using the tsp/concorde software (23). The number of markers assigned to each autosome ranged from 156 markers at 147 unique positions on chromosome 1 (Canis familiaris, CFA 1) to a minimum of 25 markers at 24 positions (CFA 38). The smallest canine chromosome, the Y, has 10 markers (Table 1). tsp/concorde (23) provides distances between markers in arbitrary units. For each chromosome, we converted the sum of the arbitrary units into kb, with a mean value of 1 unit corresponding to 11.8 kb, as calculated from a subset of well covered chromosomes (Table 1).
Table 1.
Map statistics by chromosome
| CFA | CFA size*, Mb | RH map size, TSP units | Ratio, TSP units/kb | Total no. of positions | Intermarker average distance†, Mb | Total no. of markers | No. of microsatellite markers | No. of gene-based markers | No. of BAC-end markers | No. of STS markers‡ | No. of CFA-specific markers§ | No. of Zoo-FISH CS¶ | No. of RHCS‖ | No. of singletons** |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 137 | 12,353 | 11 | 147 | 0.93 | 156 | 79 | 29 | 42 | 6 | 2 | 4 | 4 | 3 |
| 2 | 99 | 8,233 | 12 | 105 | 0.94 | 114 | 60 | 29 | 20 | 5 | 3 | 4 | 3 | 1 |
| 3 | 105 | 9,165 | 11 | 112 | 0.94 | 116 | 60 | 23 | 27 | 6 | 3 | 3 | 3 | |
| 4 | 100 | 7,587 | 13 | 100 | 1.00 | 115 | 52 | 33 | 23 | 7 | 3 | 3 | 3 | |
| 5 | 99 | 7,896 | 13 | 111 | 0.89 | 123 | 60 | 30 | 30 | 3 | 7 | 4 | 4 | 1 |
| 6 | 87 | 6,056 | 14 | 84 | 1.04 | 99 | 41 | 37 | 19 | 2 | 6 | 3 | 3 | |
| 7 | 94 | 6,834 | 14 | 116 | 0.81 | 136 | 53 | 46 | 31 | 6 | 3 | 2 | 2 | |
| 8 | 86 | 6,399 | 13 | 87 | 0.99 | 92 | 39 | 34 | 18 | 1 | 2 | 1 | 1 | |
| 9 | 77 | 7,557 | 10 | 100 | 0.77 | 111 | 40 | 48 | 16 | 7 | 6 | 2 | 2 | |
| 10 | 80 | 6,722 | 12 | 87 | 0.92 | 96 | 43 | 27 | 21 | 5 | 3 | 3 | 3 | |
| 11 | 86 | 7,257 | 12 | 104 | 0.83 | 107 | 53 | 23 | 25 | 6 | 3 | 2 | 2 | |
| 12 | 85 | 9,002 | 9 | 119 | 0.71 | 133 | 73 | 34 | 23 | 3 | 2 | 1 | 1 | |
| 13 | 75 | 4,392 | 17 | 59 | 1.27 | 68 | 36 | 20 | 11 | 1 | 1 | 2 | 2 | |
| 14 | 72 | 6,867 | 10 | 80 | 0.90 | 86 | 39 | 22 | 22 | 3 | 2 | 1 | 2 | |
| 15 | 75 | 7,523 | 10 | 81 | 0.93 | 82 | 43 | 22 | 13 | 4 | 5 | 3 | 5 | |
| 16 | 73 | 6,863 | 11 | 65 | 1.12 | 69 | 34 | 18 | 17 | 0 | 1 | 2 | 3 | |
| 17 | 80 | 6,081 | 13 | 85 | 0.94 | 93 | 47 | 32 | 12 | 2 | 3 | 2 | 2 | |
| 18 | 66 | 6,967 | 9 | 82 | 0.80 | 89 | 40 | 29 | 17 | 3 | 4 | 2 | 2 | |
| 19 | 66 | 4,468 | 15 | 60 | 1.10 | 70 | 50 | 8 | 11 | 1 | 2 | 2 | 2 | |
| 20 | 66 | 4,219 | 16 | 98 | 0.67 | 107 | 49 | 35 | 19 | 4 | 5 | 2 | 2 | |
| 21 | 61 | 8,045 | 8 | 93 | 0.66 | 95 | 49 | 29 | 16 | 1 | 1 | 1 | 1 | 1 |
| 22 | 61 | 5,349 | 11 | 74 | 0.82 | 82 | 51 | 15 | 16 | 0 | 2 | 1 | 1 | |
| 23 | 61 | 5,382 | 11 | 61 | 1.00 | 62 | 31 | 13 | 17 | 1 | 3 | 1 | 1 | |
| 24 | 73 | 5,341 | 14 | 57 | 1.28 | 63 | 31 | 12 | 17 | 3 | 2 | 1 | 1 | |
| 25 | 60 | 5,833 | 10 | 70 | 0.86 | 72 | 32 | 19 | 17 | 4 | 2 | 3 | 4 | |
| 26 | 48 | 3,256 | 15 | 53 | 0.91 | 57 | 28 | 18 | 11 | 0 | 2 | 2 | 2 | 1 |
| 27 | 57 | 7,116 | 8 | 72 | 0.79 | 78 | 39 | 30 | 8 | 1 | 2 | 1 | 1 | 1 |
| 28 | 55 | 3,332 | 17 | 58 | 0.95 | 61 | 27 | 18 | 14 | 2 | 2 | 1 | 1 | |
| 29 | 51 | 5,230 | 10 | 59 | 0.86 | 59 | 39 | 6 | 9 | 5 | 2 | 1 | 1 | |
| 30 | 47 | 3,740 | 13 | 45 | 1.04 | 48 | 21 | 14 | 10 | 3 | 2 | 1 | 1 | |
| 31 | 50 | 2,779 | 18 | 37 | 1.35 | 38 | 19 | 9 | 8 | 2 | 2 | 2 | 2 | |
| 32 | 51 | 2,477 | 21 | 29 | 1.76 | 30 | 15 | 6 | 8 | 1 | 1 | 1 | 1 | |
| 33 | 41 | 3,561 | 12 | 45 | 0.91 | 49 | 19 | 16 | 14 | 0 | 1 | 1 | 1 | |
| 34 | 50 | 3,863 | 13 | 51 | 0.98 | 58 | 33 | 10 | 13 | 2 | 2 | 1 | 2 | |
| 35 | 38 | 2,406 | 16 | 28 | 1.36 | 33 | 20 | 6 | 7 | 0 | 1 | 1 | 1 | |
| 36 | 41 | 3,968 | 10 | 46 | 0.89 | 47 | 19 | 16 | 11 | 1 | 1 | 1 | 1 | |
| 37 | 40 | 2,877 | 14 | 48 | 0.83 | 52 | 29 | 10 | 12 | 1 | 2 | 1 | 1 | |
| 38 | 38 | 1,783 | 21 | 24 | 1.58 | 25 | 9 | 8 | 8 | 0 | 1 | 1 | 1 | |
| X | 139 | 5,225 | 25 | 53 | 2.62 | 59 | 25 | 20 | 14 | 0 | 1 | 1 | 1 | |
| Y | 27 | 1,714 | 10 | 10 | 2.70 | 10 | 8 | 1 | 1 | 0 | 1 | 0 | 0 | 1 |
| Total assigned | 225,718 | 2,895 | 3,140 | 1,535 | 855 | 648 | 102 | 99 | 71 | 76 | 9 | |||
| Average | 11,8‡‡ | 0.96 | ||||||||||||
| Orphan groups | 1,409 | 20 | 22 | 15 | 0 | 6 | 1 | 0 | ||||||
| Unlinked | 106 | 108 | 46 | 45 | 14 | 3 | 0 | |||||||
| Total | 2,797 | 227,127 | 3,021 | 3,270 | 1,596 | 900 | 668 | 106 | 99 | 71 | 76 | 9 |
Chromosome sizes are given in Mb from cytofluorimetry measurements (25).
Average intermarker distances (in Mb) are calculated by dividing the size of the chromosome by the number of unique positions.
SNP-containing STS and CFA-specific STS markers.
Markers derived from clones for FISH experiments in Breen et al. (11). These markers are included in other marker categories and are not counted in the total number of markers.
Human/dog conserved segments identified from the RH map; a CS comprises two or more markers.
Putative CS identified by RH mapping but containing only one marker.
This value is calculated from the subset of well covered chromosomes (all but CFA5, 32, 35, 38, X, and Y).
The total map size for individual autosomes ranges from 12,353 units (CFA 1) to 1,783 units (CFA 38) (Table 1). The total size of the complete RH map is 227,127 units. The 3,270 markers map to 3,021 unique positions; 249 markers (8%) are copositioned. In one case, CFA 35, five independent markers colocalize to a unique position. The average intermarker distance of the map is 78 units, or ≈900 kb. The present map, therefore, represents a global 2-fold increase in marker density compared with previous iterations of the map (11), with a concomitant 1.5-fold increase in the number of microsatellite markers, a 2.8-fold increase in EST/gene markers and a novel set of mapped BAC end sequences. With this current data set of markers the RHDF5000-2 panel has yet to reach saturation; the resolution of the resulting canine RH map, however, now stands at <1 Mb.
Map Coverage.
We used a variety of different methods to estimate a coverage of 90–95% for the previously reported 1,500-marker RH map (11). In the present effort we have more than doubled the number of markers on the map and, as expected, significantly better genome coverage is now attained. By taking advantage of the fact that some markers placed on the RH map were previously localized by fluorescence in situ hybridization (FISH) (11), we conclude that coverage is now complete or nearly complete for most chromosomes. This is easiest to ascertain when markers corresponding to FISH probes localized to telomeres were then found to map to the extremities of RH groups, or when additional markers were mapped between a FISH probe and a telomeric end; i.e., CFA 34, where six markers were added to the terminal portion of the RH group, and CFA 10 and 23. For chromosomes with complete coverage, one arbitrary unit corresponds to 10–15 kb. We do note, however, that coverage is not absolutely complete for some chromosomes. For instance, comparison of RH and FISH mapping data suggests that CFA 32 and 35 are covered by smaller RH groups than expected; for those chromosomes the arbitrary unit corresponds to 21 and 16 kb, respectively. Also, in the case of CFA 5, we know that a region including and proximal to the p53 gene was not retained when the hybrid lines were constructed (9, 27). In the case of CFA 13 and 38, the number of marker positions (59 and 24, respectively) appears low considering the size of each chromosome (75 and 38 Mb, respectively). Consequently, either marker density is low for these chromosomes and/or coverage is incomplete. The presence of only a single FISH marker located near the middle of the chromosome does not allow us to distinguish between these possibilities.
Despite its large size, a relatively small number of markers have been placed on the canine X chromosome, which can be partly explained by the reported paucity of genes on mammalian X chromosomes. Thus, the existence of several unlinked RH groups of unknown spacing on the X chromosome is not surprising and reported distances probably underestimate true interval size. We did investigate use of a lod threshold of 6.0 rather than 8.0 to see whether the overall X map could be improved. That adjustment does result in generation of two large linkage groups, rather than seven obtained when a lod of 8.0 was used. But the ordering of these two groups was suboptimal and only the map constructed at lod 8.0 is presented.
Microsatellite Characteristics.
In addition to the previously placed 1,078 microsatellites, 518 microsatellite based markers have been added to the map and a total of 1,005, 20, and 571 microsatellite markers based on di-, tri-, and tetranucleotide repeats, respectively, are now mapped. Markers are randomly distributed throughout the chromosomes, ranging from the fewest (9 on CFA 38) to the most (79 on CFA 1). Polymorphism was evaluated by estimation of Het and/or PIC values for markers with 12 or more repeat units. Of these, 77% had Het or PIC values >0.5 and 480 had values >0.7. Because polymorphism levels have not been assessed for every marker on the map, the actual number is likely to be higher.
Minimal Screening Set of Microsatellite-Based Markers for Genome- Wide Scans.
We developed a minimal screening set (MSS-2) of 325 markers with an average spacing of 9 Mb to be used for genome-wide scans in the dog. Criteria for marker selection, in order of preference, included spacing (interval distribution >800 kb and <12,000 kb), informativeness, cleanliness of PCR product, and amplicon size. Preference was given to markers generating PCR products <500 bp. When possible, for chromosomes in which multiple RH groups were present, markers were selected that defined the ends of each RH group. Markers mapping to CFA Y were also selected, as they may prove useful for forensic studies, paternity testing, and for defining pseudoautosomal regions on the sex chromosomes. The final minimal screening set spans 81 RH groups and all chromosomes. The average Het is 0.73, with 171 tetra-, 151 di-, and 3 markers based on trinucleotide repeats. The largest known interval, located on CFA 8 between FH3241 and REN204K13, is 17.1 Mb. Fifty-six markers were also part of the MSS-1 set (28).
A Framework of RH Mapped BAC Clones.
From a selected set of 2,016 BACs we obtained high-quality sequences from either one or both ends of 1,504 BACs (766 for one end only, 738 for both ends). The 4,032 sequences generated had an average of 342 bases with phred scores ≥20. Markers were designed for several hundred clones, and 668 have now been genotyped across the RHDF5000-2 panel. BAC ends are randomly distributed throughout all chromosomes; ranging from one on CFA Y to 42 on CFA 1. These 668 mapped BAC ends constitute an initial framework of clones for anchoring the canine physical map and provide a format for positional cloning studies. A subset of 39 mapped BAC clones also contained microsatellites within the end sequences. These are indicated in Fig. 1 and all associated figures found at www-recomgen.univ-rennes1.fr/doggy.html and www.fhcrc.org/science/dog_genome/dog.html.
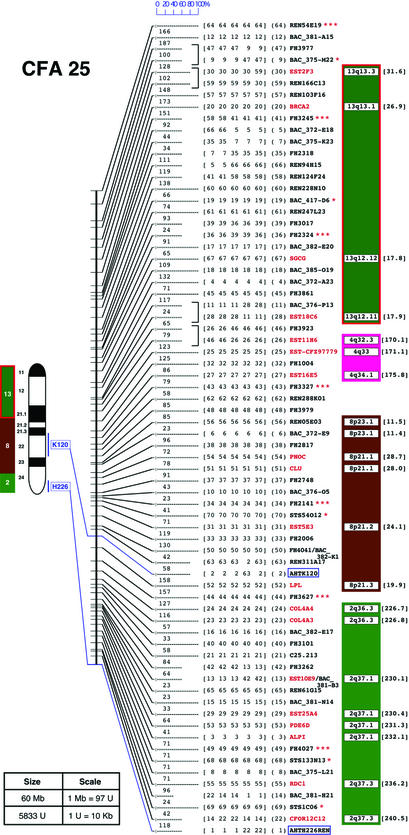
Figure 1.
RH map of CFA25. The position of each marker is reported along the RH map, symbolized by a vertical bar. The RH map shows the five maps generated by TSP/CONCORDE. Maps are highlighted by horizontal bars of variable lengths. When a marker is present on all five maps at the same position, the horizontal bar has a maximum length indicating high confidence; shorter bars reflect a lower confidence level. The scale of 0–100% reflects the relative confidence level. Marker number, as it appears in the consensus map, is indicated in parentheses. In scrambled regions, markers occupying several positions are bracketed to narrow the problematic region into smaller intervals. Marker names indicated in red correspond to gene-based markers (type I); other markers are black (see Table 1 for nomenclature). MSS-2 markers have three asterisks; polymorphic STS, genes, or BAC ends have one. Colored boxes to the right of the markers display human segments with the chromosomal band position. The corresponding position in nucleotide (Mb) of human putative orthologs is indicated between brackets. Data are based on NCBI Build 31. At the left of the RH map, a 4′,6-diamidino-2-phenylindole-banded ideogram is drawn. Markers assigned to chromosomes by FISH are linked to their RH map positions by colored lines (11). Colored bars correspond to the human evolutionary CS. Numbers indicate HSA origin as determined by reciprocal chromosome painting (30, 31). Distances between RH markers are reported in TSP units between horizontal bars. The physical size of each chromosome (in Mb), as determined by flow sorting (25), and the RH group total size (in TSP units) are reported in the frame. The correspondences between TSP unit and kb are also reported in the frame. Figures for all chromosomes are available at www-recomgen.univ-rennes1.fr/doggy.html and www.fhcrc.org/science/dog_genome/dog.html.
STS-Containing SNP Markers.
A total of 200 STS were isolated and sequenced from a canine genomic DNA library. Seventy-eight SNPs were found by sequencing each STS in 20 dogs belonging to different breeds and 72 STSs, containing one to six SNPs, were RH mapped. These are distributed on 29 of 38 canine autosomes. Relevant characteristics including sequence context, minor allele frequency, and heterozygosity can be found at www-recomgen.univ-rennes1.fr/doggy.html and www.fhcrc.org/science/dog_genome/dog.html. These polymorphic markers are indicated by a star in Fig. 1 and all figures found at the web sites listed above.
Gene-Based Markers and Comparative Mapping.
A total of 900 gene based markers were incorporated into the present version of the map, of which 580 are novel, representing a 2.8-fold increase over that presented previously (11). Four hundred forty-one represent novel ESTs for which localization of the human ortholog is known. The remaining 139 are canine gene markers of diverse origins (see the table on the web sites cited above). Some have been shown previously to be polymorphic (29) as indicated by a star in Fig. 1. The distribution of gene-based markers averages one per 3 Mb, with such markers now well distributed across all chromosomes (Table 1). CFA 32 and CFA 36, which lacked any gene-based markers on the previous map (11) now contain 6 and 16 mapped ESTs, respectively.
From the total set of 900 markers, 820 have a known orthologous localization in the human genome. This provides 780 unique positions for comparison with the human genome map. For BAC ends, microsatellites, and STS markers located in regions between conserved segments, the sequences of the original clones were tested by blat searches against the Human “NCBI Build 31” sequence. Of 380 sequences, 50 (13%) gave reliable localizations. Thus, a total of 870 canine mapped sequences occupying 830 unique positions have a known human localization, allowing anchorage of the canine and human genomes.
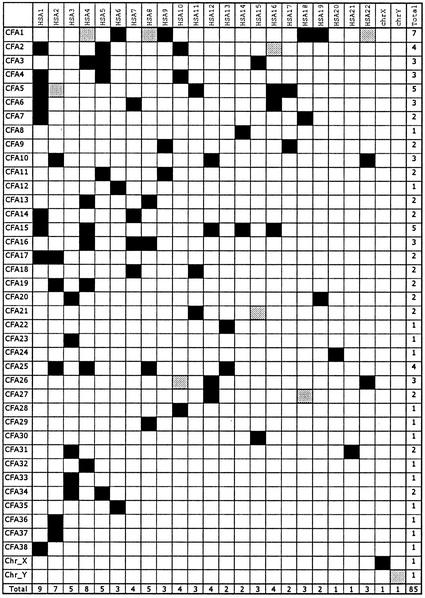
The mapping of these 870 markers allows us to confirm all but one of the conserved segments (CS) detected by human-on-dog chromosome paint studies (30, 31), or those previously identified by RH mapping as singletons (fragment containing only one gene) (11). Only the human chromosome 19 (Homo sapiens, HSA19p13) singleton containing UBA52 was discarded during the present RH map construction. Moreover, five novel CS, containing between two and four mapped genes, all with a high level of sequence analogy with their human counterparts (see Methods) have been detected: CFA14/HSA1, CFA15/HSA14 and HSA16, CFA25/HSA4, and CFA34/HSA5. In addition, five novel singletons (CFA1/HSA8, HSA4, HSA22; CFA5/HSA2; CFA21/HSA15) sharing a high level of sequence identity with their human counterpart (>91% for more than 190 nt) and two with a lower support CFA26/HSA10 (86% over 1,148 nt) and CFA27/HSA18 (85% over 139 nt) are detected. Until other mapped genes confirm their status as CS, the singletons should be interpreted with caution. We believe they are likely to be correct, however, as 16 of the 18 singletons detected previously by using the same criteria (11) have been confirmed by RH mapping of additional markers as conserved segments in this study. In total, therefore, 85 human/dog orthologous fragments corresponding to 76 CS plus 9 singletons, are presently observed by RH mapping (Fig. 2).
Figure 2.
Schematic view of RH conserved segments and singletons between dog and human. CS between both species are illustrated by black squares; singletons are illustrated by gray squares. For each CFA and HSA, the total number of CS is reported in the last column and the last line, respectively.
Conserved syntenic fragments between dog and human are shown for CFA25 on Fig. 1 and illustrated in Fig. 2. A total of 16 dog chromosomes appear to correspond to only one human fragment (CFA8 = most of HSA14q; CFA12 = most of HSA6; CFA22–24, 28–30, 32, 33, and 35–38 plus X and Y). The 24 remaining correspond to between two and seven unique human chromosomal fragments (singletons included) (Fig. 2). Only one human autosome, HSA20, shares exclusive synteny with a unique dog chromosome, CFA24. Gene order at G-banding resolution is also conserved. All other human chromosomes contain from two to nine conserved canine segments with HSA1 containing most. In addition, the size of most previously described chromosomal segments are now substantially extended. Consider, for instance, CFA3 (limits between HSA15 and HSA4) and CFA6 (limits between HSA16 and HSA1) or CFA25, where the limits between human conserved segments HSA13, HSA4, HSA8, and HSA2 are more accurately defined (Fig. 1).
Discussion
Significant progress has been made in the development of the canine genetic system recently (9–11, 32). In recent years we, and others, have demonstrated the genetic power of canines by mapping and/or cloning several disease genes, as summarized in our White Paper Proposal for Sequencing the Canine Genome (www.genome.gov/page.cfm?pageID=10002154). This has led to an increased utilization of the canine system for the development of gene therapy protocols (33–35) or in vivo targeted repair (36). Moreover, the utilization of the map to identify quantitative trait loci appears promising, as demonstrated by the recent study identifying loci for canine morphology and development (8).
This most recent iteration of the map features three major advances: (i) the presentation of a second minimal screening set of markers to be used for undertaking genome-wide scans; (ii) the placement of an initial set of BAC end sequences to facilitate positional cloning studies; and (iii) refinement of the canine/human comparative map.
The first advance featured herein is the presentation of a well characterized minimal screening set of markers (MSS-2) for undertaking genome-wide scans. The density and overall informativeness of this set surpasses that presented previously; the overall Het values are higher, and the coverage across the genome is, at a minimum, 25% denser (28). If we consider that the 325 markers define 253 intervals of known size within RH groups, only 21 of those are ≥12 Mb and a majority (166) are ≥8 Mb and ≤12 Mb. The smallest intervals appear at the ends of radiation hybrid groups, where additional markers were picked to ensure that areas bordering unknown distances beyond RH groups were appropriately covered.
One issue of ongoing concern is the degree to which any set of starting markers will be useful for genome scans in purebred dogs. Some breeds appear as outbred as the general human population, whereas others, because of popular sire effects, bottlenecks, and selective breeding, display limited genetic heterogeneity (5). A key task for the future is the development of markers that are polymorphic in multiple breeds.
A second major advance in the current map is the initial placement of a large set of BAC end sequences. Although this initial data set includes only 668 mapped BAC ends, the resultant density is sufficient that any mapped locus is likely to be close enough to multiple BACs that the construction of minimum tiling paths can be initiated.
The final major advance is summarized by the now detailed information available regarding evolutionary relationships between the human and canine genomes. The First International Workshop on Comparative Genome Organization has suggested several levels of conservation to consider when comparing genomes of two different species. The most relevant at this point in map development are conserved segments, i.e., the syntenic association of two or more contiguous, homologous genes in separate species (37). Previous human-on-dog chromosome painting studies identified 68 conserved chromosome segments, including the X chromosome (30), or 73 excluding the X (31). Conversely, 90 independent segments were identified with dog-on-human chromosome paints (31, 38). The present work is comparable in principle to human-on-dog chromosome paints and the number of conserved segments presented here are best compared with the 68 and 73 reported by Breen et al. (30) and Yang et al. (31). The analysis presented here allowed us to identify all but one previously detected conserved segments (30, 31). In addition we detect 12 novel orthologous fragments, i.e., five chromosomal segments and seven singletons (Table 1). In total, therefore, 85 human/dog orthologous fragments, 76 CS plus 9 singletons, are presently observed by RH mapping (Fig. 2).
When considering the conservation of gene order between human and dog at the human G banding level, for CS harboring more than 10 genes, three interesting situations are observed: (i) CS in which the gene order is very well conserved between the two species, i.e., CFA8/HSA14; CFA12/HSA6, CFA27/HSA12, CFA30/HSA15, CFA33/HSA3, and CFA36/HSA2. (ii) CS that can be split into several blocks where gene order is conserved. This is the case for CFA 4/HSA5, CFA14/HSA7 CFA17/HSA2, CFA21/HSA11, CFA22/HSA13, and CFA24/HSA20. This is often observed when the human orthologous segments span the centromeres. (iii) CS in which the gene order is not conserved, as for CFA 9/HSA17. To more precisely map such CS, denser gene maps built with higher-resolution RH panels will be needed.
Finally, this work highlights the utility and major advantages of using the tsp/concorde algorithm. Recalling that RH maps are not physical maps per se, but rather based on a statistical treatment of a set of mapping vectors, the tsp/concorde algorithm allows an unbiased representation of the data, rather than favoring any single interpretation. In addition, by assigning a level of confidence with which each marker can be assigned to a given position, map users can more appropriately adapt cloning strategies to fit specific needs. Recombination intervals defined by markers positioned with high confidence can reduce the overall workload associated with building a physical map across a region of interest. BACs and ESTs mapped with a high degree of confidence facilitate orientation of the map with the corresponding region of the human genome. The work presented here, therefore, provides a refined set of resources for using comparative approaches to map and clone genes of interest in the canine system.
Acknowledgments
We acknowledge the American Kennel Club Canine Health Foundation, the Burroughs Wellcome Fund, U.S. Army Grant DAAD19-01-1-0658, and National Institutes of Health Grant R01CA-92167 (to E.A.O. and F.G.). R.G. is partly supported by an American Kennel Club and Centre National de la Recherche Scientifique fellowship, and P.Q. is partly supported by a Conseil Regional de Bretagne fellowship. L.K. is supported by a Nestle Purina fellowship, H.G.P. is supported by Public Health Service Grant T32 HG00035, and J.K.L. is supported by Public Health Service National Research Service Award T32 GM07270 from the National Institute of General Medical Sciences.
Abbreviations
- BAC
bacterial artificial chromosome
- CS
conserved segments
- RH
radiation hybrid
- TSP
traveling salesman problem
- SNP
single-nucleotide polymorphism
- FISH
fluorescence in situ hybridization
- STS
sequence-tagged sites
- CFA
Canis familiaris
- HSA
Homo sapiens
- Het
heterozygosity
- lod
logarithm of odds
References
- 1.Ostrander E A, Giniger E. Am J Hum Genet. 1997;61:475–480. doi: 10.1086/515522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrander E A, Galibert F, Patterson D F. Trends Genet. 2000;16:117–123. doi: 10.1016/s0168-9525(99)01958-7. [DOI] [PubMed] [Google Scholar]
- 3.Patterson D. J Vet Intern Med. 2000;14:1–9. [PubMed] [Google Scholar]
- 4.Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. Neptune City, NJ: T.H.F.; 1995. [Google Scholar]
- 5.Ostrander E A, Kruglyak L. Genome Res. 2000;10:1271–1274. doi: 10.1101/gr.155900. [DOI] [PubMed] [Google Scholar]
- 6.Patterson D F. Canine Genetic Disease Information System: A Computerized Knowledgebase of Genetic Diseases in Dogs. St. Louis: Mosby-Elsevier; 2002. [Google Scholar]
- 7.Galibert F, Andre C, Cheron A, Chuat J C, Hitte C, Jiang Z, Jouquand S, Priat C, Renier C, Vignaux F. Bull Acad Natl Med. 1998;182:811–821. [PubMed] [Google Scholar]
- 8.Chase K, Carrier D R, Adler F R, Jarvik T, Ostrander E A, Lorentzen T D, Lark K G. Proc Natl Acad Sci USA. 2002;99:9930–9935. doi: 10.1073/pnas.152333099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priat C, Hitte C, Vignaux F, Renier C, Jiang Z, Jouquand S, Cheron A, Andre C, Galibert F. Genomics. 1998;54:361–378. doi: 10.1006/geno.1998.5602. [DOI] [PubMed] [Google Scholar]
- 10.Mellersh C S, Hitte C, Richman M, Vignaux F, Priat C, Jouquand S, Werner P, André C, DeRose S, Patterson D F, et al. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- 11.Breen M, Jouquand S, Renier C, Mellersh C S, Hitte C, Holmes N G, Cheron A, Suter N, Vignaux F, Bristow A E, et al. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vignaux F, Hitte C, Priat C, Chuat J C, Andre C, Galibert F. Mamm Genome. 1999;10:888–894. doi: 10.1007/s003359901109. [DOI] [PubMed] [Google Scholar]
- 13.Ostrander E A, Sprague G F, Jr, Rine J. Genomics. 1993;16:207–213. doi: 10.1006/geno.1993.1160. [DOI] [PubMed] [Google Scholar]
- 14.Jouquand S, Chéron A, Galibert F. Biotechniques. 1999;26:902–905. doi: 10.2144/99265st03. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Mignot E, Faraco J, Kadotani H, Cantanese J, Zhao B, Lin X, Hilton L, Ostrander E A, Patterson D F, et al. Genomics. 1999;58:9–17. doi: 10.1006/geno.1999.5772. [DOI] [PubMed] [Google Scholar]
- 16.Mahairas G G, Wallace J C, Smith K, Swartzell S, Holzman T, Keller A, Shaker R, Furlong J, Young J, Zhao S, et al. Proc Natl Acad Sci USA. 1999;96:9739–9744. doi: 10.1073/pnas.96.17.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 18.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 19.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kent W J. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matise T C, Perlin M, Chakravarti A. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 23.Agarwala R, Applegate D L, Maglott D, Schuler G D, Schaffer A A. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hitte, C., Lorentzen, T. D., Guyon, R., Kim, L., Cadieu, E., Parker, H., Quignon, P., Lowe, J., Gelfenbeyn, B., Andre, C., et al. (2003) J. Hered., in press. [DOI] [PubMed]
- 25.Langford C F, Fischer P E, Binns M M, Holmes N G, Carter N P. Chromosome Res. 1996;4:115–123. doi: 10.1007/BF02259704. [DOI] [PubMed] [Google Scholar]
- 26.Vignaux F, Priat C, Jouquand S, Hitte C, Jiang Z, Cheron A, Renier C, Andre C, Galibert F. J Hered. 1999;90:62–67. doi: 10.1093/jhered/90.1.62. [DOI] [PubMed] [Google Scholar]
- 27.Jonasdottir T J, Mellersh C S, Moe L, Heggebo R, Gamlem H, Ostrander E A, Lingaas F. Proc Natl Acad Sci USA. 2000;97:4132–4137. doi: 10.1073/pnas.070053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richman M, Mellersh C S, Andre C, Galibert F, Ostrander E A. J Biochem Biophys Methods. 2001;47:137–149. doi: 10.1016/s0165-022x(00)00160-3. [DOI] [PubMed] [Google Scholar]
- 29.Parker H G, Yuhua X, Mellersh C S, Khan S, Shibuya H, Johnson G S, Ostrander E A. Mamm Genome. 2001;12:713–718. doi: 10.1007/s00335-001-2057-3. [DOI] [PubMed] [Google Scholar]
- 30.Breen M, Thomas R, Binns M M, Carter N P, Langford C F. Genomics. 1999;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- 31.Yang F, O'Brien P C, Milne B S, Graphodatsky A S, Solanky N, Trifonov V, Rens W, Sargan D, Ferguson-Smith M A. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- 32.Mellersh C S, Langston A A, Acland G M, Fleming M A, Ray K, Wiegand N A, Francisco L V, Gibbs M, Aguirre G D, Ostrander E A. Genomics. 1997;46:326–336. doi: 10.1006/geno.1997.5098. [DOI] [PubMed] [Google Scholar]
- 33.Acland G M, Aguirre G D, Ray J, Zhang Q, Aleman T S, Cideciyan A V, Pearce-Kelling S E, Anand V, Zeng Y, Maguire A M, et al. Nat Genet. 2001;28:92–95. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- 34.Ponder K P, Melniczek J R, Xu L, Weil M A, O'Malley T M, O'Donnell P A, Knox V W, Aguirre G D, Mazrier H, Ellinwood N M, et al. Proc Natl Acad Sci USA. 2002;99:13102–13107. doi: 10.1073/pnas.192353499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herzog R W, Yang E Y, Couto L B, Hagstrom J N, Elwell D, Fields P A, Burton M, Bellinger D A, Read M S, Brinkhous K M, et al. Nat Med. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett R J, Stockinger S, Denis M M, Bartlett W T, Inverardi L, Le T T, thi Man N, Morris G E, Bogan D J, Metcalf-Bogan J, Kornegay J N. Nat Biotechnol. 2000;18:615–622. doi: 10.1038/76448. [DOI] [PubMed] [Google Scholar]
- 37.Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Mamm Genome. 1996;7:717–734. doi: 10.1007/s003359900222. [DOI] [PubMed] [Google Scholar]
- 38.Sargan D R, Yang F, Squire M, Milne B S, O'Brien P C, Ferguson-Smith M A. Genomics. 2000;69:182–195. doi: 10.1006/geno.2000.6334. [DOI] [PubMed] [Google Scholar]