Abstract
ATP-sensitive potassium channels (KATP channels) regulate cell excitability in response to metabolic changes. KATP channels are formed as a complex of a sulfonylurea receptor (SURx), a member of the ATP-binding cassette protein family, and an inward rectifier K+ channel subunit (Kir6.x). Membrane phospholipids, in particular phosphatidylinositol (PI) 4,5-bisphosphate (PIP2), activate KATP channels and antagonize ATP inhibition of KATP channels when applied to inside-out membrane patches. To examine the physiological relevance of this regulatory mechanism, we manipulated membrane PIP2 levels by expressing either the wild-type or an inactive form of PI-4-phosphate 5-kinase (PIP5K) in COSm6 cells and examined the ATP sensitivity of coexpressed KATP channels. Channels from cells expressing the wild-type PIP5K have a 6-fold lower ATP sensitivity (K1/2, the half maximal inhibitory concentration, ≈ 60 μM) than the sensitivities from control cells (K1/2 ≈ 10 μM). An inactive form of the PIP5K had little effect on the K1/2 of wild-type channels but increased the ATP-sensitivity of a mutant KATP channel that has an intrinsically lower ATP sensitivity (from K1/2 ≈ 450 μM to K1/2 ≈ 100 μM), suggesting a decrease in membrane PIP2 levels as a consequence of a dominant-negative effect of the inactive PIP5K. These results show that PIP5K activity, which regulates PIP2 and PI-3,4,5-P3 levels, is a significant determinant of the physiological nucleotide sensitivity of KATP channels.
The pancreatic ATP-sensitive potassium channel (KATP channel) is formed by association of a sulfonylurea receptor (1) with an inward rectifier K channel subunit (2–6). KATP channels serve as molecular sensors of blood glucose levels to regulate insulin secretion in pancreatic β-cells by coupling cell metabolism to cell excitability. The hallmark feature of KATP channels is their inhibition by intracellular ATP, with half-maximal inhibitory concentration (K1/2) measured by inside-out patch-clamp techniques being ≈10 μM (7, 8). Additionally, in the presence of Mg2+, ADP antagonizes the inhibitory effect of ATP and stimulates channel activity (7, 9). It has long been recognized that activation of KATP channels occurs under conditions where the cytoplasmic ATP is much higher than that required to inhibit channels in excised membrane patches (7, 10), and accumulating evidence suggests that in vivo activation of KATP channels is, at least in part, due to ADP stimulation of channels after glucose starvation (11). Hilgemann and Ball (12) and Fan and Makielski (13) showed that application of phosphoinositides (PIPs), in particular phosphatidylinositol (PI) 4,5-bisphosphate (PIP2), to inside-out membrane patches activates KATP channels. We recently showed that PIP2 dramatically reduced the sensitivity of KATP channels to inhibition by ATP (14, 15). These observations raise the possibility that, in addition to ADP, membrane PIPs may play a key role in physiological activation of KATP channels.
PIPs are phosphorylated derivatives of PI. The PIPs that have been detected in cells include PI-3-P, PI-4-P (PIP), PI-5-P, PI-3,4-P2, PI-4,5-P2 (PIP2), PI-3,5-P2, and PI-3,4,5-P3 (PIP3). PIP and PIP2 are the most abundant forms, comprising ≈60% of total PIPs (16, 17). Comparing the potency of PIP, PIP2, and PIP3 in their ability to modulate the nucleotide sensitivity of KATP channels, we find that PIP2 is much more effective than PIP and about equally effective as PIP3 (14). To examine whether endogenous membrane PIP2 and/or PIP3 can indeed regulate KATP channel activity, we perturbed the levels of these PIPs by manipulating the activity of a lipid kinase involved in their synthesis. Phosphatidylinositol-4-phosphate 5-kinases (PIP5Ks) are enzymes involved in the synthesis of PIP2, PI-3,5-P2, and PIP3. Two isoforms of PIP5Ks were characterized from erythrocytes; these were denoted type I and type II PIP5K based on their biochemical properties (18–20). In mammalian cells, three isoforms of PIP5Ks (α, β, and γ) have been cloned (21–24). Although these isoforms share a marked sequence homology in their central portion (the putative kinase catalytic domain), the N- and C-terminal regions differ significantly. Little is known about the cellular distribution of the various PIP5K isoforms or the cellular processes that are regulated by these kinases (17, 25). In a search for proteins that complement growth factor receptor-dependent mitogenic signaling, Davis et al. (26) isolated a truncated form of the murine type I PIP5K β-isoform that lacks the N-terminal 238 amino acids (PIP5K:Δ1–238) and is inactive in converting PIP to PIP2. The authors showed that overexpression of PIP5K:Δ1–238 resulted in stabilization of a growth factor receptor that normally undergoes endocytosis from the cell surface on ligand stimulation, suggesting a role of PIP5K in receptor endocytosis, possibly by regulating the level of PIP2 and/or PIP3 in the membrane.
In the present study, we overexpressed the wild-type type Iβ PIP5K (PIP5K:WT) or the N-terminally deleted mutant kinase PIP5K:Δ1–238 and examined the effects of these manipulations on nucleotide sensitivity of KATP channels. We report that PIP5K activity plays an important role in determining the ATP sensitivity of these channels.
Materials and Methods
Molecular Biology.
Constructs containing point mutations were prepared by overlap extension at the junctions of the relevant residues by sequential PCR. Resulting PCR products were subcloned into pECE or pCMV6b vector and sequenced to verify the correct mutant construct before transfection.
Expression of KATP Channels in COSm6 Cells.
COSm6 cells were plated at a density of ≈2.5 × 105 cells per well (30-mm six-well dishes) and cultured in DMEM plus 10 mM glucose supplemented with 10% (vol/vol) FCS. The next day, cells were transfected by incubation for 4 h at 37°C in DMEM containing 10% (vol/vol) Nuserum, 0.4 mg/ml DEAE-dextran, 100 μM chloroquine, and 5 μg each of pCMV6b-Kir6.2 or mutant isoforms and pECE-SUR1 cDNA. Cells were subsequently incubated for 2 min in Hepes-buffered salt solution containing 10% (vol/vol) DMSO and returned to DMEM plus 10 mM glucose and 10% (vol/vol) FCS. Cells were assayed for KATP currents by inside-out patch-clamp measurements 2–4 days after transfection.
Construction of Recombinant Sindbis Viruses Expressing the PIP5K:WT and PIP5K:Δ1–238 Mutant.
The two cDNAs were amplified from the pCDNA3 GFP-PIP5K:WT construct (GFP, green fluorescent protein; ref. 26) by PCR (25 cycles). The primers used had a 5′ XbaI linker followed by, first, 15 nucleotides for 5′ PIP5K:WT; second, 24 nucleotides for 3′ PIP5K:WT; and third, 26 nucleotides for the Δ1–238 mutant. The PCR products were subcloned first in the Sindbis shuttle vector (pH2J1; ref. 27). After the correct sequences and orientation of the inserts were confirmed by DNA sequencing, the inserts were subcloned (ApaI and XhoI sites) into the Sindbis virus vector (pTOTO3′2J). RNA transcripts were produced in vitro from pTOTO3′2J PIP5K:WT and PIP5K:Δ1–238 constructs with the SP6-DNA-dependent RNA polymerase. Recombinant viruses were generated from these transcripts for transfection of BHK cells. Viruses were harvested 24 h after transfection and titered (≈108 plaque-forming units/ml) on BHK monolayers.
Preparation of Membrane and Cytosolic Fractions from Cells Expressing GFP-PIP5K:WT and GFP-PIP5K:Δ1–238.
Cells transfected with either PIP5K:WT or PIP5K:Δ1–238 were used to prepare membrane and cytosolic fractions as described (28). Briefly, the cells were homogenized in 20 mM Hepes-NaCl, pH 7.0/0.25 M Sucrose/0.5 mM EGTA and pelleted for 5 min at 600 × g to eliminate nuclei and intact cells. The supernatant was centrifuged for 15 min at 50,000 × g. The pellet (membrane fraction) was resuspended in homogenization buffer containing 1% Triton X-100, 0.5 μM para-PMSF, and 1 μM leupeptin and then frozen in liquid nitrogen and stored at −80°C. Aliquots were used to determine the distribution of GFP-PIP5K:WT and GFP-PIP5K:Δ1–238 mutant by Western blotting by using anti-GFP antibodies (polyclonal antisera from CLONTECH).
Expression of PIP5K in COSm6 Cells.
COSm6 cells transfected with KATP channel protein subunits were subsequently infected with recombinant Sindbis viruses expressing either the wild-type or the mutant PIP5K. High multiplicity of infection (50 plaque-forming units per cell) was employed to ensure high levels of infection. Virus adsorption was performed at 4°C for 1 h with rocking. Cells were incubated in regular medium for another 10–16 h at 37°C before being used for patch-clamp experiments.
Measurement of PIP Levels.
COSm6 cells were plated in 35-mm dishes. At 2 days before use, the medium was aspirated and replaced with inositol-free DMEM containing 3% (vol/vol) FCS and 1 μCi/ml [3H]myo-inositol. Cells were infected with Sindbis viruses expressing various PIP5K isoforms 12 h before the experiment. To measure PIPs, cells were washed in cold PBS, scraped into 1 ml of methanol:concentrated HCl (10:1, vol/vol). One ml of water was added, and the samples were extracted with 2 ml of chloroform. The upper, aqueous layer was removed, and the organic layer was reextracted with methanol:1 M HCl (1:1, vol/vol). Samples were evaporated to dryness, and PIPs were separated on thin layer plates as described (29). Plates were sprayed with En3Hance (NEN) and exposed to x-ray film. PI, PIP, and PIP2 were identified by comigration with standards. Bands corresponding to these lipids were scraped from the plates and counted for 3H.
Patch-Clamp Measurements.
Patch-clamp experiments were made at room temperature in an oil-gate chamber that allowed the solution bathing the exposed surface of the isolated patch to be changed in less than 50 ms. Micropipettes were pulled from thin-walled glass (WPI Instruments, New Haven, CT) on a horizontal puller (Sutter Instruments, Novato, CA). Electrode resistance was typically 0.5–1 MΩ when filled with K-INT solution (see below). Microelectrodes were “sealed” by applying light suction to the rear of the pipette to cells that fluoresced green under UV illumination. Inside-out patches were obtained by lifting the electrode and then passing the electrode tip through the oil-gate. Membrane patches were voltage-clamped with an Axopatch 1B patch-clamp (Axon Instruments, Foster City, CA). The standard bath (intracellular) and pipette (extracellular) solution used in these experiments (K-INT) had the following composition: 140 mM KCl/10 mM K-Hepes/1 mM K-EGTA, pH 7.3. PIP2 was bath sonicated in ice for 30 min before use. All currents were measured at a membrane potential of −50 mV (pipette voltage = +50 mV). Inward currents at this voltage are shown as upward deflections. Data were normally filtered at 0.5–3 kHz; signals were digitized at 22 kHz (Neurocorder, Neurodata, New York) and stored on video tape. Experiments were replayed onto a chart recorder or digitized into a microcomputer by using axotape software (Axon Instruments). Off-line analysis was performed with microsoft excel. Wherever possible, data are presented as means ± SEM. microsoft solver was used to fit ATP-dose response curves by a least-square algorithm. Goodness of fit for the individual fits was tested with the χ2 test of goodness of fit. The χ2 value of all curves is <χ0.052. Test of significance was performed by Student's unpaired t test. P values of <0.05 were considered significantly different.
Results
Expression Level of PIP5K:WT and PIP5K:Δ1–238.
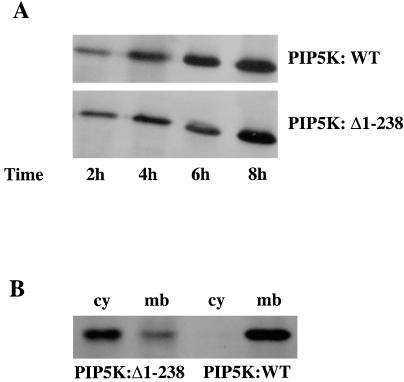
To monitor the expression of PIP5K, we used viral constructs with the GFP attached to the N terminus of PIP5K (Fig. 1). At 10 h after infection, more than 90% of COSm6 cells were green (data not shown). Western blot analysis with anti-GFP antibodies showed that PIP5K:WT and PIP5K:Δ1–238 were expressed at a similar level (Fig. 1), although, interestingly, the subcellular distribution of PIP5K:WT is different from that of PIP5K:Δ1–238. Although the majority of the wild-type kinase is found in the membrane fraction, most of the Δ1–238 kinase is in the cytosolic fraction.
Figure 1.
Expression and distribution of the PIP5K:WT and PIP5K:Δ1–238 mutant. (A) Cells were transfected with Sindbis virus expressing GFP-PIP5K:WT or GFP-PIP5K:Δ1–238 for different times as indicated, and PIP5K proteins were analyzed by Western blotting with anti-GFP antibodies. (B) Distribution of PIP5K:WT and PIP5K:Δ1–238 mutant in membrane and cytosol. After infection, cytosolic (cy) and membrane (mb) fractions were prepared as described in Materials and Methods, and the distributions of PIP5K protein were analyzed by Western blotting with anti-GFP antibodies.
Overexpression of PIP5K:WT Decreases ATP Sensitivity of KATP Channels.
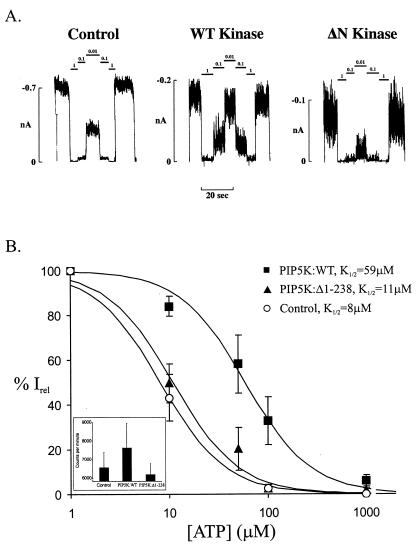
The ATP-sensitivity of KATP channels was measured 10–16 h after viral infection by rapid application of solutions of different ATP concentrations, after excision of inside-out membrane patches. The activity of KATP channels in membrane patches typically decreases within a few minutes, a phenomenon known as channel “rundown.” This phenomenon has been attributed to a decrease in the levels of membrane PIPs (12, 13). To minimize time-dependent changes of PIP concentrations in the membrane patch, which could obscure our results, the ATP-sensitivity was measured immediately (within 1 min) after patch excision. Patches that showed significant channel rundown within the first minute were excluded from the analysis. Representative current traces recorded from control cells and cells infected with PIP5K:WT or with PIP5K:Δ1–238 are shown in Fig. 2A. The dose-response relationships of channel inhibition by ATP were fit by the Hill equation {Irel = 1/[1 + ([ATP]/K1/2)H]; Irel = current in [ATP]/current in zero ATP} to averaged data. For channels from control cells, the K1/2 ([ATP] causing half-maximal inhibition) is 8 μM, with a Hill coefficient (H) of 1.3 (n = 26), similar to the K1/2 values reported previously under identical conditions (30). For cells overexpressing PIP5K:WT, the K1/2 increased by ≈6-fold to a value of 59 μM (n = 15; Fig. 2), significantly higher than that seen in control cells (P < 0.05). Channels in cells infected with a control virus that does not carry the PIP5K:WT gene have ATP sensitivity similar to the sensitivities in uninfected cells (data not shown).
Figure 2.
Overexpression of PIP5K:WT decreases the sensitivity of KATP channels to inhibition by ATP. (A) Representative wild-type KATP channel currents recorded in inside-out membrane patches from control cells, cells overexpressing PIP5K:WT (WT Kinase), or cells overexpressing PIP5K:Δ1–238 (ΔN Kinase). Currents were recorded at −50 mV and are shown as upward deflections. The patch was exposed to differing [ATP] as indicated by the bars above the record. (B) K1/2 estimated for wild-type KATP channels in cells overexpressing PIP5K:WT or cells overexpressing PIP5K:Δ1–238, from fits of the Hill equation {Irel = 1/[1 + ([ATP]/K1/2)H]}, with Irel being the current in ATP, relative to the current in the absence of ATP. The Hill coefficient H was fixed at 1.3. Each data point represents the average of 15–26 patches. The error bar is the SEM. (Inset) PIP2 levels measured in control cells, cells overexpressing PIP5K:WT, or cells overexpressing PIP5K:Δ1–238. Results represent the means ± SD of triplicate determinations.
Overexpression of PIP5K:Δ1–238 Increases the ATP Sensitivity of a Mutant KATP Channel.
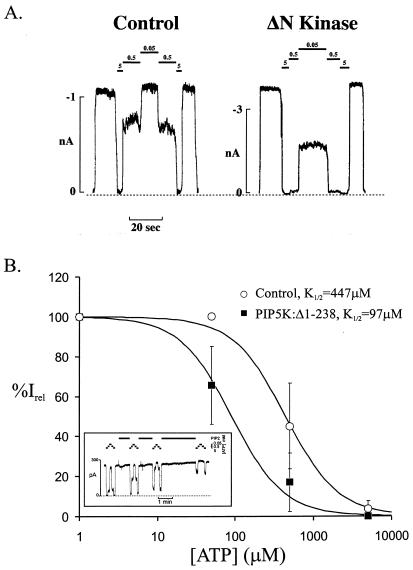
The PIP5K:Δ1–238 mutant lacks kinase activity (26). In addition, it functions as a dominant-negative mutant and blocks the activity of the wild-type PIP5K (J.N.D., unpublished work; A.B. and P.D.S., unpublished work). Overexpression of PIP5K:Δ1–238 is therefore expected to lower the PIP2 levels in the membrane. Consequently, the KATP channels are expected to have a decreased open state stability. Based on experimental data collected from several studies, a positive but nonlinear correlation exists between the channel open probability in the absence of ATP (Po zero) and ATP-sensitivity (K1/2) (D. Enkvetchakul, G. Loussouarn, E. Makhina, S.-L.S., and G.G.N., unpublished work). As open state stability increases above that of wild-type channels, Po zero saturates, and the K1/2, ATP rises. As open state stability falls toward and below that of the wild-type channel (Po zero ≈ 0.4), ATP-sensitivity approaches a minimum (ref. 4; D. Enkvetchakul, G. Loussouarn, E. Makhina, S.-L.S., and G.G.N., unpublished work). Consistent with these observations, we have previously shown that raising the PIP2 in the membrane (or making mutations that intrinsically stabilize the open state of the channel) resulted in reduced ATP-sensitivity. Conversely, lowering the level of accessible PIP2 by treatment with poly-l-lysine or introducing mutations that reduce PIP2 binding (as for the Kir6.2 mutation Arg-176 to Ala) did not increase ATP-sensitivity beyond that of the wild-type channel (14). Accordingly, expression of PIP5K:Δ1–238 had no effect on the ATP-sensitivity of wild-type KATP channels (K1/2 = 11 μM; n = 16; Fig. 2B). To determine whether overexpression of PIP5K:Δ1–238 did decrease membrane PIP2 levels and had an effect on KATP channel activity, we made use of a mutant KATP channel (Kir6.2[I154C, C166S] + SUR1) that has a lower ATP sensitivity than that of the wild-type channels because of an increased intrinsic open state stability (D. Enkvetchakul, G. Loussouarn, E. Makhina, S.-L.S., and G.G.N., unpublished work). Because the ATP-sensitivity is a better indicator of higher channel open state stabilities, the mutant channel is expected to be a better indicator of reductions in PIP2 levels. In control uninfected cells, this mutant channel has a K1/2 of 447 μM (Fig. 3). In cells infected with the PIP5K:Δ1–238 virus, ATP sensitivity of the mutant channel increased by ≈5 fold, with a K1/2 of 97 μM (Fig. 3B), significantly lower than that seen in control uninfected cells (P < 0.05). These results are consistent with the expression of PIP5K:Δ1–238 actually resulting in a decrease in the membrane PIP2 level. To confirm that overexpression of PIP5K:WT results in an increase in membrane PIP2 levels, whereas overexpression of PIP5K:Δ1–238 results in a decrease, we directly measured the amount of cellular PIP2. Whereas cells infected with wild-type kinase (PIP5K:WT) show modest elevation of total PIP2 levels compared with control cells, cells infected with the dominant-negative construct (PIP5K:Δ1–238) show reduced total PIP2 levels (Fig. 2B Inset). Because there is no available assay for surface membrane PIP2 levels, these data provide a lower-limit estimate of PIP2 levels in the surface membrane. The left shift in ATP sensitivity of a mutant KATP channel (Kir6.2[I154C, C166S] + SUR1) coexpressed with kinase lacking enzymatic activity can be restored by subsequent addition of exogenous PIP2. In fact, the ATP-sensitivity can be right shifted to levels well beyond those seen in control cells (Fig. 3B Inset).
Figure 3.
Overexpression of PIP5K:Δ1–238 increases the sensitivity of a mutant KATP channel (Kir6.2[I154C, C166S] + SUR1) to inhibition by ATP. (A) Representative (Kir6.2[I154C, C166S] + SUR1) KATP channel currents recorded in inside-out membrane patches from control cells or cells overexpressing PIP5K:Δ1–238 (ΔN Kinase). Currents were recorded at −50 mV and are shown as upward deflections. The patch was isolated and exposed to differing [ATP] as indicated by the bars above the record. The dashed line indicates zero current. (B) To estimate the K1/2 in control cells or cells overexpressing PIP5K:Δ1–238, data points were fitted to the Hill equation {Irel = 1/[1 + ([ATP]/K1/2)H]}, with Irel being the current relative to the current in the absence of ATP and with H being fixed at 1.3. Each data point represents the average of 4–6 patches, with the error bar being the SEM. (Inset) Response of (Kir6.2[I154C, C166S] + SUR1) KATP channels in an isolated inside-out membrane patch from cells overexpressing PIP5K:Δ1–238 to exogenous PIP2. Currents were recorded at −50 mV and are shown as upward deflections. The patch was exposed to differing [ATP] and 5 μg/ml PIP2 as indicated by the bars above the record. The K1/2 increased from 219 μM to >5 mM with continued PIP2 application.
Discussion
ATP inhibition is the hallmark feature of KATP channels (31). The finding that the ATP sensitivity of KATP channels can be modulated over orders of magnitude by exogenous application of PIP2 on the cytoplasmic side of membrane patches (14, 15) suggests the important possibility that the ATP sensitivity of KATP channels is not a fixed physiological parameter but a parameter controlled by the PIPs in the plasma membrane. The physiological relevance of KATP channel regulation by PIPs has been implicated by our demonstration that a mutant KATP channel (Kir6.2 Arg-176 to Ala), which is predicted to have a lower affinity for PIP2 binding, has very low activity in intact cells (14). Using a Xenopus oocyte expression system, Baukrowitz et al. (15) showed that activation of the P2Y2 receptor, which activates phospholipase C and thus decreases PIP2 concentration, reduced KATP channel activity and rendered channels more sensitive to ATP. The present study shows that the sensitivity of KATP channels to ATP inhibition can be modulated over an order of magnitude in either direction through direct manipulation of the activity of a lipid kinase that is responsible for the synthesis of PIP2 and PIP3. These results provide direct evidence for the hypothesis that PIPs play an important role in the regulation of KATP channel activity in intact cells.
KATP Channels as Molecular Indicators of the PIP2/PIP3 Levels in the Plasma Membrane.
Assuming sufficient supplies of the substrates, overexpression of PIP5K:WT is expected to increase the levels of PIP2, whereas the expression of PIP5K:Δ1–238 is expected to decrease the levels of PIP2. Overexpression of PIP5K:WT or PIP5K:Δ1–238 is correlated with an increase or decrease in the cellular PIP2 levels, respectively. Because we are measuring total cellular PIP2 levels, it is not clear how many of these changes occur at the plasma membrane where the channels are located. The present results suggest that KATP channels might in fact be useful as bioindicators of plasma membrane PIP2/PIP3.
Overexpression of PIP5K:Δ1–238 caused a decrease in the membrane PIP2 or PIP3 levels and increased the ATP sensitivity of a mutant KATP channel with a low intrinsic ATP sensitivity. How the PIP5K:Δ1–238 exerts a dominant-negative effect on the PIP5K:WT activity to lower PIP2/PIP3 levels is not yet clear. It is possible that the PIP5K:Δ1–238 competes for the biosynthesis of the endogenous wild-type PIP5K. It is also possible that wild-type kinases exist as multimers, and the truncated mutant kinase interferes with the normal function of the wild-type PIP5K by competing with the wild-type PIP5K for incorporation into the multimers. The fact that most PIP5K:Δ1–238 is found in the cytosolic fraction, whereas the wild-type kinase is found in the membrane fraction (Fig. 1), suggests the possibility that association of the mutant kinase with the wild-type kinase may lead to mislocalization of the enzyme. No studies have yet been performed to examine the likelihood of multimeric assembly of lipid kinases, but the present results suggest that this distinct possibility warrants investigation.
The Roles of Other Enzymes in the PIP Metabolic Pathway in Regulating KATP Channel Activity.
The metabolic pathways of cellular PIPs are very complex (16, 17). The amount of each PIP is dynamically regulated by the various lipid kinases, lipid phosphatases, and phospholipases. To have a comprehensive understanding of how KATP channel activity is regulated by PIPs, the role of individual kinase, phosphatase, and lipase needs to be evaluated, potentially by using approaches like the one presented herein. The enzyme activities of lipid kinases, phosphatases, and lipases are acutely modulated by hormones and growth factors. An important future direction is to identify physiological stimuli that use the PIP signaling pathway to regulate KATP channel activity.
Acknowledgments
The original Kir6.2 clone was a gift from Dr. S. Seino. We thank Dr. G. Loussouarn for providing technical assistance. This work was supported by a Career Development Award from the American Diabetes Association (to S.-L.S), by Grants HL45742 (to C.G.N.) and GM42559-27 (to P.D.S.) from the National Institutes of Health, and by the Department of Biochemistry and Molecular Biology and the Feist–Weiller Cancer Center, Louisiana State University Health Sciences Center (to J.N.D.). We are grateful to the Diabetes Research and Training Center at Washington University for continued support with oligonucleotide synthesis.
Abbreviations
- KATP channel
ATP-sensitive potassium channel
- PI
phosphatidylinositol
- PIP
phosphoinositide
- PIP2
PI 4,5-bisphosphate
- PIP3
PI-3,4,5-P3
- PIP5K
PI-4-phosphate 5-kinase
- K1/2
half-maximal inhibitory concentration
- GFP
green fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Aguilar-Bryan L, Nichols C G, Wechsler S W, Clement J P, IV, Boyd A E, III, Gonzalez G, Herrera-Sosa H, Nguy K, Bryan J, Nelson D A. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 2.Inagaki N, Tsuura Y, Namba N, Masuda K, Gonoi T, Horie M, Seino Y, Mizuta M, Seino S. J Biol Chem. 1995;270:5691–5694. doi: 10.1074/jbc.270.11.5691. [DOI] [PubMed] [Google Scholar]
- 3.Inagaki N, Gonoi T, Clement J P, IV, Namba N, Inazawa J, Gonzales G, Aguilar-Bryan L, Seino S, Bryan J. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 4.Shyng S L, Nichols C G. J Gen Physiol. 1997;110:655–664. doi: 10.1085/jgp.110.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement J P, IV, Kunjilwar K, Gonzalez G, Schwanstecher M, Panten U, Aguilar-Bryan L, Bryan J. Neuron. 1997;18:827–838. doi: 10.1016/s0896-6273(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 6.Inagaki N, Gonoi T, Seino S. FEBS Lett. 1997;409:232–236. doi: 10.1016/s0014-5793(97)00488-2. [DOI] [PubMed] [Google Scholar]
- 7.Ashcroft F M. Annu Rev Neurosci. 1988;11:97–118. doi: 10.1146/annurev.ne.11.030188.000525. [DOI] [PubMed] [Google Scholar]
- 8.Inagaki N, Gonoi T, Clement J P, IV, Wang C Z, Aguilar-Bryan L, Bryan J, Seino S. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 9.Nichols C G, Lederer W J. Am J Physiol. 1991;261:H1675–H1686. doi: 10.1152/ajpheart.1991.261.6.H1675. [DOI] [PubMed] [Google Scholar]
- 10.Ashcroft F M. Science. 1998;282:1059–1060. doi: 10.1126/science.282.5391.1059. [DOI] [PubMed] [Google Scholar]
- 11.Nichols C G, Shyng S-L, Nestorowicz A, Glaser B, Clement J, IV, Gonzalez G, Aguilar-Bryan L, Permutt A M, Bryan J P. Science. 1996;272:1785–1787. doi: 10.1126/science.272.5269.1785. [DOI] [PubMed] [Google Scholar]
- 12.Hilgemann D W, Ball R. Science. 1996;273:956–959. doi: 10.1126/science.273.5277.956. [DOI] [PubMed] [Google Scholar]
- 13.Fan Z, Makielski J C. J Biol Chem. 1997;272:5388–5395. doi: 10.1074/jbc.272.9.5388. [DOI] [PubMed] [Google Scholar]
- 14.Shyng S-L, Nichols C G. Science. 1998;282:1138–1141. doi: 10.1126/science.282.5391.1138. [DOI] [PubMed] [Google Scholar]
- 15.Baukrowitz T, Schulte U, Oliver D, Herlitze S, Krauter T, Tucker S J, Ruppersburg J P, Fakler B. Science. 1998;282:1141–1144. doi: 10.1126/science.282.5391.1141. [DOI] [PubMed] [Google Scholar]
- 16.Carpenter C L, Cantley L C. Curr Opin Cell Biol. 1996;8:153–158. doi: 10.1016/s0955-0674(96)80060-3. [DOI] [PubMed] [Google Scholar]
- 17.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 18.Ling L E, Schulz J T, Cantley L C. J Biol Chem. 1989;264:5080–5088. [PubMed] [Google Scholar]
- 19.Bazenet C E, Ruano A R, Brockman J L, Anderson R A. J Biol Chem. 1990;265:18012–18022. [PubMed] [Google Scholar]
- 20.Jenkins G H, Fisette P L, Anderson R A. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- 21.Ishihara H, Shibasaki Y, Kizuki N, Katagiri H, Yazaki Y, Asano T, Oka Y. J Biol Chem. 1996;271:23611–23614. doi: 10.1074/jbc.271.39.23611. [DOI] [PubMed] [Google Scholar]
- 22.Castellino A M, Parker G J, Boronenkov I V, Anderson R A, Chao M V. J Biol Chem. 1997;272:5861–5870. doi: 10.1074/jbc.272.9.5861. [DOI] [PubMed] [Google Scholar]
- 23.Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. J Biol Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 24.Itoh T, Ijuin T, Takenawa T. J Biol Chem. 1998;273:20292–20299. doi: 10.1074/jbc.273.32.20292. [DOI] [PubMed] [Google Scholar]
- 25.Anderson R A, Boronenkov I V, Doughman S D, Kunz J, Loijens J C. J Biol Chem. 1999;274:9907–9910. doi: 10.1074/jbc.274.15.9907. [DOI] [PubMed] [Google Scholar]
- 26.Davis J N, Rock C O, Cheng M, Watson J B, Ashmun R A, Kirk H, Kay R J, Roussel M F. Mol Cel Biol. 1997;17:7398–7406. doi: 10.1128/mcb.17.12.7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Stahl P D. J Biol Chem. 1993;268:24475–24480. [PubMed] [Google Scholar]
- 28.Mayorga L S, Diaz R, Stahl P D. J Biol Chem. 1989;264:5392–5399. [PubMed] [Google Scholar]
- 29.Pike L J, Eakes A T. J Biol Chem. 1987;262:1644–1651. [PubMed] [Google Scholar]
- 30.Shyng S-L, Ferrigni T, Nichols C G. J Gen Physiol. 1997;110:141–153. doi: 10.1085/jgp.110.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noma A. Nature (London) 1983;305:147–148. doi: 10.1038/305147a0. [DOI] [PubMed] [Google Scholar]