Abstract
PD-L1 and PD-L2 are ligands for PD-1, a costimulatory molecule that plays an inhibitory role in regulating T cell activation in the periphery. We find that PD-L1 is highly expressed on inflammatory macrophages as compared with resident peritoneal macrophages but can be induced on resident macrophages by classical activation stimuli such as lipopolysaccharide, IFN-γ, and polyinosinic-polycytidylic acid. Further up-regulation of PD-L1 on inflammatory macrophages can also be induced by subsequent exposure to lipopolysaccharide and IFN-γ. In contrast, PD-L2 is not expressed on inflammatory macrophages but can be induced by alternative activation via IL-4. Although PD-L1 is highly inducible on a variety of antigen-presenting cell lines as well as resident macrophages, PD-L2 is most significantly inducible only on inflammatory macrophages. PD-L1 up-regulation depends on TLR4 and STAT1, whereas PD-L2 expression depends on IL-4Rα and STAT6. Consistent with these results, T helper 1/T helper 2 (Th1/Th2) cells also differentially up-regulate PD-L1 and PD-L2 expression on inflammatory macrophages. Hence, Th1 cells as well as microbial products can enhance PD-L1 expression on many different macrophage populations, whereas Th2 cells instruct only inflammatory macrophages to up-regulate PD-L2. These results suggest that PD-L1 and PD-L2 might have different functions in regulating type 1 and type 2 responses.
Keywords: IL-4‖Stat6‖Stat1‖B7-DC‖B7-H1
Inflammation in the peripheral tissues is the site of important interactions between effector T cells and innate immune cells. Although interactions of the classical costimulatory molecules CD28/CTLA-4 and their ligands B71/B72 are crucial in the primary activation of naïve T cells by dendritic cells (1), the more recently described members of this family (ICOS:B7H, PD-1:PD-L1/PD-L2) have been implicated in the regulation of effector T cell function, especially in peripheral tissues (2). Because inflammatory monocyte/macrophages often are the major innate immune cell type in peripheral tissue inflammation, their interactions with effector T cells should be a crucial determinant of the course of the immune response. However, this macrophage/T cell interaction is not well understood, especially with regard to the recently described costimulatory molecules and ligands.
PD-1 (for programmed death 1) was originally identified by subtractive hybridization in T cells undergoing apoptosis (3) but has recently been recognized as a costimulatory molecule that is thought to provide an inhibitory signal. Interestingly, mice deficient in PD-1 suffer from a lupus-like glomerulonephritis and arthritis on the C57BL/6 background (4), whereas they have a fatal autoimmune dilated cardiomyopathy in BALB/c mice (5). These observations suggest a role for PD-1 in regulating peripheral tolerance. Two ligands that belong to the B7 family have been identified for PD-1, PD-L1 (also B7-H1; refs. 6 and 7), and PD-L2 (also B7-DC; refs. 8 and 9). PD-L1 is expressed on macrophages and tumor cells (10), whereas PD-L2 is largely restricted to dendritic cells (9). A more recent study showed that PD-L1 was up-regulated further on macrophages by lipopolysaccharide (LPS), IFN-γ, granulocyte/macrophage colony-stimulating factor, and IL-4, whereas PD-L2 was induced on macrophages by IFN-γ, granulocyte/macrophage colony-stimulating factor, and IL-4 (11). Although the phenotype of PD-1-deficient mice clearly suggests an inhibitory role for this molecule, its ligands have been reported to have both positive (9) as well as negative capabilities (6, 8).
Macrophages are classically activated by engagement of Toll-like receptors (TLRs) (12) or the binding of IFN receptors by IFN-α/β or IFN-γ (13). Classically activated macrophages are critically involved in the destruction of intracellular pathogens and often are regulated by T helper 1 (Th1) cells or CD8+ cells through IFN-γ (14). Recently, it has become recognized that macrophages also can be activated by an alternative pathway involving the type 2 cytokines IL-4 and IL-13 (15, 16). Although these macrophages have been implicated in performing immunoregulatory roles through their in vitro activity, the in vivo function of these alternatively activated macrophages remains unclear. However, the prevalence of these alternatively activated macrophages as one of the major inflammatory cell types in chronic Th2 inflammatory conditions such as allergy (17) and parasitic infection (18) strongly suggests that they are important in type 2 responses. Here, we show that when inflammatory macrophages are activated by IL-4 or Th2 cells, they strongly up-regulate PD-L2, a costimulatory ligand that could be important in regulating T cell function. Apart from being a potential indicator of IL-4 activity on macrophages, PD-L2 might be playing an important role in the immunoregulatory effects of alternatively activated macrophages.
In recent years, much emphasis has been placed on how antigen-presenting cells “instruct” T cells through costimulatory ligands. Here, we show that differentiated effector T cells instruct macrophages to express different costimulatory ligands. Interestingly, induction of PD-L2 is most pronounced on inflammatory macrophages, whereas PD-L1 is inducible on all cell types. We examined the mechanisms regulating PD-L1 and PD-L2 expression and found that, although PD-L1 is stably expressed on inflammatory macrophages, increased expression as a result of stimuli is associated with rapid turnover of this molecule. The up-regulation of both PD-L1 and PD-L2 depends on transcription and translation. Furthermore, we show that up-regulation of PD-L1 by LPS is TLR4-dependent and that Stat1 signaling also is critical in regulating PD-L1 expression, whereas expression of PD-L2 is regulated by IL-4Rα and Stat6. The differential regulation of PD-L1 and PD-L2 suggests that they might have different functions under different Th1/Th2 inflammatory situations, although they both bind PD-1.
Materials and Methods
Animals and Treatment.
C57BL/6, BALB/c, C3H/HeJ, and IL-4Rα−/− mice and Stat6−/− mice on the BALB/c background were obtained from The Jackson Laboratory. OT2 Rag−/− and DO11.10 Rag−/− mice were bred in-house. Stat1−/− mice on a 129S6/SvEv background were obtained from Taconic Farms. Mice used in experiments were 6–8 weeks old. Both male and female mice were used with identical results. To classically activate resident macrophages, mice were injected i.p. with 500 μg of polyinosinic-polycytidylic acid (Poly IC). To induce inflammatory macrophages, mice were injected with 3–4 ml of 4% thioglycolate. Forty-eight to 72 h later, peritoneal macrophages were washed out with RPMI 1640 medium.
Macrophage Activation.
Peritoneal macrophages were adhered onto 12-well plates (1–2 × 106 cells) or six-well plates (2 × 106 cells) for between 2 and 12 h. Nonadherent cells were removed by washing two to three times with PBS. Unless otherwise stated, adherent macrophages then were cultured with medium alone or activated with IL-4 (20 ng/ml) or LPS + IFN-γ (100 ng/ml each) for 24 h. Recombinant mouse IL-4 and IFN-γ were purchased from R & D Systems. LPS was purchased from Sigma. Activated macrophages were harvested with a cell scraper for FACS analysis. In time course studies as well as some of the other experiments, harvested cells were frozen (in FCS and 10% DMSO) and stained together at the end of the experiment. Nitric oxide production was measured by incubating an equal volume of supernatant with Griess Reagent (Sigma), using NaNO2 as a standard.
Antibodies and Flow Cytometry.
FITC-conjugated α-F4/80 was purchased from Caltag (South San Francisco, CA). Phycoerythrin-labeled α-PD-L1, α-PD-L2, and rat IgG2A isotype control were purchased from eBioscience (San Diego). For FACS analysis, cells (2–5 × 105) were preincubated with unlabeled α-CD16/32 (24G2) and then incubated with relevant antibodies. After washing three times with PBS, cells were fixed with paraformaldehyde (2%) before FACS analysis.
T Cell Activation and Differentiation.
To generate Th1/Th2 cells, lymph node cells from OT2 RAG−/− and DO11.10 RAG−/− mice were stimulated with 10 μg/ml OVA (323–329) peptide (ISQAVHAAHAEINEAGR) and irradiated splenocytes from C57/BL6 and BALB/C mice, respectively, in the presence of IL-12 (5 ng/ml) and α-IL-4 (11B11) (20 μg/ml) for Th1 cells and IL-4 (50 ng/ml) and α-IFN-γ (50 mg/ml) for Th2 cells. T cells were expanded after 48 h and rested for 7–10 days before being purified over Histopaque 1119 (Sigma) and restimulated or used in assays. To activate inflammatory macrophages, 1 × 105 rested T cells were cocultured with 1 × 106 macrophages in 12-well plates for 24 h in the presence or absence of OVA peptide. All cell culture was performed in RPMI 1640 medium supplemented with 10% FCS, l-glutamine (2 μm), penicillin (100 units/ml), streptomycin (100 units/ml), and 2-mercaptoethanol (2 μm).
Results
Differential Induction of PD-L1 and PD-L2 by Classical vs. Alternative Activation.
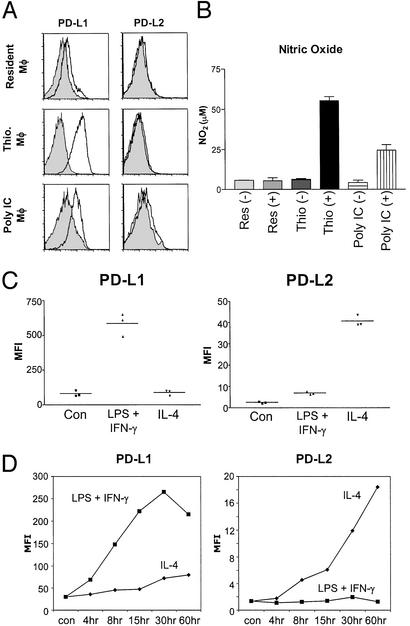
Because PD-1 ligands have been implicated in regulating effector T cells in the periphery (2), we decided to investigate the expression of PD-L1 and PD-L2 in macrophage populations under different conditions in peripheral tissues. When resident macrophages in the peritoneal cavity were analyzed ex vivo, we found that they expressed low levels of PD-L1 and no PD-L2 (Fig. 1A). Inflammatory macrophages that were recruited into the peritoneal cavity by thioglycolate injection expressed high levels of PD-L1 ex vivo, but no PD-L2. Resident macrophages could be induced to up-regulate PD-L1 expression by in vivo exposure to double-stranded RNA in the form of Poly IC, which stimulates IFN-α/β production (19). Poly IC treatment did not significantly increase the number of macrophages in the peritoneal cavity, suggesting that it is inducing PD-L1 expression on resident macrophages. Importantly, Poly IC treatment did not induce PD-L2. When we investigated the sensitivity of these different macrophage populations to subsequent LPS and IFN-γ stimulation, we found that PD-L1 expression correlated with the ability of macrophages to produce NO after stimulation in vitro (Fig. 1B). Thioglycolate-recruited macrophages that express the highest levels of PD-L1 produced the highest amount of NO, as compared with Poly IC-activated resident macrophages. Resident macrophages did not produce detectable levels of NO when activated by LPS and IFN-γ. These results suggest that macrophages have to be in a “primed” state before they can be responsive to subsequent stimulation to produce nitric oxide. This result is consistent with recently published work by Hu et al. (20), which showed that different populations of monocyte/macrophages have different sensitivities to IFN-γ stimulation and that subthreshold concentrations of IFN-γ will increase sensitivity to subsequent IFN-γ stimulation.
Figure 1.
Expression of PD-L1 and PD-L2 on macrophages. (A) PD-L1 but not PD-L2 is expressed on inflammatory and activated resident macrophages. Peritoneal macrophages from untreated mice (Resident MΦ), mice injected with thioglycolate (Thio. MΦ), and mice injected with Poly IC (Poly IC MΦ) were stained for PD-L1 and PD-L2 expression (solid line), ex vivo, 72 h after treatment and compared with rat IgG2A isotype control antibody staining (shaded histogram). Data shown are representative of at least three separate mice in different experiments. (B) NO production after LPS and IFN-γ stimulation by macrophages expressing PD-L1. PD-L1-expressing macrophages produce more NO. Adherent peritoneal macrophages from treated mice were activated with LPS and IFN-γ (100 ng/ml) for 24 h before the supernatants were assayed for NO2. SD bars indicate variation between animals in treatment groups. (C) Differential up-regulation of PD-L1 and PD-L2 by type 1 and type 2 stimuli on inflammatory macrophages. Inflammatory macrophages from thioglycolate-treated mice were activated with IL-4 (20 ng/ml) or LPS + IFN-γ (100 ng/ml each) or cultured in medium alone for 24 h, after which cells were harvested and stained for PD-L1 and PD-L2 expression as well as for F4/80. Mean fluorescence intensity (MFI) of staining results shown are from F4/80+ gated cells alone (typically >90%). Data shown are representative of >10 experiments with similar results. (D) Time course of PD-L1 and PD-L2 up-regulation after stimulation. Macrophages were activated as described above and harvested at the indicated time points. Harvested cells were frozen (in FCS + 10% DMSO) and stained together at the end of the experiment. Time course data shown are representative of three separate experiments.
We then decided to investigate how PD-L1 and PD-L2 were regulated on inflammatory macrophages in response to subsequent stimulation. When inflammatory macrophages were classically activated by LPS and IFN-γ, they strongly up-regulated PD-L1, far above the constitutively high levels of expression by these macrophages. Interestingly, when they were activated by the type 2 cytokine IL-4, they strongly up-regulated PD-L2 instead (Fig. 1C). The induction of PD-L1 and PD-L2 by classical vs. alternative stimuli was not mutually exclusive, because IL-4 also stimulates some PD-L1 up-regulation and LPS and IFN-γ also stimulate some PD-L2 expression, especially after longer periods of time (Fig. 1D). This is consistent with the observations of Yamazaki et al. (11). However, there is a dramatic difference in the level of PD-L1 and PD-L2 up-regulation depending on classical vs. alternative activation. It remains to be determined whether the effects of IL-13 are the same or similar to IL-4.
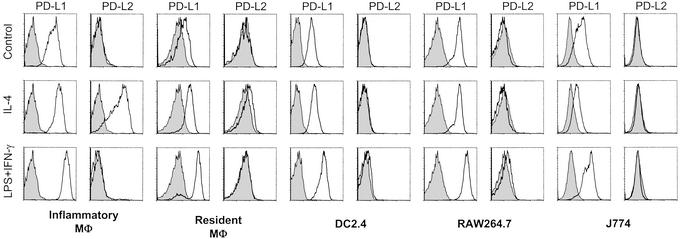
It was important to determine whether this differential regulation by classical vs. alternative activation was restricted to inflammatory macrophages or was common to other macrophage populations, including antigen-presenting cell lines (Fig. 2). We found that the macrophage cell lines J774 and RAW264.7 and the dendritic cell line DC2.4 could be induced to express higher levels of PD-L1 after stimulation with LPS and IFN-γ but could not be induced to express PD-L2 by IL-4. Resident macrophages from the peritoneal cavity showed the same pattern of inducible expression, although there is a slight increase in PD-L2 expression upon IL-4 activation. Therefore, PD-L1 is easily inducible on a number of different cell types, whereas PD-L2 expression is much more restricted and can be highly up-regulated by IL-4 only on inflammatory macrophages.
Figure 2.
PD-L1 is inducible on cell lines and resident macrophages but PD-L2 is inducible only on inflammatory macrophages. Peritoneal macrophages from untreated mice (Resident MΦ) and thioglycolate-injected mice (Inflammatory MΦ) were activated together with cell lines with IL-4 (20 ng/ml) or LPS + IFN-γ (100 ng/ml each) for 24 h and stained for PD-L1/L2 expression. Data shown are representative of at least two experiments with each cell type.
Mechanisms of PD-L1 and PD-L2 Induction.
Very little is known about the mechanisms that regulate expression of PD-L1 and PD-L2. Our first goal was to determine whether transcription and translation were required for increased expression after stimulation. Although the kinetics of PD-L1 and PD-L2 up-regulation (Fig. 1D) suggested that protein synthesis was involved, it was important to exclude changes in the trafficking and differential cellular localization of these molecules. It was also important to determine the turnover rate of PD-L1 and PD-L2, because this could have important consequences for the engagement of their receptor(s) on T cells or other cells.
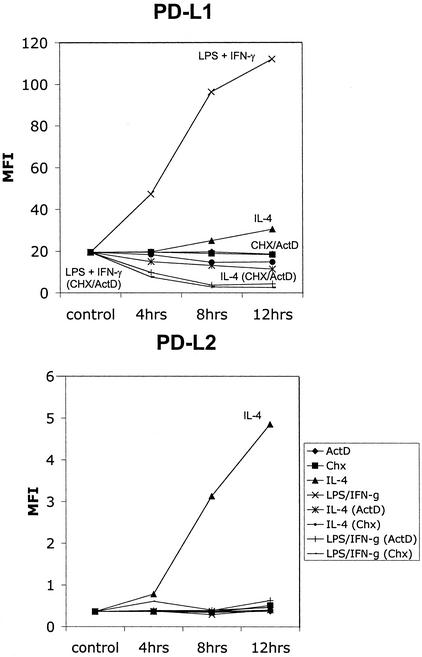
Inflammatory macrophages were activated classically or alternatively in the presence of either actinomycin D or cycloheximide to inhibit transcription and translation. Interestingly, the expression of PD-L1 rapidly decreased in classically activated macrophages in the absence of transcription and translation, whereas PD-L1 expression was stable on unactivated control macrophages (Fig. 3). In alternatively activated macrophages, there was a slight decrease in PD-L1 expression that was consistent with the slight up-regulation in PD-L1 expression by IL-4. These results showed that the expression of PD-L1 is stable on inflammatory macrophages and that increased expression as a result of stimulation is associated with increased turnover. The expression of PD-L1 is especially dynamically regulated by classical stimulation. PD-L2 is not expressed on the surface of unactivated macrophages, and its up-regulation by alternative activation with IL-4 is inhibited by blocking of transcription and translation. Protein synthesis is therefore necessary for PD-L2 expression.
Figure 3.
PD-L1 and PD-L2 induction depend on transcription and translation. PD-L1 is turned over rapidly after activation with LPS + IFN-γ. Thioglycolate-induced macrophages were activated with LPS + IFN-γ and IL-4 in the presence or absence of actinomycin D (1 μg/ml) or cycloheximide (10 μg/ml) and harvested at various time points. MFI of staining results shown is from F4/80+ gated cells. Data shown are representative of two experiments.
PD-L1 Expressions Is Regulated by TLR4 and Stat1.
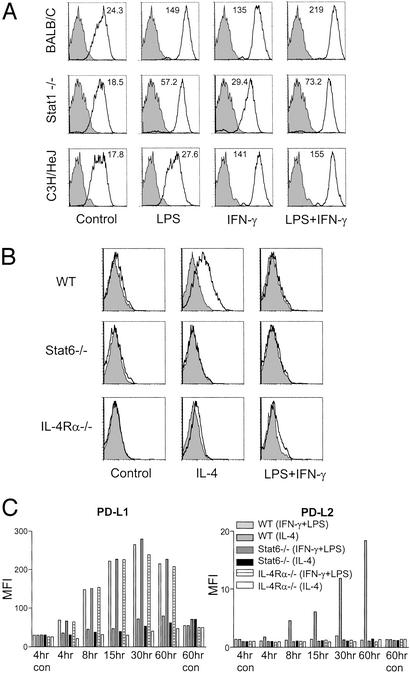
We decided to examine further the mechanism behind the induction of PD-L1 by LPS and IFN-γ. LPS and IFN-γ alone can separately activate inflammatory macrophages to up-regulate PD-L1 (Fig. 4A), although not as effectively as LPS and IFN-γ together, regardless of the dose (from 10 to 200 ng/ml). To determine whether the LPS response is mediated via TLR4, we injected thioglycolate to recruit inflammatory macrophages from C3H/HeJ mice that have a mutation in TLR4 (21). As expected, inflammatory macrophages from C3H/HeJ mice do not up-regulate PD-L1 in response to LPS but do up-regulate PD-L1 in response to IFN-γ. This shows that up-regulation of PD-L1 on macrophages is yet another downstream effect of TLR4 engagement by LPS.
Figure 4.
PD-L1 and PD-L2 up-regulation are regulated by Stat1 and Stat6. (A) PD-L1 up-regulation by LPS and IFN-γ depends on Stat1. Thioglycolate-induced macrophages from BALB/c, Stat1−/−, and C3H/HeJ mice were cultured in medium alone (control) or activated for 24 h with LPS (10–200 ng/ml) and IFN-γ (10–200 ng/ml) or LPS and IFN-γ together (10–200 ng/ml). Representative histograms shown are for activation at 100 ng/ml. (B) PD-L2 induction depends on IL-4Rα signaling and the Stat6 pathway. Thioglycolate-induced macrophages from BALB/c, Stat6−/−, and IL-4Rα−/− mice were activated for 24 h as described before and stained for PD-L1 and PD-L2 expression. Data shown are representative of at least three experiments with both Stat6−/− and IL-4Rα−/− mice. (C) Time course of PD-L1 and PD-L2 up-regulation on IL-4Rα- and Stat6-deficient macrophages. Macrophages were activated and harvested as described before and stained together at the end of the experiment. Time course data are representative of two experiments.
Stat1 is a critical transcription factor in IFN-dependent responses (13, 22). Stat-deficient mice cannot up-regulate many IFN-inducible genes and are highly susceptible to microbes, viruses, and tumors. However, microarray analysis has shown that there are also STAT1-independent genes that can be induced by IFN-γ (22). To investigate whether PD-L1 up-regulation is mediated by Stat1 signaling, we injected thioglycolate to recruit inflammatory macrophages from Stat1-deficient mice and examined their response to LPS and IFN-γ. Stat1-deficient macrophages express normal levels of PD-L1 on inflammatory macrophages, showing that Stat1 is not required for constitutive expression of PD-L1 on these macrophages. Activation of Stat1-deficient macrophages with IFN-γ increases PD-L1 expression very slightly, and the response to LPS also was diminished. This indicates that Stat1 is not required for the constitutive expression of PD-L1 on inflammatory macrophages but is involved in the up-regulation of PD-L1 by subsequent inflammatory factors such as LPS and IFN-γ.
PD-L2 Expression Is Regulated by IL-4Rα and Stat6.
Like the interferons, IL-4 acts through the Jak/Stat pathway, although it is Stat6 that is critical for regulating downstream changes in gene expression (23). To determine whether the effects of IL-4 on macrophage expression of PD-L2 also are mediated through this pathway, we decided to examine inflammatory macrophages from the IL-4Rα- and Stat6-deficient mice. PD-L2 expression on these macrophages could not be induced by IL-4 (Fig. 4B), and the slight up-regulation of PD-L1 by IL-4 also was eliminated (Fig. 4C). However, IL-4Rα−/− and Stat6−/− macrophages were indistinguishable from WT macrophages in their ability to up-regulate PD-L1 in response to classical stimulation, showing a clear separation in the pathways regulating PD-L1 and PD-L2 up-regulation by classical vs. alternative activation (Fig. 4C).
Differential Induction of PD-L1 and PD-L2 by Th1 vs. Th2 Cells.
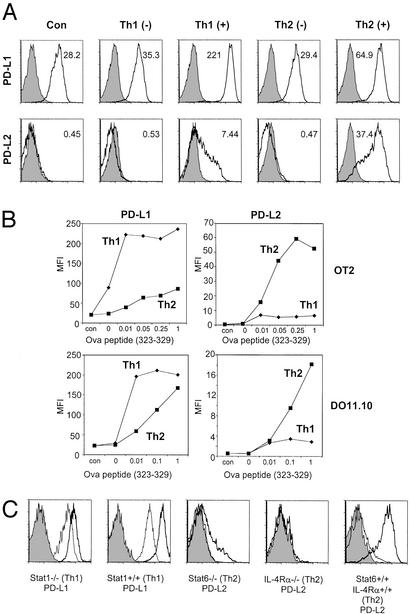
Having determined that classical vs. alternative activation differentially regulate PD-L1 and PD-L2 expression, we wanted to determine whether Th1 and Th2 cells also had this differential effect through antigen presented by inflammatory macrophages. Because different genetic backgrounds are known to influence Th1 and Th2 differentiation (24), we investigated T cell/macrophage interactions with polarized T cells derived from OT2 RAG−/− mice on the C57/BL6 background, as well as the DO11.10 RAG−/− mice on the BALB/c background. These T cells then were presented with different concentrations of peptide on inflammatory macrophages from C57/BL6 or BALB/c mice (Fig. 5). In repeated experiments, Th1 cells alone even in the absence of peptide would increase slightly the expression of PD-L1 on macrophages. In the presence of even very low concentrations of peptide, Th1 cells would induce extremely high levels of PD-L1 on macrophages. Th2 cells also would induce PD-L1 on the macrophage surface, although only at much higher doses of peptide. The reciprocal situation was observed with regard to PD-L2 expression on macrophages and Th2 cells. Th1 and Th2 cells did not exclusively induce only PD-L1 and PD-L2; however, the differences in the levels of induction were very significant. To determine whether the effects of Th1/Th2 cells are dependent on Stat1 (for Th1) and IL-4Rα/Stat6 (for Th2), we repeated these experiments with inflammatory macrophages derived from mice deficient for these molecules. Stat1-deficient mice (129S6/SvEv) on a H2b background were activated with OT2 Th1 cells, whereas Stat6- and IL-4Rα-deficient mice on the BALB/c background were activated with DO11.10 Th2 cells. The up-regulation of PD-L1 expression by Th1 cells was reduced severely (although not completely ablated) in the Stat1-deficient macrophages, consistent with previous experiments with IFN-γ treatment. The up-regulation of PD-L2 expression was completely ablated in the IL-4Rα−/− macrophages, although a very small number of Stat6-deficient macrophages were able to up-regulate PD-L2 expression. However, PD-L2 expression is still severely diminished in these Stat6−/− macrophages. These results showed that Th1/Th2 cells could replace classical/alternative activation stimuli and differentially regulate PD-L1/PD-L2 expression through Stat1/Stat6.
Figure 5.
Differential regulation of PD-L1 and PD-L2 expression by Th1 and Th2 cells. (A) Differentiated Th1 and Th2 cells were generated from the OT2 RAG−/− mice and DO11.10 RAG−/− mice stimulated under polarizing conditions. Rested T cells (after two to three rounds of polarizing stimulation) were cocultured with thioglycolate-induced macrophages in the presence (+; from 0.0001 to 1 μg/ml) or absence (−) of peptide (Ova 323–329). Twenty-four hours later, all cells were harvested and stained for PD-L1 and PD-L2 expression as well as for F4/80. Representative histograms shown are gated on F4/80+ cells to distinguish macrophages from T cells. (B) Induction of PD-L1 and PD-L2 by Th1/Th2 cells at different peptide concentrations. MFI of staining results shown is for F4/80+ gated cells. Results shown for A and B are representative of three to four experiments, each with DO11.10 and OT2 T cells, all with similar results. (C) Induction of PD-L1 expression by Th1 cells is Stat1-dependent, and PD-L2 expression by Th2 cells is IL-4Rα- and Stat6-dependent. Differentiated Th1 cells from OT2 RAG−/− mice were cocultured with macrophages from Stat1−/− mice, and differentiated DO11.10 RAG−/− Th2 cells were cocultured with macrophages from Stat6 and IL-4Rα−/− mice for 24 h in the presence of peptide (heavy line) or the absence of peptide (thin line). Histograms shown are representative of several different peptide concentrations.
Discussion
There are two distinct phases of an immune response that require important interactions between the innate and the adaptive immune system. During the priming phase of an immune response, dendritic cells leave the site of tissue inflammation carrying foreign antigens into the secondary lymphoid organs to activate naïve T cells. Apart from presenting antigenic peptide–MHC complexes to specifically activate naïve T cells, dendritic cells also provide costimulation by engaging CD28 on the T cells through its ligands (B71/B72). During the effector phase of a response, activated and differentiated effector T cells leave the secondary lymphoid organs, penetrate the sites of tissue inflammation, and coordinate the effector arms of the innate immune system. Macrophages are one of the major innate inflammatory cell types that can present antigen and interact with effector T cells there. When inflammatory monocytes encounter and present antigen to effector T cells, they would be exposed to the cytokine microenvironment that is created by the activated T cells. Hence, the most likely determinant of whether macrophages are classically or alternatively activated is their encounter with Th1 or Th2 cells. Indeed, an important but previously unappreciated function of Th2 cells could be the alternative activation of macrophages. Here, we find that Th1 and Th2 cells can differentially up-regulate PD-L1 and PD-L2 on macrophages in the periphery; hence, they are the major determinant of the costimulatory signals that are received by subsequent T cells that interact with macrophages expressing PD-L1 or PD-L2.
We found that, whereas PD-L1 is easily inducible on a number of different cell types, PD-L2 expression is much more restricted and can be highly up-regulated by IL-4 only on inflammatory macrophages. Previously, the expression of PD-L2 has been reported to be restricted mainly to dendritic cells (9). PD-L1 can also be induced on endothelial cells (25). This suggests that PD-L1 might play a more general role in down-regulating activated T cells in the periphery as opposed to a more specific role for PD-L2. Interestingly, we found that on macrophages, PD-L1 expression correlated with the amount of NO production upon subsequent stimulation. These results suggest that PD-L1 expression could be an important marker of “primed” macrophages. This “primed” state can be induced by low doses of IFN-γ on monocyte/macrophages (20) as well as type 1 IFNs (26). Whereas PD-L2 might be a marker of alternatively activated macrophages, PD-L1 could be a marker for macrophages “primed” to be sensitive to subsequent IFN-γ signaling.
Our results also show that the up-regulation of PD-L1 and PD-L2 expression is mediated by the Jak/Stat pathway. However, the expression of constitutive PD-L1 on inflammatory macrophages is not Stat1-dependent. PD-L1 expression can be enhanced further on inflammatory macrophages by classical activation via LPS and IFN-γ in a Stat1-dependent manner. LPS and interferons are known to induce a common set of genes in macrophages. Whereas LPS signals through TLR4 with adapter molecules such as Myd88 and TIRAP and a cascade that eventually leads to nuclear translocation of NFKB and AP-1 (12), interferons signal through the Janus kinases and Stat family of transcription factors (13). Just recently, it has become clear that a subset of genes induced by LPS is actually a result of secondary autocrine production of IFN-β (27). This response depends on Stat1 signaling as opposed to Stat1-independent genes directly induced through TLR4 signaling. The fact that PD-L1 expression on Stat1-deficient macrophages has a diminished response to LPS suggests that LPS could be enhancing PD-L1 expression through a secondary mechanism involving autocrine production of IFN-β (27).
The effects of IL-4 and downstream signaling pathways have been studied most carefully in T and B cells. Whereas IL-4 has been shown to inhibit gene transcription by subsequent classical stimulation (28), little was known about the direct effects of IL-4 on inflammatory macrophage gene transcription until very recently, when a microarray study revealed that IL-4 can act on macrophages through the Stat6 pathway to induce a gene known as Ym1 (29). Here, we find that PD-L2 is regulated through the same pathway as Ym1. However, examination of the upstream region of PD-L2 did not reveal any Stat6-binding domains. How Stat6 regulates PD-L2 expression remains to be established.
Our findings that Th1 cells induce PD-L1 whereas Th2 cells induce PD-L2 are the first examples of costimulatory ligands differentially regulated by T helper subsets. An important question raised by this study is why PD-L1 and PD-L2 should be differentially regulated by type 1 and type 2 conditions because they both bind PD-1. There are a number of possibilities, especially because PD-L1 and PD-L2 have been reported to be stimulatory or inhibitory under different experimental conditions. One is a positive feedback loop, whereby higher PD-L1 levels on macrophages preferentially stimulate Th1 cells and inhibit Th2 cells, and vice versa with PD-L2. The other possibility is a negative feedback loop, whereby PD-L1 inhibits Th1 cells and stimulates Th2 cells, and vice versa with PD-L2. Different levels of PD-L1 and PD-L2 also could have different effects on subsequent interaction of macrophages with naïve vs. effector and memory T cells. A most intriguing possibility also is raised by recent papers showing that costimulatory ligands can deliver signals back into the antigen-presenting cell that influence the function of dendritic cells (30, 31). Perhaps, PD-L1 and PD-L2 are transducing signals that influence the way that macrophages are activated upon subsequent interaction with T cells. Regardless of these possibilities, our results suggest that PD-L1 and PD-L2 might have different roles in regulating type 1 and type 2 responses in the periphery. Analysis of mice deficient for these molecules should reveal whether this is indeed the case. It would also be interesting to determine whether blocking antibodies against PD-L1 and PD-L2 could be beneficial in the treatment of diseases such as allergy and experimental autoimmune encephalomyelitis, where the Th1/Th2 balance is involved in pathogenesis.
Acknowledgments
We thank Xingxing Zang for helpful advice and critical review of the manuscript. This work was supported by the Howard Hughes Medical Institute and the Wellcome Trust. P.L. is a recipient of the Wellcome International Research Fellowship.
Abbreviations
- Th
T helper
- LPS
lipopolysaccharide
- Poly IC
polyinosinic-polycytidylic acid
- PD-1
programmed death 1
- TLR
Toll-like receptor
- MFI
mean fluorescence intensity
References
- 1.Lanzavecchia A, Sallusto F. Cell. 2001;106:263–266. doi: 10.1016/s0092-8674(01)00455-x. [DOI] [PubMed] [Google Scholar]
- 2.Liang L, Sha W C. Curr Opin Immunol. 2002;14:384–390. doi: 10.1016/s0952-7915(02)00342-4. [DOI] [PubMed] [Google Scholar]
- 3.Ishida Y, Agata Y, Shibahara K, Honjo T. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 5.Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N, et al. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 6.Freeman G J, Long A J, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz L J, Malenkovich N, Okazaki T, Byrne M C, et al. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong H, Zhu G, Tamada K, Chen L. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 8.Latchman Y, Wood C R, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long A J, Brown J A, Nunes R, et al. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 9.Tseng S Y, Otsuji M, Gorski K, Huang X, Slansky J E, Pai S I, Shalabi A, Shin T, Pardoll D M, Tsuchiya H. J Exp Med. 2001;193:839–846. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong H, Strome S E, Salomao D R, Tamura H, Hirano F, Flies D B, Roche P C, Lu J, Zhu G, Tamada K, et al. Nat Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, Shin T, Tsuchiya H, Pardoll D M, Okumura K, et al. J Immunol. 2002;169:5538–5545. doi: 10.4049/jimmunol.169.10.5538. [DOI] [PubMed] [Google Scholar]
- 12.Janeway C A, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 13.O'Shea J J, Gadina M, Schreiber R D. Cell. 2002;109,Suppl.:S121–S131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 14.Seder R A, Hill A V. Nature. 2000;406:793–798. doi: 10.1038/35021239. [DOI] [PubMed] [Google Scholar]
- 15.Goerdt S, Orfanos C E. Immunity. 1999;10:137–142. doi: 10.1016/s1074-7613(00)80014-x. [DOI] [PubMed] [Google Scholar]
- 16.Gordon S. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 17.Lee S C, Jaffar Z H, Wan K S, Holgate S T, Roberts K. J Immunol. 1999;162:6867–6879. [PubMed] [Google Scholar]
- 18.Loke P, MacDonald A S, Robb A, Maizels R M, Allen J E. Eur J Immunol. 2000;30:2669–2678. doi: 10.1002/1521-4141(200009)30:9<2669::AID-IMMU2669>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs B L, Langland J O. Virology. 1996;219:339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 20.Hu X, Herrero C, Li W P, Antoniv T T, Falck-Pedersen E, Koch A E, Woods J M, Haines G K, Ivashkiv L B. Nat Immunol. 2002;3:859–866. doi: 10.1038/ni828. [DOI] [PubMed] [Google Scholar]
- 21.Poltorak A, He X, Smirnova I, Liu M Y, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 22.Ramana C V, Gil M P, Schreiber R D, Stark G R. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 23.Nelms K, Keegan A D, Zamorano J, Ryan J J, Paul W E. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh C S, Macatonia S E, O'Garra A, Murphy K M. J Exp Med. 1995;181:713–721. doi: 10.1084/jem.181.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazanet M M, Hughes C C. J Immunol. 2002;169:3581–3588. doi: 10.4049/jimmunol.169.7.3581. [DOI] [PubMed] [Google Scholar]
- 26.Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S, Tanaka N, Taniguchi T. Science. 2000;288:2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 27.Toshchakov V, Jones B W, Perera P Y, Thomas K, Cody M J, Zhang S, Williams B R, Major J, Hamilton T A, Fenton M J, et al. Nat Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 28.Hamilton T A, Ohmori Y, Tebo J. Immunol Res. 2002;25:229–245. doi: 10.1385/IR:25:3:229. [DOI] [PubMed] [Google Scholar]
- 29.Welch J S, Escoubet-Lozach L, Sykes D B, Liddiard K, Greaves D R, Glass C K. J Biol Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen L T, Radhakrishnan S, Ciric B, Tamada K, Shin T, Pardoll D M, Chen L, Rodriguez M, Pease L R. J Exp Med. 2002;196:1393–1398. doi: 10.1084/jem.20021466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Grohmann U, Orabona C, Fallarino F, Vacca C, Calcinaro F, Falorni A, Candeloro P, Belladonna M L, Bianchi R, Fioretti M C, et al. Nat Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]