Abstract
Mutations have been described in the ataxia telangiectasia mutated (ATM) gene in small numbers of cases of lymphoid neoplasia. However, surveys of the ATM mutation status in lymphoma have been limited due to the large size (62 exons) and complex mutational spectrum of this gene. We have used microarray-based assays with 250,000 oligonucleotides to screen lymphomas from 120 patients for all possible ATM coding and splice junction mutations. The subtypes included were diffuse large B cell, mantle cell, immunoblastic large B cell, follicular, posttransplant lymphoproliferative disorder, and peripheral T cell lymphoma. We found the highest percentage of ATM mutations within the mantle cell (MCL) subtype (43%, 12 of 28 cases), followed by a lower level (10% of cases) in the other subtypes. A frame-shift ATM mutation was found in one peripheral T cell lymphoma patient. In six MCL cases examined, four ATM variants were due to somatic mutation in the tumor cells whereas two others seemed to be germ-line in origin. There was no difference in p53 mutation status in the ATM mutant and wild-type groups of MCL. There was no statistically significant difference in the median overall survival of patients with wild-type vs. mutated ATM in MCL. Additional mutational and functional analyses are needed to determine whether ATM mutations contribute to the development and progression of MCL or are just the consequence of genomic instability in MCL.
Nucleic-acid based biomarkers are powerful tools used in the classification, prognosis, and selection of treatments for lymphomas. High-throughput technologies have been developed to rapidly screen for specific cytogenetic (1), gene expression (2), and DNA methylation profiles (3) with clinical relevance to specific disease states. Nevertheless, it is still a significant challenge to rapidly screen for disease-causing point mutations (4) that occur in distinct lymphoma subtypes.
The high incidence of leukemias and lymphomas in ataxia telangiectasia patients (5) and the presence of genomic deletions spanning the ataxia telangiectasia mutated (ATM) gene in sporadic forms of lymphoid neoplasia (6–10) have prompted investigations into the role that ATM mutations play in these diseases (11). However, the technical challenges associated with screening the entire 9.45-kb ATM coding region for all possible mutations have limited the number of lymphomas investigated. Deleterious ATM mutations have been found in significant numbers of T cell prolymphocytic leukemias (12–17), B cell chronic lymphocytic leukemias (B-CLLs; refs. 18–22), and mantle cell lymphomas (MCLs; refs. 23–26). However, the clinical significance of these ATM mutations is not completely understood.
We have developed oligonucleotide microarray-based assays (27–29) to screen tumors from 120 lymphoma patients for all possible ATM coding and splice junction mutations. These include 38 diffuse large B cell (DLBCL), 28 MCL, 19 immunoblastic large B cell (IBL), 18 follicular (FL), 12 posttransplant lymphoproliferative disorder (PTLD), and 10 peripheral T cell lymphoma (PTCL) patients. We sought to identify lymphoma subtypes having an excess of ATM mutations and correlate ATM mutation status with disease prognosis in the MCL subtype.
Materials and Methods
Patient Materials.
Diagnostic biopsies from 125 patients with lymphoma diagnosed from 1986 to 1996 were retrieved from the registry of the Nebraska Lymphoma Study Group with Institutional Review Board approval. The patients included were all deceased before 1996. For a case to be eligible, tumor cells had to comprise the majority (>70%) of cells present by review of morphology. A subset of cases also had Southern blots of Ig heavy chain gene rearrangements, which confirmed that tumor DNA content exceeded normal cell DNA.
Microdissection of Normal Cells.
Benign tissue cells (blood vessels, adipose tissue, histiocytes) were retrieved onto a microtube cap by using a Pixcell II laser capture microdissection instrument (Arcturus Engineering, Mountain View, CA). Afterward, the cap was incubated in 30 μl of digestion buffer (1 mg/ml proteinase K/10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/1% Tween 20) for 20 h at 50°C. Later, the digestion buffer containing the eluted DNA was incubated at 95°C for 10 min to inactivate the proteinase K.
ATM sequences were amplified by using 5–10 μl of lysate with ATM primers containing M13 sequences at the 5′ end, in 50 μl of PCR buffer I (Perkin–Elmer), 100 μM of each dNTP, 2.5 units Taq Gold Polymerase (Perkin–Elmer), and 0.5 μM of each primer pair. Cycling conditions were 94°C for 9 min, then 94°C for 75 s, 54°C for 15 s, 68°C for 1 min for 45 cycles, with a final extension of 7 min. After confirming the presence of amplicons by agarose gel electrophoresis, 5 μl of the PCR reaction was reamplified for 35 cycles. These amplicons were purified by using QIAquick PCR purification kit (Qiagen, Valencia, CA).
p53 Mutation Analysis in MCL.
DNA in 22 of 25 cases was previously screened for p53 mutations in exons 5–8 by using denaturing gradient gel electrophoresis as described (4). Samples with shifted bands were isolated and sequenced with fluorescently labeled dye terminators (Applied Biosystems) as described (4).
Survival Analysis.
Death from any cause was analyzed on the 21 of 28 cases of MCL with clinical information available by using the product-limit method of Kaplan and Meier (30) and compared by using the log-rank test (31).
Oligonucleotide Microarray Design.
A pair of oligonucleotide microarrays (Affymetrix, Santa Clara, CA) containing >250,000 probes (25 nt in length) were designed to interrogate both ATM strands for all possible sequence variations in the 62 coding exons and their splice junctions. Subsets of probes are complementary to all possible ATM single nucleotide substitutions, single base pair insertions, and 1- and 2-bp deletions in this gene. A series of perfect match (PM) probes complementary to every 25-nt segment of the ATM coding region are present in five copies across the array. This redundancy increases assay sensitivity and specificity by providing multiple measurements of target hybridization.
Target Preparation and Data Acquisition.
Single-stranded RNA target for microarray-based hybridization analysis was prepared as described (29). Individual ATM-coding exons were amplified from lymphoma DNA (without knowledge of the morphologic subtype) by using primers containing T3 and T7 RNA polymerase tails, pooled, and then in vitro transcribed by using T3 or T7 RNA polymerase to create biotin-labeled sense and antisense strand targets, respectively. Fluorescein-labeled reference target was made by using genomic DNA from an unaffected individual. Reference and test sample targets were fragmented, diluted in hybridization buffer, and hybridized to the ATM microarrays for 4 h at 42°C as described (29). Afterward, the microarray surface was stained with a phycoerythrin-streptavidin conjugate to label the hybridized test target. Digitized hybridization images from both reference and test targets were acquired by using the Gene Array Scanner (Hewlett–Packard, Palo Alto, CA) equipped with the appropriate 515- to 545-nm bandpass and 560-nm longpass emission filters. genechip software (Affymetrix) was used to quantify hybridization signals for each probe. Custom software was used to correct for spectral overlap between the reference and test channels and perform two-color hybridization analysis (29).
Two-color cohybridization experiments were used to screen for all possible heterozygous ATM sequence changes. In this procedure, the ratio of reference and test target occupancy to each PM probe is measured. Test targets with altered sequence tracts are predicted to have reduced affinity toward specific sets of PM probes. Localized losses of hybridization signals are displayed by plotting the ratio of reference to test sample hybridization signals for all PM probes. Peak ratio values should be centered nearby PM probes interrogating mutant sequences due to expected localized decreases in sample hybridization. Experimental noise was reduced by using a series of multiplicative correction factors determined by target sequence composition and structure (29). Afterward, the dataset was normalized against PM probe signal ratios obtained from similar experiments. These corrected hybridization signal ratios were plotted against their respective nucleotide positions.
True sequence variants are distinguished from noise based on peak width, height, and the reproducibility of data from duplicated PM probes. Exons were sequenced if either strand produced peak heights >1.3 units and the other strand showed a peak of any height at the same sequence tract. Exons were also sequenced if the average peak height of the sense and antisense strand data was >1.25 units. Peaks where the apex was centered three or more nucleotides upstream of the 3′-AG acceptor or downstream of the 5′-GT donor sequences were not considered for further analysis because they represent sequence changes located further into the intron. In some cases, experiments were repeated and a composite data set was used to discriminate signals from noise by searching for common perturbations in baseline ratio values.
Dideoxysequencing Analysis.
Sample exons were amplified by using the protocols stated above, purified on agarose gels, and eluted from gel slices by using the GFX PCR and Gel Band Purification Kit (Amersham Biosciences). Amplicons were sequenced by using the BigDye Terminator Kit (Applied Biosystems) and analyzed on an ABI 377 DNA sequencer. Excess primers and dNTPs were removed with a Centra Sep column (Princeton Separations, Adelphia, NJ). The DNA sequences were analyzed on a 377 fluorescent sequencing systems (Applied Biosystems) by using analysis 1.2 and sequence navigator software (Applied Biosystems) as well as sequencer software (GeneCodes, Ann Arbor, MI) to compare with wild-type ATM sequence. Amplicons were subcloned by using the Zero Blunt TOPOII PCR Cloning Kit (Invitrogen), and individual subclones were sequenced to confirm the identity of mutations.
Results
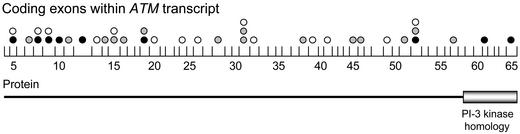
The sequence variants uncovered were evenly distributed across the ATM gene (Fig. 1). We classified these sequence variants based on their putative effect on ATM function and allele frequency. Deleterious mutations refer to sequence changes that adversely affect ATM function by producing a truncated protein or altering critical amino acids (Table 1). Unclassified missense changes refer to amino acid substitutions for which the affect on ATM activity is currently unknown (Table 2). As the effect that these sequence variants have on ATM function is elucidated in the future, these variants will be classified as deleterious mutations, rare variants, or polymorphisms. Herein, polymorphisms refer to silent substitutions or to previously reported variants with allele frequencies greater than 0.01 that do not affect ATM function (Table 3).
Figure 1.
Distribution of 38 ATM sequence variants in lymphoma samples. Vertical lines show the boundaries of the 62 coding exons within the ATM transcript. The spacing between these lines is proportional to the size of each exon. Black, gray, and white circles above each exon indicate the presence of deleterious mutations, unclassified missense changes, and polymorphisms, respectively. The location of the PI3-kinase homology domain is listed under the appropriate exons.
Table 1.
Deleterious ATM mutations detected
| Sample | Type | Change | Result | State* |
|---|---|---|---|---|
| W | MCL | 171G/A | W57X | ND |
| T | MCL | 503del6 | F168_V170delinsL | ND |
| D24 | MCL | 781insA | 273Stop | ND |
| M | MCL | 923G/A | W308X | Acq |
| A29 | PTCL | 1562delAG | Frame-shift | ND |
| D21 | MCL | [T/C 19+2] | 5′-GT donor altered | ND |
| R | MCL | 7327C/T | R2443X | Acq |
| N | MCL | 8663T/C | I2888T | Acq |
| N | MCL | 9023G/A | R3008H | ND |
Mutation is acquired (Acq) or germ-line (GL), or its status is not determined (ND).
Table 2.
Unclassified missense changes detected
| Sample | Type | Change | Result | Mmu* | State† |
|---|---|---|---|---|---|
| D4 | MCL | 378 T/A | D126E | D | ND |
| D6 | FL | 1229 T/C | V410A | V | ND |
| C7, C22 | IBL | 1810 C/T | P604S | P | ND |
| B10 | PTLD | 2149 C/T | R717W | R | ND |
| A6, A11 | DLBCL | 2572 T/C | F858L | S | ND |
| A32 | FL | 3295 G/A | A1309T | S | ND |
| A15 | DLBCL | 4362 A/C | K1454N | Q | ND |
| C22 | IBL | 4388 T/C | F1463C | F | ND |
| A19 | DLBCL | 4424 A/G | Y1475C | Y | ND |
| C13 | DLBCL | 5401 A/T | N1801Y | S‡ | ND |
| N | MCL | 6227 T/G | I2076S | I§ | ND |
| A2 | MCL | 6374 A/G | H2125R | H¶ | GL |
| D24 | MCL | 7267 G/A | E2423L | E¶ | ND |
| A1 | MCL | 7334 T/C | L2445P | L¶ | Acq |
| S | MCL | 8089 A/G | N2697D | N¶ | GL |
Residue in orthologous mouse ATM gene.
Mutation is acquired (Acq) or germ-line (GL), or its status is not determined (ND).
Residue in African clawed frog ATM genes is N.
Residue in African clawed frog ATM genes is M.
Same residue in African clawed frog ATM gene.
Table 3.
ATM polymorphisms detected
| Sample | Type | Change | Result |
|---|---|---|---|
| C1 | IBL | 1746C/T | F582F |
| B5 | DLBCL | 2119 T/C | S707P |
| F | DLBCL | 2805 G/C | T935T |
| A6, A11, H, I | DLBCL | 3161 C/G | P1054R |
| A32, D6 | FCL | 3161 C/G | P1054R |
| A14 | DLBCL | 4258 C/T | L1420F |
| E, G, L, A33 | MCL | 4578 C/T | P1526P |
| A, A15, A19 | DLBCL | 5557 G/A | D1853N |
| A2, Z, A21, A22 | MCL | 5557 G/A | D1853N |
| A28 | PTCL | 5557 G/A | D1853N |
| C1, D15 | IBL | 5557 G/A | D1853N |
| P, Q | MCL | 5793 T/C | A1931A |
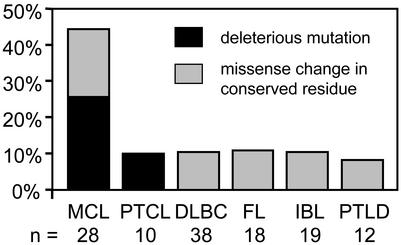
We found the highest percentage of ATM mutations within the mantle cell subtype (43%, 12 of 28 cases), followed by a lower level (10% of cases) in the other subtypes of lymphoma. Of 28 MCL cases analyzed, seven contained deleterious ATM mutations (Figs. 1 and 2, Table 1). These included three nonsense mutations (W57X, W308X, and R2443X) as well as one frame-shift (781insA) that led to a truncated ATM protein (Table 1). In sample D21 (T/C 19 + 2), the exon 19 5′-splice donor sequence is altered and is predicted to produce a nonfunctional protein (Table 1). In sample T, the 503del6 mutation substitutes amino acids 168–170 (Phe-Ser-Val), which are conserved in mouse with leucine. In sample N, two missense changes (I2888T and R3008H) occur in the conserved phosphatidylinositol 3-kinase (PI3-kinase) domain (Fig. 3). One (R3008H) was previously found in a B-CLL (23), DLBCL (32), and MCL (26) case. A related mutation (R3008C) was reported in two T cell prolymphocytic leukemia (12, 14) cases and one MCL (24) case. Interestingly, a germ-line R3008C missense change was found in an ataxia telangiectasia patient (29). Nevertheless, it should be noted that there is no direct evidence that the R3008H or I2888T alleles compromise ATM function. Five samples contained missense changes (D126E, H2125R, E2423L, L2445P, and N2697D) that altered conserved amino acids in mouse and African clawed frog (Xenopus laevis) (33), where sequence information for the latter species exists (Table 2). For all these deleterious and missense changes, sequencing analysis also detected the presence of wild-type alleles. It is likely this result is due in part to the presence of normal cell DNA in the MCL tumor DNA samples.
Figure 2.
Frequency of deleterious and missense ATM mutations in 124 lymphomas. The number of cases investigated is listed below each subtype.
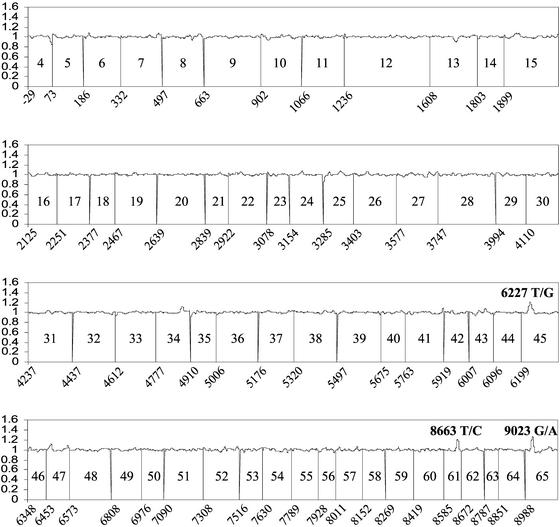
Figure 3.
Detection of mutations in a MCL sample by using the two-color loss of signal assay. Fluorescein-labeled wild-type reference and biotinylated test MCL sample N targets were cohybridized to the ATM microarray. To correct for reproducible differences in the hybridization efficiencies of reference and test targets, the ratio of reference-to-test target signal at each wild-type PM probe was normalized against ratios derived from 10 separate chip cohybridization experiments. Averaged sense and antisense strand ratios are plotted on the y axis against the nucleotide position of the corresponding PM probe on the x axis. The identity of each exon is listed between the vertical lines depicting their boundaries in the ATM transcript. Peaks at the mutated 6227 T/G, 8663 T/C, and 9023 G/A positions (giving rise to I2076S, I2888T, and R3008H missense changes, respectively) are labeled.
Only one deleterious ATM mutation was found in the 10 (10%) PTCL cases. This case involved a frame-shift mutation (1562delAG) located in the first quarter of the coding region. As was the case with MCL, sequencing analysis detected the presence of the wild-type allele in the PTCL tumor DNA sample. No unclassified missense mutations were detected in the remaining PTCL cases.
Although no deleterious mutations were found in the IBL, FL, DLBCL, and PTLD cases, unclassified missense mutations appeared in each subtype. Three of 19 (15.8%) IBL cases had such missense mutations (P604S and F1463) in residues conserved in mouse, with the P604S variant appearing in two cases. This variant has also been found in a DLBCL case (32). The two missense mutations in 18 cases (11.1%) of FL involved conserved (V410A) and nonconserved (A1309T) residues in mouse (Table 2). Five of 38 (13.2%) DLBCL cases contained missense mutations. One involved a residue (Y1475C) conserved in mouse. Two variants (F858L and K1454N) occurred in a nonconserved amino acid in mouse, with the F858L mutation appearing in two separate cases. The N1801Y missense mutation involves an amino acid conserved in frog but not in mouse. Only one missense mutation (R717W) was identified in the 12 (8%) PTLD cases.
The germ-line status of ATM sequence variants in six MCL cases was determined by sequencing amplicons from microdissected stromal cells surrounding the tumors (Tables 1 and 2) because no normal tissue biopsies were available. Four variants were the consequence of somatic mutation in the tumor cells. Two variants (H2125R and N2697D) seemed to be heterozygous in the microdissected material and are most likely germ-line in origin. However, contaminating tumor cells could falsely indicate the presence of the mutant alleles in the stromal cells.
There was no difference in the rate of p53 mutations in the ATM mutant and wild-type groups. MCLs with ATM mutations had p53 mutations in two of eight cases (25%). On the other hand, 5 of 14 (35%) ATM wild-type cases had p53 mutations.
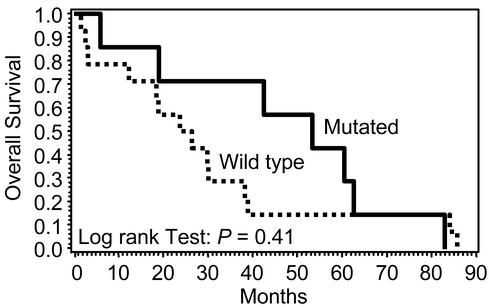
To evaluate the clinical relevance of ATM mutation status, the overall survival times of MCL patients having tumors with and without ATM mutations were compared (Fig. 4). The median overall survival of patients with wild-type ATM in MCL was 25.1 mo, whereas those with mutant ATM (deleterious or missense mutations) had a median survival of 53.3 mo. However, these differences did not reach statistical significance (P = 0.41), nor was a difference seen in stage, response to chemotherapy, or progression of disease between the two groups.
Figure 4.
Overall survival by ATM mutational status. The median overall survival of mutant ATM cases (solid line) of 53.3 mo was not significantly different from the median survival of 25.1 mo in the wild-type ATM cases (dashed line).
Discussion
In agreement with previous studies, deleterious or unclassified missense mutations in the ATM gene were found most commonly in the MCL subtype where 12 of 28 (43%) cases were mutation positive (Fig. 2; refs. 11, 24, and 26). Eight cases (W, T, D24, M, D21, R, and N) had deleterious mutations, whereas four cases (D4, A2, A1, and S) had unclassified missense mutations (Tables 1 and 2). We investigated the germ-line status of mutations in six cases of MCL by sequencing DNA obtained from stromal cells surrounding the tumors. Four mutations (W308X, R2443X, I2888T, and L2445P) were of somatic origin and acquired in the tumor. We found evidence that two missense mutations (H2125R and N2676D) may have been germ-line in origin. This finding is in agreement with other studies where most mutant ATM alleles examined were acquired in MCL patients (24, 26).
Patients with MCL exhibit a poor overall survival, with a median of 3–4 yr, and p53 mutations are known to be a prognostic factor in our series of MCL (4). In the MCL subtype, we found no preferential association of p53 mutations with ATM mutations status, similar to a previous study in MCL (26). This finding would be compatible with the lack of any reported association of 17p13 deletions with 11q22-23 deletions in MCL (34–36). Although there was a difference in the median overall survival between patients with tumors having wild-type (25.1 mo) and mutated ATM (53.3 mo) sequences, the difference did not reach statistical significance (Fig. 4). Although ATM mutations were not predictive of overall survival in the MCL patients in this study and others (26), it is possible that ATM mutations have a subtle effect on tumor biology that will require studies of a larger number of patients. The role of the high incidence of ATM mutations in MCL is unknown.
The significance of the low level of ATM mutations in the other subtypes of lymphoma (28 DLBC, 19 IBL, 18 FL, 12 PTBC, and 10 PTCL) also remains to be determined. Grønbaek (32) reported a somewhat higher frequency of missense mutations (20%) in DLBCL than our series and no ATM mutations were seen in PTCL. The lack of deleterious ATM mutations in FL is in agreement with other reports examining 17 FL cases (37), and the 10% missense mutation rate concurs with another series (32). Although an unequivocally deleterious ATM mutation occurred in one PTCL case in our series, it is not possible to assess the clinical significance of this finding. It can be speculated that distinct subsets of PTCL cases are enriched for ATM mutations. Currently, there is no clear evidence of deleterious ATM mutations in chronic myeloid leukemia (CML; ref. 38) or T cell lymphoblastic leukemia (T-ALL; ref. 39).
The relevance of missense mutations outside the conserved PI3-kinase domain is difficult to assess without functional assays. If the missense variants were acquired, this would indicate a background level of genomic instability in the ATM gene in most subtypes of lymphoma. On the other hand, they may represent rare germ-line variants that do not have a significant effect on ATM function and thus are of no biological significance. Whereas MCL, T cell prolymphocytic leukemia, and B-CLL cases tend to have mutations that truncate the ATM protein or perturb amino acids in the PI3-kinase domain, breast cancers have numerous ATM missense mutations (40). Dominant negative ATM missense mutations naturally exist and may confer an increased risk of developing breast cancer in some families (41, 42).
Both copies of the ATM gene are frequently inactivated in MCL cases due to deleterious point mutations and deletions in the 11q22-q23 genomic region (7, 9, 23–26). Except for sample N, only one deleterious mutation was found in each MCL case. However, it was not possible to determine whether these mutations occurred in cis or in trans in sample N. Although genomic deletions are likely to have inactivated the other ATM allele in these samples, several other mechanisms are possible. Aberrant DNA methylation could inactivate the other ATM allele (39, 43), ATM haploinsufficiency may be responsible for the disease, or certain ATM mutations could act in a dominant negative fashion. Furthermore, the microarray assays may not have detected the other allele due to false negatives, especially in repetitive sequence tracts (28), or the location of the mutation (intronic, 5′- or 3′-UTRs, or promoter regions).
The oligonucleotide microarray-based assays distinguished MCL as the lymphoma subtype with the highest incidence of ATM mutations. Although the sensitivity of the assays had been previously determined in blinded analysis of ATM carriers (17 of 18 heterozygous sequence changes detected; ref. 29), this study provides direct evidence of its utility in examining tumor samples for mutations in the presence of wild-type alleles. However, detecting all possible ATM mutations in complex mixtures of tumor and normal cells is still a significant challenge. By relying on “gain of hybridization” signals to mutation-specific oligonucleotide probes, it may be possible to detect alleles present in lesser abundance (44–46). However, such assays depend on representing all mutations in the microarray, not just single nucleotide substitutions and single base pair deletions. It is not currently feasible to represent all possible multiple base pair insertions in the microarray. Furthermore, the single base pair insertion and deletion probes showed greater cross-hybridization when compared with the single nucleotide substitution probes (data not shown). It will be even more problematic to efficiently detect larger insertions and deletions by using this strategy due to sequence composition effects. Therefore, the loss of signal approach is a more consistent way of detecting all possible mutations but will begin to rapidly lose sensitivity as the mutant DNA comprises less that 40% of the cellular population. Oligonucleotide microarray-based minisequencing assays (47) could provide an alternative hybridization-based approach to these problems.
In the two lymphoid malignancies with the highest rate of ATM mutations, B-CLL and MCL, recent gene-expression profiling highlighted basic fundamental differences between the two disorders (25, 48). In B-CLL, there seem to be defects in apoptotic pathways that prevent tumor cell death, but there is no promotion of the cell cycle. In contrast, defects in the control of cell proliferation seem to be more important than apoptotic defects in MCL cases. Differential expression of genes involved in lymphocytic trafficking and differentiation, growth factors and receptors, metastasis and angiogenesis, and neurotransmitter receptors have also been noted in MCL cases (49). If ATM mutations play a role in both diseases, it may be through the modulation of different pathways.
Acknowledgments
We thank Larry Brody of the National Human Genome Research Institute for thoughtful discussion and Deb Lytle for technical assistance. This study was partially funded by the Margaret E. Early Foundation (J.G.H.), the Donald E. and Delia B. Baxter Foundation (J.G.H.), the V Foundation for Cancer Research (J.G.H.), National Cancer Institute Grant U01-CA84967 and U.S. Public Health Service Grant CA 36727 (to W.C.C., T.C.G., J.O.A., J.M.V., and D.D.W.), and Leukemia and Lymphoma Society Grant 6605-01 (to T.C.G. and W.C.C.).
Abbreviations
- B-CLL
B cell chronic lymphocytic leukemia
- MCL
mantle cell lymphoma
- DLBCL
diffuse large B cell lymphoma
- IBL
immunoblastic large B cell lymphoma
- FL
follicular lymphoma
- PTLD
posttransplant lymphoproliferative disorder
- PTCL
peripheral T cell lymphoma
- PM
perfect match
- PI3-kinase
phosphatidylinositol 3-kinase
References
- 1.Carpenter N J. Semin Pediatr Neurol. 2001;8:135–146. doi: 10.1053/spen.2001.26447. [DOI] [PubMed] [Google Scholar]
- 2.Rosenwald A, Staudt L M. Semin Oncol. 2002;29:258–263. doi: 10.1053/sonc.2002.32901. [DOI] [PubMed] [Google Scholar]
- 3.Fraga M F, Esteller M. Biotechniques. 2002;33:632. doi: 10.2144/02333rv01. , 634, 636–649. [DOI] [PubMed] [Google Scholar]
- 4.Greiner T C, Moynihan M J, Chan W C, Lytle D M, Pedersen A, Anderson J R, Weisenburger D D. Blood. 1996;87:4302–4310. [PubMed] [Google Scholar]
- 5.Taylor A M, Metcalfe J A, Thick J, Mak Y F. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 6.Panayiotidis P, Kotsi P. Semin Hematol. 1999;36:171–177. [PubMed] [Google Scholar]
- 7.Stilgenbauer S, Winkler D, Ott G, Schaffner C, Leupolt E, Bentz M, Moller P, Muller-Hermelink H K, James M R, Lichter P, et al. Blood. 1999;94:3262–3264. [PubMed] [Google Scholar]
- 8.Haidar M A, Kantarjian H, Manshouri T, Chang C Y, O'Brien S, Freireich E, Keating M, Albitar M. Cancer. 2000;88:1057–1062. doi: 10.1002/(sici)1097-0142(20000301)88:5<1057::aid-cncr16>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 9.Monni O, Knuutila S. Leuk Lymphoma. 2001;40:259–266. doi: 10.3109/10428190109057924. [DOI] [PubMed] [Google Scholar]
- 10.Cuneo A, Bigoni R, Rigolin G M, Roberti M G, Bardi A, Cavazzini F, Milani R, Minotto C, Tieghi A, Della Porta M, et al. Haematologica. 2002;87:44–51. [PubMed] [Google Scholar]
- 11.Boultwood J. J Clin Pathol. 2001;54:512–516. doi: 10.1136/jcp.54.7.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stilgenbauer S, Schaffner C, Litterst A, Liebisch P, Gilad S, Bar-Shira A, James M R, Lichter P, Dohner H. Nat Med. 1997;3:1155–1159. doi: 10.1038/nm1097-1155. [DOI] [PubMed] [Google Scholar]
- 13.Vorechovsky I, Luo L, Dyer M J, Catovsky D, Amlot P L, Yaxley J C, Foroni L, Hammarstrom L, Webster A D, Yuille M A. Nat Genet. 1997;17:96–99. doi: 10.1038/ng0997-96. [DOI] [PubMed] [Google Scholar]
- 14.Yuille M A, Coignet L J, Abraham S M, Yaqub F, Luo L, Matutes E, Brito-Babapulle V, Vorechovsky I, Dyer M J, Catovsky D. Oncogene. 1998;16:789–796. doi: 10.1038/sj.onc.1201603. [DOI] [PubMed] [Google Scholar]
- 15.Yuille M A, Coignet L J. Recent Res Cancer Res. 1998;154:156–173. doi: 10.1007/978-3-642-46870-4_9. [DOI] [PubMed] [Google Scholar]
- 16.Stoppa-Lyonnet D, Soulier J, Lauge A, Dastot H, Garand R, Sigaux F, Stern M H. Blood. 1998;91:3920–3926. [PubMed] [Google Scholar]
- 17.Stankovic T, Taylor A M, Yuille M R, Vorechovsky I. Blood. 2001;97:1517–1518. doi: 10.1182/blood.v97.5.1517. [DOI] [PubMed] [Google Scholar]
- 18.Starostik P, Manshouri T, O'Brien S, Freireich E, Kantarjian H, Haidar M, Lerner S, Keating M, Albitar M. Cancer Res. 1998;58:4552–4557. [PubMed] [Google Scholar]
- 19.Bevan S, Catovsky D, Marossy A, Matutes E, Popat S, Antonovic P, Bell A, Berrebi A, Gaminara E, Quabeck K, et al. Leukemia. 1999;13:1497–1500. doi: 10.1038/sj.leu.2401531. [DOI] [PubMed] [Google Scholar]
- 20.Bullrich F, Rasio D, Kitada S, Starostik P, Kipps T, Keating M, Albitar M, Reed J C, Croce C M. Cancer Res. 1999;59:24–27. [PubMed] [Google Scholar]
- 21.Stankovic T, Weber P, Stewart G, Bedenham T, Murray J, Byrd P J, Moss P A, Taylor A M. Lancet. 1999;353:26–29. doi: 10.1016/S0140-6736(98)10117-4. [DOI] [PubMed] [Google Scholar]
- 22.Stankovic T, Stewart G S, Fegan C, Biggs P, Last J, Byrd P J, Keenan R D, Moss P A, Taylor A M. Blood. 2002;99:300–309. doi: 10.1182/blood.v99.1.300. [DOI] [PubMed] [Google Scholar]
- 23.Schaffner C, Stilgenbauer S, Rappold G A, Dohner H, Lichter P. Blood. 1999;94:748–753. [PubMed] [Google Scholar]
- 24.Schaffner C, Idler I, Stilgenbauer S, Dohner H, Lichter P. Proc Natl Acad Sci USA. 2000;97:2773–2778. doi: 10.1073/pnas.050400997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korz C, Pscherer A, Benner A, Mertens D, Schaffner C, Leupolt E, Dohner H, Stilgenbauer S, Lichter P. Blood. 2002;99:4554–4561. doi: 10.1182/blood.v99.12.4554. [DOI] [PubMed] [Google Scholar]
- 26.Camacho E, Hernandez L, Hernandez S, Tort F, Bellosillo B, Bea S, Bosch F, Montserrat E, Cardesa A, Fernandez P L, et al. Blood. 2002;99:238–244. doi: 10.1182/blood.v99.1.238. [DOI] [PubMed] [Google Scholar]
- 27.Hacia J G, Brody L C, Chee M S, Fodor S P, Collins F S. Nat Genet. 1996;14:441–447. doi: 10.1038/ng1296-441. [DOI] [PubMed] [Google Scholar]
- 28.Hacia J G, Edgemon K, Fang N, Mayer R A, Sudano D, Hunt N, Collins F S. Hum Mutat. 2000;16:354–363. doi: 10.1002/1098-1004(200010)16:4<354::AID-HUMU8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 29.Hacia J G, Sun B, Hunt N, Edgemon K, Mosbrook D, Robbins C, Fodor S P, Tagle D A, Collins F S. Genome Res. 1998;8:1245–1258. doi: 10.1101/gr.8.12.1245. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan E L, Meier P. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 31.Peto R, Pike M C, Armitage P, Breslow N E, Cox D R, Howard S V, Mantel N, McPherson K, Peto J, Smith P G. Br J Cancer. 1977;35:1–39. doi: 10.1038/bjc.1977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grønbaek K, Worm J, Ralfkiaer E, Ahrenkiel V, Hokland P, Guldberg P. Blood. 2002;100:1430–1437. doi: 10.1182/blood-2002-02-0382. [DOI] [PubMed] [Google Scholar]
- 33.Robertson K, Hensey C, Gautier J. Oncogene. 1999;18:7070–7079. doi: 10.1038/sj.onc.1203194. [DOI] [PubMed] [Google Scholar]
- 34.Weisenburger D D, Vose J M, Greiner T C, Lynch J C, Chan W C, Bierman P J, Dave B J, Sanger W G, Armitage J O. Am J Hematol. 2000;64:190–196. doi: 10.1002/1096-8652(200007)64:3<190::aid-ajh9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 35.Swerdlow S H, Williams M E. Hum Pathol. 2002;33:7–20. doi: 10.1053/hupa.2002.30221. [DOI] [PubMed] [Google Scholar]
- 36.Decaudin D. Leuk Lymphoma. 2002;43:773–781. doi: 10.1080/10428190290016881. [DOI] [PubMed] [Google Scholar]
- 37.Lossos I S, Thorstenson Y R, Wayne T L, Oefner P J, Levy R, Chu G. Leuk Lymphoma. 2002;43:1079–1085. doi: 10.1080/10428190290021623. [DOI] [PubMed] [Google Scholar]
- 38.Melo J V, Kumberova A, van Dijk A G, Goldman J M, Yuille M R. Leukemia. 2001;15:1448–1450. doi: 10.1038/sj.leu.2402223. [DOI] [PubMed] [Google Scholar]
- 39.Luo L, Lu F M, Hart S, Foroni L, Rabbani H, Hammarstrom L, Yuille M R, Catovsky D, Webster A D, Vorechovsky I. Cancer Res. 1998;58:2293–2297. [PubMed] [Google Scholar]
- 40.Vorechovsky I, Luo L, Ortmann E, Steinmann D, Dork T. Lancet. 1999;353:1276. doi: 10.1016/s0140-6736(05)75199-0. [DOI] [PubMed] [Google Scholar]
- 41.Scott S P, Bendix R, Chen P, Clark R, Dork T, Lavin M F. Proc Natl Acad Sci USA. 2002;99:925–930. doi: 10.1073/pnas.012329699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chenevix-Trench G, Spurdle A B, Gatei M, Kelly H, Marsh A, Chen X, Donn K, Cummings M, Nyholt D, Jenkins M A, et al. J Natl Cancer Inst. 2002;94:205–215. doi: 10.1093/jnci/94.3.205. [DOI] [PubMed] [Google Scholar]
- 43.Kim W J, Vo Q N, Shrivastav M, Lataxes T A, Brown K D. Oncogene. 2002;21:3864–3871. doi: 10.1038/sj.onc.1205485. [DOI] [PubMed] [Google Scholar]
- 44.Ahrendt S A, Halachmi S, Chow J T, Wu L, Halachmi N, Yang S C, Wehage S, Jen J, Sidransky D. Proc Natl Acad Sci USA. 1999;96:7382–7387. doi: 10.1073/pnas.96.13.7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wikman F P, Lu M L, Thykjaer T, Olesen S H, Andersen L D, Cordon-Cardo C, Orntoft T F. Clin Chem. 2000;46:1555–1561. [PubMed] [Google Scholar]
- 46.Wen W H, Bernstein L, Lescallett J, Beazer-Barclay Y, Sullivan-Halley J, White M, Press M F. Cancer Res. 2000;60:2716–2722. [PubMed] [Google Scholar]
- 47.Kristensen V N, Kelefiotis D, Kristensen T, Borresen-Dale A L. Biotechniques. 2001;30:318–322. doi: 10.2144/01302tt01. , 324, 326. [DOI] [PubMed] [Google Scholar]
- 48.Faderl S, Keating M J, Do K A, Liang S Y, Kantarjian H M, O'Brien S, Garcia-Manero G, Manshouri T, Albitar M. Leukemia. 2002;16:1045–1052. doi: 10.1038/sj.leu.2402540. [DOI] [PubMed] [Google Scholar]
- 49.Ek S, Hogerkorp C M, Dictor M, Ehinger M, Borrebaeck C A. Cancer Res. 2002;62:4398–4405. [PubMed] [Google Scholar]