Abstract
The control of lipid and glucose metabolism is closely linked. The nuclear receptors liver X receptor (LXR)α and LXRβ have been implicated in gene expression linked to lipid homeostasis; however, their role in glucose metabolism is not clear. We demonstrate here that the synthetic LXR agonist GW3965 improves glucose tolerance in a murine model of diet-induced obesity and insulin resistance. Analysis of gene expression in LXR agonist-treated mice reveals coordinate regulation of genes involved in glucose metabolism in liver and adipose tissue. In the liver, activation of LXR led to the suppression of the gluconeogenic program including down-regulation of peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1), phosphoenolpyruvate carboxykinase (PEPCK), and glucose-6-phosphatase expression. Inhibition of gluconeogenic genes was accompanied by an induction in expression of glucokinase, which promotes hepatic glucose utilization. In adipose tissue, activation of LXR led to the transcriptional induction of the insulin-sensitive glucose transporter, GLUT4. We show that the GLUT4 promoter is a direct transcriptional target for the LXR/retinoid X receptor heterodimer and that the ability of LXR ligands to induce GLUT4 expression is abolished in LXR null cells and animals. Consistent with their effects on GLUT4 expression, LXR agonists promote glucose uptake in 3T3-L1 adipocytes in vitro. Thus, activation of LXR alters the expression of genes in liver and adipose tissue that collectively would be expected to limit hepatic glucose output and improve peripheral glucose uptake. These results outline a role for LXRs in the coordination of lipid and glucose metabolism.
Liver X receptor (LXR)α and LXRβ have emerged as important regulators of lipid and lipoprotein metabolism. The LXRs are activated by physiological concentrations of oxidized derivatives of cholesterol such as 22(R)-hydroxycholesterol, 27-hydroxycholesterol, and 24(S),25-epoxycholesterol (1–3). LXRα is expressed at particularly high levels in liver, adipose tissue, and macrophages, whereas LXRβ is expressed ubiquitously. These ligand-activated transcription factors form obligate heterodimers with the retinoid X receptor (RXR) and regulate the expression of target genes containing LXR response elements (LXREs). All the LXREs identified thus far are DR-4 hormone response elements (direct repeat of the consensus AGGTCA separated by four nucleotides) (4).
To date, more than a dozen LXR target genes have been identified (5). In the liver, LXRs regulate expression of a number of proteins involved in cholesterol and fatty acid metabolism, including CYP7A and sterol regulatory binding element protein 1c (SREBP-1c) (6, 7). In macrophages and other peripheral cells, LXRs have been implicated in the reverse cholesterol transport pathway. LXRs control the transcription of several genes involved in cellular cholesterol efflux including ATP-binding cassette (ABC)A1, ABCG1, and apolipoprotein E (8–11). LXRs also seem to influence lipoprotein metabolism through the control of modifying enzymes such as lipoprotein lipase, cholesteryl ester transfer protein, and phospholipid transfer protein (12–16).
Here we outline a previously unrecognized role for LXRs in the control of glucose metabolism in liver and adipose tissue. We demonstrate that activation of LXR in the liver leads to the induction of glucokinase expression and to the down-regulation of peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1) and genes involved in gluconeogenesis. We also show that the insulin-sensitive glucose transporter GLUT4 is a direct target for regulation by the LXR/RXR heterodimer in adipose tissue. Finally, we demonstrate that a synthetic LXR agonist improves glucose tolerance in a model of diet-induced obesity and insulin resistance. These observations outline a role for the LXRs in the coordination of lipid and glucose metabolism and suggest that LXR agonists may have utility in the modulation of glucose homeostasis.
Materials and Methods
Reagents and Cell Culture.
pCMX expression plasmids for LXRα, LXRβ, and RXRα have been described (11). GW3965 was kindly provided by Jon Collins and Timothy Willson (GlaxoSmithKline). T1317 was from Cayman Chemical (Ann Arbor, MI). Ligands were dissolved in ethanol or DMSO before use in cell culture. For gene-expression studies, 3T3-L1 cells were cultured in DMEM containing 10% FBS and differentiated into adipocytes by treatment with dexamethasone, methylisobutylxanthine, and 50 nM GW7845. Peritoneal macrophages were obtained from thioglycolate-injected mice as described (11) and cultured in DMEM containing 5% lipoprotein-deficient serum and receptor ligands for 24 h as indicated. For stable promoter studies, undifferentiated 3T3-L1 cells were infected with self-inactivating retroviruses carrying various truncations of the human GLUT4 promoter linked to a luciferase reporter. Stable clones were selected and induced to differentiate into mature adipocytes. On day 8 of differentiation, cells were incubated in serum-free medium supplemented with T1317 as indicated. Luciferase activity was measured 24 h later. Transient transfections of NIH 3T3 cells were performed as described (17). All experiments were performed in triplicate.
RNA and DNA Analysis.
Real-time quantitative PCR assays were performed by using an Applied Biosystems 7700 sequence detector. Total RNA was reverse-transcribed with random hexamers by using TaqMan reverse-transcription reagents kit (Applied Biosystems) according to manufacturer protocol. RNA expression was determined by Sybr green or TaqMan assays as described (18). Statistical analysis of mRNA expression data were performed by using the Student's t test. Probe and primer sequences are available on request. For gel-shift assays, in vitro-translated RXRα and LXRα were generated from pCMX-RXRα and pCMX-hLXRα plasmids (19) by using the TNT quick-coupled transcription/translation system (Promega). Gel-shift assays were performed as described (20) by using in vitro-translated proteins and the following oligonucleotides (only one strand is shown): mGLUT4 LXRE, 5′-gatcctccgggttacttcggggcataca-3′; hGLUT4 LXRE, 5′-gatcccccgggttactttggggcattgc-3′; and GLUT4 mut, 5′-gatcctccggaatacttcggaacataca-3′.
Glucose Uptake.
Glucose-uptake assays were performed in triplicate in differentiated 3T3-L1 adipocytes. Briefly, on day 6 postdifferentiation, 3T3-L1 cells were plated into 96-well Cytostar-T microplates (Amersham Pharmacia) at a density of 50,000 cells per well in 10% FBS-supplemented DMEM. On day 10, the medium was replaced with serum-free medium supplemented with T1317 as indicated. Twenty hours after compound addition, cells were washed twice with KRH buffer (125 mM NaCl/5 mM KCl/1.8 mM CaCl2/2.6 mM MgSO4/5 mM Hepes, pH 7.2) and incubated with FCS buffer (KRH buffer plus 2 mM sodium pyruvate/2% BSA). After 1 h, PBS or insulin (as a control) was added to the cells and incubated for 15 min. In some wells, 20 μM cytochalasin B was also added for background subtraction of nonspecific glucose uptake. Cells were incubated with glucose mixture [0.125 μCi per well 14C-labeled deoxyglucose/100 μM deoxyglucose in PBS (1 Ci = 37 GBq)] for 20 min at 37°C and quenched on ice. Cells were washed with ice-cold KRH buffer, and the radioactivity was quantitated.
Animals and Diets.
For gene-expression studies, 10-week-old female C57BL/6 mice were maintained on standard rodent chow and gavaged daily with GW3965 (20 mg/kg per day) or vehicle (0.5% methylcellulose) for 3 days before being killed. LXRαβ −/− mice on a mixed background and wild-type age-matched controls were maintained on standard chow and treated with vehicle or GW3965 (20 mg/kg per day) for 3 days before being killed. All mice were killed during midlight cycle after a 12-h fast. For diet-induced obesity studies, C57BL/6 mice were fed a high-fat diet for 3 months (Clinton/Cybulsky rodent diet, 40% kcal from fat, devoid of cholesterol; Research Diets, New Brunswick, NJ). After 3 months, mice were treated for 1 week with vehicle or 20 mg/kg per day GW3965 by gavage or supplementation in the chow as indicated. All mice received their final dose of GW3965 by gavage 2–8 h before glucose-tolerance test and/or being killed. Glucose-tolerance tests were performed by i.p. injection of glucose at 2 g/kg body weight after 8 h of fasting. Studies were conducted in accordance with the Animal Research Committee of the University of California, Los Angeles.
Results
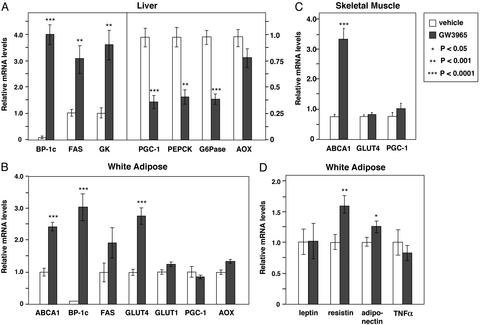
We investigated whether activation of LXRs altered the expression of genes involved in glucose metabolism. C57BL/6 mice (nine animals per group) were treated for 3 days with either vehicle or 20 mg/kg body weight of the synthetic LXR agonist GW3965 (21). At the end of the treatment period, mice were fasted overnight, and total RNA was isolated. Gene expression was analyzed by real-time quantitative PCR assays. The levels of mRNA expression were determined for each animal individually, and the average expression for each group is presented in Fig. 1. In the liver, treatment with GW3965 led to a strong induction of SREBP-1c and fatty acid synthase (FAS) mRNA, consistent with previous work (7). Moreover, LXR agonist treatment also altered expression of a number of genes linked to glucose metabolism. Treatment with GW3965 decreased expression of the transcriptional coactivator PGC-1, which was identified recently as a key regulator of gluconeogenesis (22). Consistent with the decrease in PGC-1, expression of the gluconeogenic enzymes phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase were also down-regulated by GW3965. In addition to these repressive effects, LXR agonist induced expression of glucokinase mRNA. The glucokinase gene is positively regulated by insulin, and its expression is an important determinant of hepatic glucose metabolism. In contrast, expression of the peroxisome proliferator-activated receptor α target gene acyl-CoA oxidase was not altered by LXR ligand in these animals.
Figure 1.
Coordinate regulation of genes involved in glucose metabolism by LXR agonist in vivo. (A) LXR ligand induces glucokinase expression and represses genes involved in gluconeogenesis in liver. (B) LXR ligand induces GLUT4 expression in white adipose tissue. (C) LXR ligands regulate ABCA1 but do not alter GLUT4 or PGC-1 expression in skeletal muscle. (D) Effects of LXR ligands on expression of adipocyte signaling molecules. Ten-week-old female C57BL/6 mice (nine per group) were gavaged daily with GW3965 (20 mg/kg per day) or vehicle. At the end of the treatment period, mice were fasted for 12 h and killed, and total RNA was isolated. Gene expression for individual animals was determined by real-time quantitative-PCR assays. Results are presented as the average expression for each group ± standard deviation.
In adipose tissue, treatment with GW3965 led to the induction of SREBP-1c and ABCA1 expression, consistent with previous work (Fig. 1B). In contrast to the effects observed in liver, expression of PGC-1 was not altered in white fat, indicating that the effects of LXR on this gene are tissue-specific. Interestingly, LXR agonist also stimulated expression of the insulin-sensitive glucose transporter GLUT4 in adipose tissue but had no effect on expression of GLUT1. In skeletal muscle, LXR ligand induced ABCA1 mRNA as expected (23) but did not alter GLUT4 expression (Fig. 1C). We also examined the effect of LXR agonist on the expression of adipocyte signaling molecules known to modulate glucose metabolism and insulin sensitivity (Fig. 1D). Activation of LXR led to a modest increase in expression of resistin and adiponectin but had no effect on expression of either leptin or tumor necrosis factor-α.
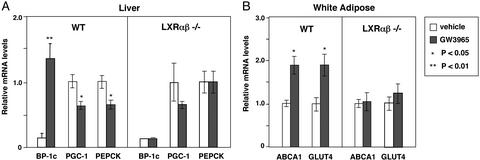
We further examined the ability of this compound to regulate gene expression in mice carrying targeted disruptions in both the lxrα and lxrβ genes. As shown in Fig. 2, the ability of GW3965 to regulate hepatic expression of SREBP-1c, PGC-1, and PEPCK is lost in mice lacking LXRs. Glucokinase expression was also not induced by LXR agonists in livers of LXR null mice (data not shown). Similarly, white adipose tissue expression of ABCA1 and GLUT4 was not significantly altered by LXR agonist in LXR null mice. Together, these observations indicate that LXRs coordinately regulate expression of genes involved in glucose homeostasis in liver and adipose tissue.
Figure 2.
The effects of GW3965 on expression of genes involved in glucose metabolism depend on LXR expression. LXR null mice or wild-type controls on a mixed background (three to five per group) were gavaged daily with GW3965 (20 mg/kg per day) or vehicle. At the end of the treatment period, mice were fasted for 12 h and killed, and total RNA was isolated. Gene expression was determined by real-time quantitative-PCR assays. (A) Regulation of PGC-1 and PEPCK expression by GW3965 is abolished in livers of LXR null mice. (B) Regulation of GLUT4 expression is lost in adipose tissue of LXR null mice. Data are presented as expression relative to vehicle control for each genotype.
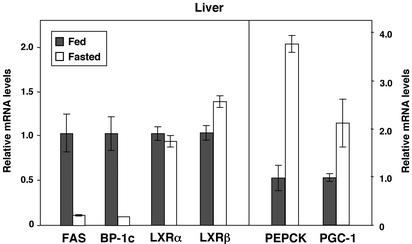
The effects of LXR activation on hepatic gene expression are similar to the effects of insulin in this tissue. We therefore investigated whether expression of LXRα or LXRβ was altered by fasting. As shown in Fig. 3, expression of LXRα and LXRβ was not different between fasted and fed animals. In contrast, SREBP-1c, FAS, and PEPCK gene expression was markedly regulated in these same animals (Fig. 3). These observations suggest that although LXR is an important regulator of SREBP-1c expression, alteration of LXR mRNA expression is unlikely to play a significant role in the induction of SREBP-1c expression in response to the fed state.
Figure 3.
Expression of LXRs is not altered by fasting. C57BL/6 mice (four per group) were fasted for 12 h and killed, and total RNA was isolated. Gene expression was determined by real-time quantitative-PCR assays. Data are presented as relative mRNA expression.
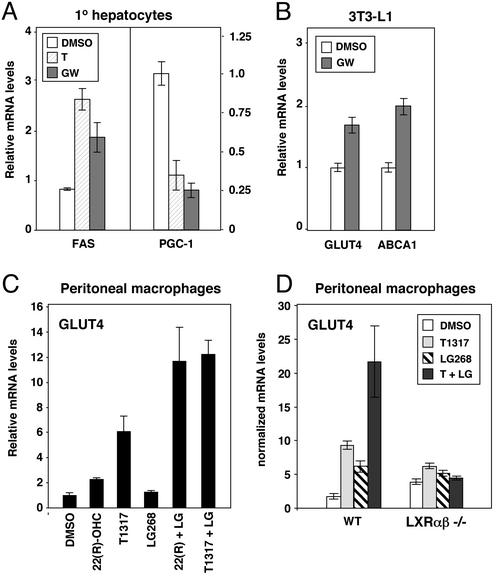
We next investigated whether LXR agonists could regulate PGC-1 and GLUT4 expression in isolated cells. Treatment of primary human hepatocytes for 24 h with 1 μM GW3965 strongly repressed expression of PGC-1 and induced expression of FAS, consistent with the in vivo effects (Fig. 4A). Treatment of differentiated 3T3-L1 adipocytes with GW3965 led to a modest but significant increase in GLUT4 and ABCA1 expression (Fig. 4B), also consistent with the results observed in vivo. To confirm the LXR-dependence of these effects, we isolated peritoneal macrophages from either wild-type or LXR null mice and cultured them in the presence of the oxysterol LXR ligand 22(R)-hydroxycholesterol, the synthetic LXR agonist T1317, and/or the RXR-specific agonist LG268. In wild-type cells, GLUT4 expression was induced by both LXR agonists, and the combination of an LXR agonist and an RXR agonist had an additive effect (Fig. 4 C and D). In contrast, the ability of these compounds to regulate GLUT4 expression was abolished in cells lacking LXRs (Fig. 4D). Similar results were obtained when GW3965 was used in place of T1317 (data not shown). Thus, the ability of LXR agonists to regulate PGC-1 and GLUT4 expression is cell-autonomous and does not seem to require the production of systemic mediators.
Figure 4.
LXR agonists regulate PGC-1 and GLUT4 expression in a cell-autonomous manner. (A) LXR ligands repress PGC-1 expression in primary human hepatocytes. (B) LXR ligand induces GLUT4 expression in 3T3-L1 adipocytes. (C) Regulation of GLUT4 expression in macrophages by synthetic and oxysterol LXR ligands. (D) Regulation of GLUT4 expression by LXR ligand is abolished in cells from LXR null mice. Cells were treated with vehicle or 1 μM T1317 (T), 1 μM GW3965 (GW), 2 μM 22(R)-hydroxycholesterol, or 50 nM LG268 for 24 h as indicated. mRNA expression was determined by real-time quantitative-PCR assays.
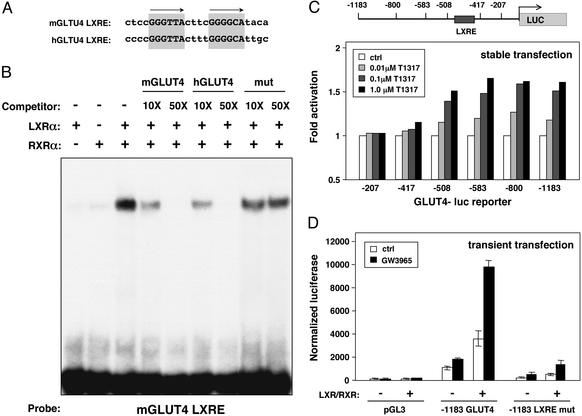
We analyzed the GLUT4 promoter region from the mouse and human genes and identified a conserved DR-4 hormone response element (GLUT4 LXRE, Fig. 5A). In the human, this sequence is located at position −463 bp relative to the transcriptional initiation site, and in the mouse it is located at position −420 bp. Electrophoretic mobility-shift assays using in vitro-translated proteins demonstrated that LXRα/RXRα heterodimers bound to an oligonucleotide spanning this site (Fig. 5B). The specificity of the shifted complex was confirmed by competition with excess unlabeled mGLUT4 or hGLUT4 LXRE oligonucleotide. In contrast, an oligonucleotide carrying a mutation in the LXRE did not compete.
Figure 5.
The GLUT4 promoter is a direct target for regulation by LXR/RXR heterodimers. (A) A conserved DR-4 hormone response element in the GLUT4 promoter. Sequence alignment of LXREs in the mouse and human GLUT4 promoters is shown. (B) A functional LXR binding site in the GLUT4 promoter. Electrophoretic mobility-shift assays were performed by using in vitro-translated proteins and radiolabeled mGLUT4 oligonucleotide. Unlabeled oligonucleotide was added as competitor as indicated. (C) LXR ligands activate the human GLUT4 promoter. Clonal populations of 3T3-L1 cells carrying stably integrated luciferase reporters under the control of various truncations of the human GLUT4 promoter were differentiated into adipocytes. On day 8 of differentiation, cells were incubated in serum-free medium with the indicated concentrations of T1317. Luciferase activity was measured 24 h later. (D) LXRE-dependent activation of the GLUT4 promoter in transient transfection assays. NIH 3T3 cells were transfected with wild-type or LXRE mutant −1,183-bp human GLUT4 luciferase reporters along with expression vectors for LXRα and RXRα. Cells were treated with 1 μM GW3965 12 h after transfection, and luciferase activity was determined 24 h later.
To determine whether this sequence was a functional LXRE, we tested the ability of LXR agonists to activate the human GLUT4 promoter. 3T3-L1 adipocytes carrying stably integrated human GLUT4 promoter-luciferase reporter constructs were cultured for 24 h in the presence of various concentrations of T1317. As shown in Fig. 5C, activity of the −1,183-bp construct was enhanced when cells were treated with T1317 in a dose-dependent manner. Deletion of sequences between −1,183 and −508 bp did not alter ligand responsiveness; however, deletion of the region between −508 and −417 bp, which contains the LXRE, abolished the response. We further tested the ability of LXRs to transactivate the GLUT4 promoter in transient transfection assays. The −1,183-bp promoter construct was cotransfected into NIH 3T3 cells along with expression vectors for LXRα and RXRα. Expression of this construct was strongly stimulated by LXR/RXR in a ligand-dependent manner (Fig. 5D). Furthermore, LXR activation of the −1,183-bp GLUT4 promoter construct was reduced dramatically when a specific mutation was introduced into the −463-bp LXRE, suggesting that this is the primary element mediating induction by LXR. Taken together, the results of Fig. 5 indicate that the GLUT4 promoter is a direct target for regulation by LXR.
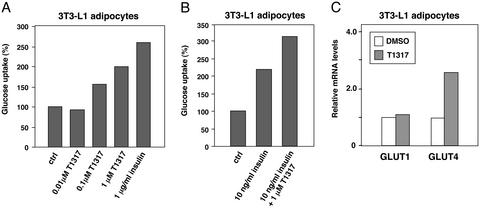
The ability of LXR to regulate GLUT4 expression suggested that LXR ligands might modulate glucose uptake in isolated adipocytes. To investigate this possibility, we performed glucose-uptake assays in differentiated 3T3-L1 adipocytes. As shown in Fig. 6A, treatment of the cells with T1317 significantly increased basal glucose uptake, consistent with previous work (24). Furthermore, LXR agonist also increased insulin-stimulated glucose uptake in 3T3-L1 cells (Fig. 6B). Parallel samples processed for RNA analysis confirmed increased expression of GLUT4 mRNA in these cells under assay conditions (Fig. 6C). Ross et al. (24) reported that GLUT1 protein expression was increased by T1317 in 3T3-L1 cells; however, GLUT1 mRNA expression was not altered by LXR agonist in our study.
Figure 6.
Synthetic LXR ligand promotes glucose uptake in 3T3-L1 adipocytes. (A and B) 3T3-L1 adipocytes at day 10 of differentiation were incubated for 24 h with serum-free medium supplemented with T1317 and/or insulin as indicated. The following day, glucose uptake of treated cells was measured in 96-well Cytostar-T microplates using 14C-labeled 2-deoxyglucose. (C) LXR agonist increases GLUT4 mRNA expression in 3T3-L1 cells. Gene expression was determined by real-time quantitative-PCR assays.
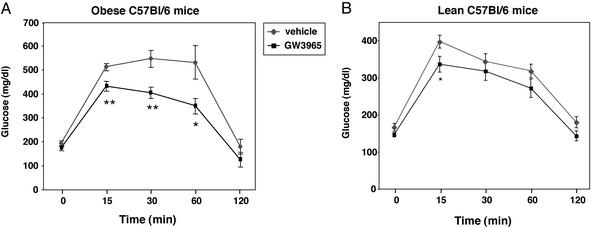
Finally, we investigated the ability of the synthetic LXR agonist GW3965 to influence glucose tolerance in a model of diet-induced obesity and insulin resistance. C57BL/6 mice were maintained on a high-fat diet for 3 months. The obese mice (six animals per group) were treated for 1 week with either vehicle or 20 mg/kg body weight GW3965. Mice were fasted overnight, glucose-tolerance tests were performed, and plasma lipid levels were determined. Fig. 7A demonstrates that treatment with the LXR agonist significantly improved glucose tolerance (P < 0.01 at 15, 30, and 60 min) in obese mice. In contrast, GW3965 had a minimal effect on the normal glucose tolerance of lean C57BL/6 mice maintained on normal chow diet (Fig. 7B). Fasting glucose and insulin levels were not different between treated and untreated groups (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). There was also no statistically significant difference in free fatty acids or triglyceride levels. These results demonstrate that activation of the LXR signaling pathway modulates glucose as well as lipid homeostasis in vivo.
Figure 7.
Synthetic LXR ligand improves glucose tolerance in a model of diet-induced obesity and insulin resistance. (A) Effect of GW3965 on glucose tolerance in obese mice. C57BL/6 mice were fed a high-fat diet for 3 months to induce obesity. After 3 months, mice were treated for 1 week with vehicle or 20 mg/kg body weight GW3965. Glucose-tolerance tests were performed by i.p. injection of glucose (2 g/kg body weight) after 8 h of fasting. (B) Effect of GW3965 on glucose tolerance in lean C57BL/6 mice. Lean C57BL/6 mice were maintained on normal chow diet and treated as described above. *, P < 0.05; **, P < 0.01.
Discussion
The control of lipid and glucose metabolism are closely linked. In the fed state, insulin stimulates peripheral glucose uptake, suppresses hepatic glucose production, and stimulates de novo lipogenesis. Disorders of lipid metabolism can lead to the impairment of peripheral glucose utilization and the development of insulin resistance and type II diabetes. Here we present data to suggest that the nuclear receptors LXRα and LXRβ play a role in the coordinated regulation of lipid and glucose metabolism. We have shown that activation of LXRs with synthetic agonist leads to the suppression of genes involved in gluconeogenesis and induction of hepatic glucokinase expression and insulin-sensitive glucose transporter GLUT4 in adipose tissue. Together, these effects would be expected to limit hepatic glucose production and promote peripheral glucose uptake. Consistent with this hypothesis, we have shown that LXR agonists promote glucose uptake in 3T3-L1 adipocytes and improve glucose tolerance in a model of diet-induced insulin resistance. Finally, we have shown that the GLUT4 promoter is a direct target for the LXR/RXR heterodimer. These observations identify the LXR signaling pathway as a potential target for the pharmacologic modulation of glucose metabolism.
The effects of LXR activation on hepatic gene expression are remarkably similar to the effects of insulin in this tissue. A principal mechanism whereby insulin alters gene expression in this tissue is through transcriptional up-regulation of SREBP-1c (25). Transgenic expression of SREBP-1c in liver induces the entire program of fatty acid synthesis (26). Moreover, the fasting/refeeding response is compromised in SREBP-1c null mice (27). Recent studies have also shown that adenoviral expression of SREBP-1c in liver induces expression of glucokinase and represses the expression of gluconeogenic genes such as PEPCK and glucose-6-phosphatase (28–30). Thus, many of the effects of insulin on both lipid and glucose metabolism may be mediated by SREBP-1c. The results presented here suggest that activation of LXR is also a mechanism for modulation of the gluconeogenic program. Because SREBP-1c is a direct target of LXR, it is possible that many of the effects of LXR agonists in liver are the result of increased expression of SREBP-1c. LXR ligands may mimic the effects of insulin in the liver, because they mimic the effects of insulin on SREBP-1c expression.
An important question, then, is whether LXRs are involved in the physiologic effects of insulin or whether endogenous LXR ligands signal through a parallel pathway. Others have reported that LXRα expression in the liver is induced by insulin (31). By contrast, in our studies expression of LXRα and LXRβ was not different between fasted and fed animals, even though SREBP-1c expression was strongly regulated in these same animals (Fig. 3). LXR expression is clearly required for the optimal expression of SREBP-1c, because SREBP-1c expression is extremely low in LXR null animals (7). However, our data suggest that it is unlikely that modulation of LXRα or LXRβ mRNA expression is the principal mechanism for regulation of SREBP-1c expression during fasting/feeding.
The results presented here raise the possibility that aspects of the LXR signaling pathway may be amenable to pharmacologic manipulation in the context of insulin resistance. A critical question in this regard is whether the beneficial effects of LXR agonists on hepatic glucose metabolism can be separated from effects on SREBP-1c. If not, then suppression of gluconeogenesis would invariably be accompanied by induction of de novo lipogenesis. Accumulation of triglycerides in the liver would offset the beneficial effects on blood glucose levels. In fact, we have observed that treatment of ob/ob or KKAy mice for 2 weeks with LXR ligand does not improve glucose tolerance but dramatically raises plasma triglyceride levels and leads to profound hepatic steatosis (data not shown). Thus, it is likely that LXR effects on glucose metabolism would need to be separated from SREBP-1c-dependent effects on lipogenesis before LXR agonists would be useful as antidiabetic agents. Additional work will be required to decipher the mechanism of LXR action on hepatic expression of genes such as PGC-1 and glucokinase. Another potential mechanism whereby LXR activity may impact hepatic glucose metabolism was reported recently by Stulnig et al. (32), who showed that 11β-hydroxysteroid dehydrogenase type 1 and PEPCK expression are reduced by LXR ligands.
In contrast to many of the effects of LXR agonists in the liver, induction of GLUT4 expression in adipose tissue seems to result from direct action of LXR on the GLUT4 promoter. Previous work has shown that adipose expression of GLUT4 is an important determinant of insulin sensitivity (33). In diabetic rodents and humans, insulin resistance leads to suppression of GLUT4 expression in adipose tissue. Transgenic expression of GLUT4 in white fat improves glucose disposal and ameliorates insulin resistance in diabetic mice (34). Conversely, selective elimination of GLUT4 expression in adipose tissue impairs insulin action in muscle and liver (35). Thus, the development of tissue or gene-selective LXR modulators may allow adipose GLUT4 expression to be regulated independent of lipogenic genes such as SREBP-1c and FAS. More research will be required to determine whether such compounds might have utility as modulators of glucose disposal and insulin resistance in syndromes of obesity/diabetes.
Supplementary Material
Acknowledgments
We thank Tim Willson for GW3965 and helpful discussions and Michael Young, Alan Koder, Russell Romeo, and Leslie Nelson for technical help. P.T. is an Assistant Investigator of The Howard Hughes Medical Institute at the University of California, Los Angeles. D.J.M. is an Associate Investigator of The Howard Hughes Medical Institute at the University of Texas Southwestern Medical Center.
Abbreviations
- LXR
liver X receptor
- RXR
retinoid X receptor
- LXRE
LXR response element
- SREBP-1c
sterol regulatory binding element protein 1c
- ABC
ATP-binding cassette
- PGC-1
peroxisome proliferator-activated receptor γ coactivator-1α
- FAS
fatty acid synthase
- PEPCK
phosphoenolpyruvate carboxykinase
References
- 1.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 2.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundseth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 3.Fu X, Menke J G, Chen Y, Zhou G, MacNaul K L, Wright S D, Sparrow C P, Lund E G. J Biol Chem. 2001;276:38378–38387. doi: 10.1074/jbc.M105805200. [DOI] [PubMed] [Google Scholar]
- 4.Repa J J, Mangelsdorf D J. Annu Rev Cell Dev Biol. 2000;16:459–481. doi: 10.1146/annurev.cellbio.16.1.459. [DOI] [PubMed] [Google Scholar]
- 5.Repa J J, Mangelsdorf D J. Nat Med. 2002;8:1243–1248. doi: 10.1038/nm1102-1243. [DOI] [PubMed] [Google Scholar]
- 6.Peet D J, Turley S D, Ma W, Janowski B A, Lobaccaro J M, Hammer R E, Mangelsdorf D J. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 7.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J M, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laffitte B A, Repa J J, Joseph S B, Wilpitz D C, Kast H R, Mangelsdorf D J, Tontonoz P. Proc Natl Acad Sci USA. 2001;98:507–512. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repa J J, Turley S D, Lobaccaro J A, Medina J, Li L, Lustig K, Shan B, Heyman R A, Dietschy J M, Mangelsdorf D J. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 10.Venkateswaran A, Laffitte B A, Joseph S B, Mak P A, Wilpitz D C, Edwards P A, Tontonoz P. Proc Natl Acad Sci USA. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkateswaran A, Repa J J, Lobaccaro J-M A, Bronson A, Mangelsdorf D J, Edwards P A. J Biol Chem. 2000;275:14700–14707. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- 12.Cao G, Beyer T P, Yang X P, Schmidt R J, Zhang Y, Bensch W R, Kauffman R F, Gao H, Ryan T P, Liang Y, et al. J Biol Chem. 2002;277:39561–39565. doi: 10.1074/jbc.M207187200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Repa J J, Gauthier K, Mangelsdorf D J. J Biol Chem. 2001;276:43018–43024. doi: 10.1074/jbc.M107823200. [DOI] [PubMed] [Google Scholar]
- 14.Luo Y, Tall A R. J Clin Invest. 2000;105:513–520. doi: 10.1172/JCI8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mak P A, Kast-Woelbern H R, Anisfeld A M, Edwards P A. J Lipid Res. 2002;43:2037–2041. doi: 10.1194/jlr.c200014-jlr200. [DOI] [PubMed] [Google Scholar]
- 16.Laffitte B A, Joseph S B, Chen M, Repa J J, Wilpitz D C, Mangelsdorf D J, Tontonoz P. Mol Cell Biol. 2003;23:2182–2191. doi: 10.1128/MCB.23.6.2182-2191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 18.Laffitte B A, Joseph S B, Walczak R, Pei L, Wilpitz D C, Collins J L, Tontonoz P. Mol Cell Biol. 2001;21:7558–7568. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 20.Laffitte B A, Kast H R, Nguyen C M, Zavacki A M, Moore D D, Edwards P A. J Biol Chem. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 21.Collins J L, Fivush A M, Watson M A, Galardi C M, Lewis M C, Moore L B, Parks D J, Plunket K D, Tippin T K, Morgan D G, et al. J Med Chem. 2002;45:1963–1966. doi: 10.1021/jm0255116. [DOI] [PubMed] [Google Scholar]
- 22.Yoon J C, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn C R, Granner D K, et al. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 23.Muscat G E, Wagner B L, Hou J, Tangirala R K, Bischoff E D, Rohde P, Petrowski M, Li J, Shao G, Macondray G, Schulman I G. J Biol Chem. 2002;277:40722–40728. doi: 10.1074/jbc.M206681200. [DOI] [PubMed] [Google Scholar]
- 24.Ross S E, Erickson R L, Gerin I, DeRose P M, Bajnok L, Longo K A, Misek D E, Kuick R, Hanash S M, Atkins K B, et al. Mol Cell Biol. 2002;22:5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horton J D, Bashmakov Y, Shimomura I, Shimano H. Proc Natl Acad Sci USA. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimomura I, Shimano H, Korn B S, Bashmakov Y, Horton J D. J Biol Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 27.Liang G, Yang J, Horton J D, Hammer R E, Goldstein J L, Brown M S. J Biol Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 28.Becard D, Hainault I, Azzout-Marniche D, Bertry-Coussot L, Ferre P, Foufelle F. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]
- 29.Ferre P, Foretz M, Azzout-Marniche D, Becard D, Foufelle F. Biochem Soc Trans. 2001;29:547–552. doi: 10.1042/bst0290547. [DOI] [PubMed] [Google Scholar]
- 30.Chakravarty K, Leahy P, Becard D, Hakimi P, Foretz M, Ferre P, Foufelle F, Hanson R W. J Biol Chem. 2001;276:34816–34823. doi: 10.1074/jbc.M103310200. [DOI] [PubMed] [Google Scholar]
- 31.Tobin K A, Ulven S M, Schuster G U, Steineger H H, Andresen S M, Gustafsson J A, Nebb H I. J Biol Chem. 2002;277:10691–10697. doi: 10.1074/jbc.M109771200. [DOI] [PubMed] [Google Scholar]
- 32.Stulnig T M, Oppermann U, Steffensen K R, Schuster G U, Gustafsson J A. Diabetes. 2002;51:2426–2433. doi: 10.2337/diabetes.51.8.2426. [DOI] [PubMed] [Google Scholar]
- 33.Stenbit A E, Tsao T S, Li J, Burcelin R, Geenen D L, Factor S M, Houseknecht K, Katz E B, Charron M J. Nat Med. 1997;3:1096–1101. doi: 10.1038/nm1097-1096. [DOI] [PubMed] [Google Scholar]
- 34.Tozzo E, Gnudi L, Kahn B B. Endocrinology. 1997;138:1604–1611. doi: 10.1210/endo.138.4.5043. [DOI] [PubMed] [Google Scholar]
- 35.Abel E D, Peroni O, Kim J K, Kim Y B, Boss O, Hadro E, Minnemann T, Shulman G I, Kahn B B. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.