Abstract
Cancer patients and tumor-bearing mice possess serum antibodies that recognize antigens expressed by cancer cells at the time of diagnosis. After diagnosis, cancers progress to more aggressive stages, most often by acquiring new genetic changes that can give rise to new proteins, some of which are antigenic. However, at these relatively later stages of tumor growth, it remains unclear whether, when, and how a host can generate de novo antibody responses against these newly appearing tumor antigens. To this end, we used a tamoxifen-regulated Cre-loxP system, MerCreMer, to induce genetic recombination in cancer cells of well-established tumors, resulting in increased enhanced green fluorescence protein (EGFP) expression. These late tumor-bearing mice generated specific IgG antibodies against EGFP within 3 wk after antigen induction. Mice generated these antibody responses in the presence of preexisting anti-tumor antibody responses. Preexisting CD4+ T cell responses to already expressed tumor antigens likely enhanced antibody responses to the induced EGFP antigen. By analogy, new antibody responses in cancer patients may identify genetic changes occurring in a growing tumor and indicate imminent tumor progression.
Keywords: Cre-loxP‖immunosurveillance‖EGFP‖SEREX‖cancer
Cancer patients, and similarly mice challenged with antigenic cancer cells, possess CD8+ T cells, CD4+ T cells, and antibodies that recognize antigens expressed by the tumor (1). Methods using tumor-specific T cells or antibodies demonstrated that the molecular nature of these antigens seems to result from mutant or aberrantly expressed proteins (2–4). Many of the genetic changes leading to the expression of these antigens seem to occur early during tumor growth (5, 6). Furthermore, detection of these existing tumor antigens has diagnostic and therapeutic potential (7).
However, genetic changes also seem to govern tumor progression during later stages of tumor growth (8), namely, by generating new subpopulations that can invade, metastasize, grow more rapidly, and/or become more resistant to previous treatments (9). Because progression does necessarily not correlate with the loss of antigenicity (10, 11), new genetic changes may also generate new proteins that may be recognized by the immune system. Currently, it remains unclear whether, when, and how antibodies in a tumor-bearing host respond to the antigenic byproducts of these new genetic changes. Such responses could be influenced by “preexisting” immune responses against other tumor antigens and/or T cell immunodeficiencies that have been reported in cancer patients and tumor-bearing animals (1, 12, 13).
To determine whether tumor-bearing hosts generate antibodies against new tumor antigens, we applied a tamoxifen (TAM) regulated Cre-loxP recombination system, MerCreMer (14, 15), to induce antigens in an established tumor. Once induced, these antigens elicited high titer IgG antibody responses within 3 wk. By analogy, new antibody responses arising in cancer patients may help identify and track genetic changes occurring in a progressing tumor, thereby enabling therapies to be tailored to these new antigenic variants.
Materials and Methods
Mice, Cell Lines, and Reagents.
C57BL/6-TgN(ACTbEGPF)Osb (enhanced GFP, EGFP) transgenic mice and B6.129S7-Rag1tm1Mom (Rag1−/−) mice were purchased from The Jackson Laboratory. (BALB/c × C57BL/6)F1 mice (F1) were purchased from the National Cancer Institute, Frederick Cancer Research Facility (Frederick, MD). P. Ohashi (University of Toronto, Toronto) provided the MC57G fibrosarcoma cell line. The 8101 PRO1A fibrosarcoma is a progressor variant of the C57BL/6 tumor 8101 (6). TAM and 4-hydroxytamoxifen (4-OHTAM) were purchased from Sigma. For Western analysis, EGFP was detected by using the Living Color A.v. peptide antibody, an anti-EGFP antibody from CLONTECH, and developed by using Enhanced Chemiluminescence (Amersham Pharmacia). All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Tumor Injection, EGFP Induction, and Delayed Type Hypersensitivity Reaction (DTH).
Cancer cells (2 × 106 to 5 × 106; in 0.2 ml) were injected s.c. into Rag1−/− mice or in F1 mice treated with anti-CD8 ascites (clone YTS 169.4.2). After 2–3 wk of growth, solid tumor fragments were harvested and implanted s.c. into the F1 mice by using a 13-gauge trochar. We observed no difference in the growth rate of uninduced tumors compared with tumors induced to express EGFP. Tumors grew in 20–50% of the F1 mice challenged with tumor fragments. For EGFP induction, 2 mg TAM (in 0.1 ml) was injected i.p. Only the sera from mice bearing 30- to 40-day established tumors were analyzed for anti-EGFP antibodies (see Fig. 4; see Fig. 5 and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org). DTH was performed as described (16).
Figure 4.
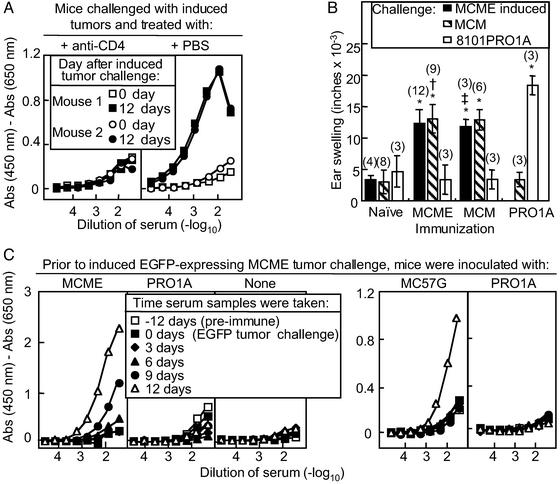
Preexisting, tumor-specific CD4+ T cell responses potentiate anti-EGFP antibody responses. (A) Mice bearing uninduced MCME tumors for 12 days were challenged with induced MCME tumors and treated with anti-CD4 ascites or PBS control i.p. (B) Mice were immunized with the induced MCME, MCM, or PRO 1A cells, and, 2–3 wk later, tested for a DTH response to induce MCME cells, MCM cells, or PRO 1A cells. *, P < 0.005 for DTH response of immunized mice compared with DTH response of naive mice. †, P = 0.4602 for DTH response of mice immunized with MCME-induced cells and challenged with MCM- or MCME-induced cells. ‡, P = 0.1469 for DTH response of mice immunized with MCM cells and challenged with MCM- or MCME-induced cells. Numbers in parentheses indicate the total number of mice used in two independent experiments. (C) Mice were challenged with uninduced MCME tumor fragments, parental MC57G tumor fragments, or PRO1A tumor fragments. Twelve days later, mice were challenged with induced MCME tumor fragments that expressed high levels of EGFP. Data are representative of two to three independent experiments, each using two mice per group.
ELISA for Measuring Anti-EGFP Antibodies.
Anti-EGFP antibodies were measured as described with certain modifications (17). The full ELISA methodology (as well as the methodology for generating the inducible EGFP vector and the inducible EGFP clone, the PCR, and detecting CD4+ and CD8+ T cell responses) is described in the Supporting Text, which is published as supporting information on the PNAS web site.
Results
A TAM-Regulated Cre-loxP System Induced EGFP Expression in Cancer Cells.
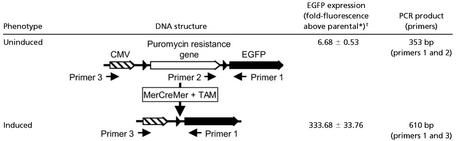
We used EGFP as a model tumor antigen because this antigen stimulates antibody responses in mice (17, 18). We constructed the inducible EGFP vector (iEGFP), in which a puromycin resistance gene flanked by loxP sites separated the cytomegalovirus promoter and the EGFP gene (Table 1). The puromycin resistance gene both inhibited EGFP expression and enabled the selection of cells that contained the unrecombined iEGFP construct. We transfected the fibrosarcoma MC57G with pAN-MerCreMer to generate the clone MC57G-MerCreMer (MCM). MerCreMer contained a Cre recombinase fused to two mutant murine estrogen receptors that regulated the recombinase activity in a TAM-dependent manner (15). MCM was then transfected with the iEGFP construct to generate an MC57G-MerCreMer-iEGFP clone (MCME). In the absence of TAM, MCME cells were uninduced and the iEGFP gene remained unrecombined (Table 1). In the presence of TAM, MerCreMer recombined the iEGFP gene, causing the removal of the floxed puromycin gene and the expression of higher levels of EGFP in MCME cells.
Table 1.
Genetic and expression characteristics of the uninduced and induced phenotypes of MCME tumor cells
Parental is MC57G-MerCreMer clone, MCM.
The fold-increase in EGFP fluorescence above parental MCM cells was determined by the MFI of MCME cells before TAM treatment (0 h) and 4 days after TAM treatment (96 h) from three independent experiments. The fold increase is the average of three independent experiments preformed in duplicate.
4-OHTAM Treatment of MCME Cells Results in Site-Specific DNA Recombination and Increased EGFP Expression in Vitro.
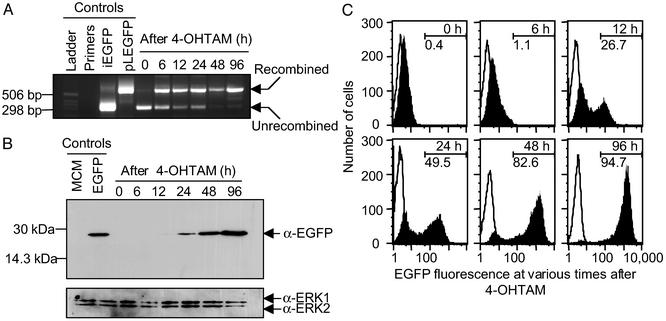
We treated MCME cells with 200 nM of 4-OHTAM. At different times, we harvested genomic DNA and performed multiplex PCR to detect the unrecombined or recombined iEGFP gene (Fig. 1A and Table 1). In the absence of 4-OHTAM, the majority of MCME cells did not recombine the iEGFP gene, whereas in the presence of 4OH-TAM, DNA recombination became apparent within 6 h. We also subjected 4-OHTAM-treated MCME cells to Western analysis for EGFP expression. Whereas multiplex PCR detected DNA recombination by 6 h, we did not detect EGFP expression until 24 h after 4-OHTAM treatment (Fig. 1B).
Figure 1.
4-OHTAM induces site-specific genetic changes in MCME cells, resulting in increased EGFP expression and tumor cell fluorescence. (A) At the indicated times after 4-OHTAM treatment, genomic DNA was subjected to PCR. The 506-bp and 298-bp fragments of the DNA ladder are indicated by the tick marks. Primers, primers only, no DNA. The expected 610-bp fragment indicating the recombined iEGFP gene (see Table 1) becomes apparent after 6 h, although the 353-bp fragment indicating the unrecombined iEGFP gene diminishes within 24 h. (B) At the indicated times after 4-OHTAM treatment, cell lysates were subjected to Western analysis for EGFP. The membrane was then stripped and reprobed with an anti-Erk antibody for loading control. (C) At the indicated times after 4-OHTAM treatment, cells were analyzed by fluorescence activated cell sorting. Shaded histogram, MCME; unshaded histogram, MCM. The numbers above the markers in each histogram plot denote the time after 4OH-TAM treatment; the numbers below the markers in each histogram plot denote the percentage of MCME cells within the demarcated region. Data are representative of three independent experiments performed in duplicate.
To quantify the frequency and level of EGFP induction, we analyzed MCME cells for EGFP fluorescence (Fig. 1C). Although we did not detect EGFP expression by Western analysis, the uninduced MCME cells [mean fluorescence intensity (MFI) 14.07] had ≈5.5-fold higher fluorescence than MCM cells (MFI 2.53). Consistent with previous observations (19), the floxed puromycin gene and the poly(A) termination sites did not completely inhibit EGFP transcription because cells transfected with only the iEGFP vector had slightly elevated levels of EGFP fluorescence (data not shown). Within 6 h of 4-OHTAM treatment, a significant population of cells increased fluorescence, and, after 96 h, >90% of the cells fluoresced above background. At 96 h, induced MCME cells (MFI 1319.2) fluoresced 388-fold above the parental MCM clone (MFI 3.4) indicating that 4-OHTAM induced ≈70-fold increase in EGFP expression. These results also showed that most cells carried the recombined iEGFP gene within 96 h of 4-OHTAM treatment.
TAM Rapidly Induces High, Stable Levels of EGFP Expression in Vivo.
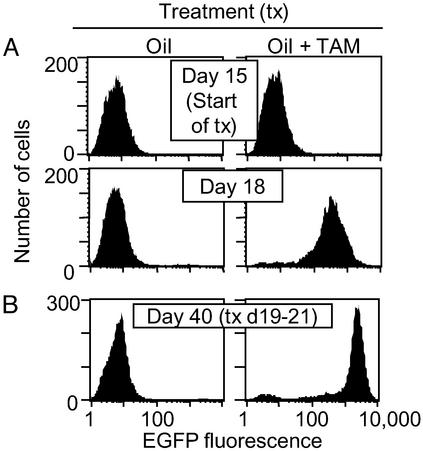
Rag1−/− mice were injected with MCME cancer cells, and, 15 days later, the mice were treated with vehicle alone, 0.25 mg, 0.5 mg, 1 mg, or 2 mg of TAM i.p. for 11 days. Mice were treated with TAM because mice could metabolize TAM into more potent derivatives, e.g., 4-OHTAM (20). We monitored changes in EGFP expression within the tumor by taking fine needle biopsies every 3 days. Fine needle biopsies were cultured in G418 for 7–10 days to kill the contaminating stroma, and EGFP induction was assessed by fluorescence cytometry. Induction of EGFP in tumors was similar in male and female mice (data not shown). In addition, we observed a dose-dependent increase in the rate at which EGFP was induced. Whereas TAM doses of 0.25–1 mg induced the greatest percentage of cells expressing high levels of EGFP at 6 days, 2 mg of TAM induced the greatest percentage of cells expressing EGFP within 3 days (Fig. 2A). To determine the stability of the uninduced or induced phenotype, mice were challenged with uninduced MCME tumor fragments and, at 19 days, treated the mice with 2 mg TAM or the vehicle alone for 3 days (Fig. 2B). Three weeks after TAM treatment, the tumor cells in mice treated with TAM still expressed high levels of EGFP, indicating that the induced phenotype was stable in a growing tumor. Conversely, MCME tumors in mice treated with the control vehicle maintained low levels of EGFP fluorescence, indicating that uninduced phenotype was also stable.
Figure 2.
TAM induces high, stable levels of EGFP in progressively growing tumors whereas the uninduced phenotype is stable for 3 wk. (A) RAG 1−/− mice were injected with 1 × 106 MCME cancer cells. On day 15, mice received 2 mg of TAM or sunflower seed oil every 24 h. Fine needle aspirates of tumors were taken immediately before (day 15) or 3 days after (day 18) the start of TAM treatment. Data are representative of three mice from two independent experiments. (B) Mice were challenged with uninduced MCME tumor fragments. Nineteen days after challenge, tumor-bearing mice were treated with TAM or oil for 3 days. Three weeks after treatment (day 40), tumors were reisolated, and EGFP expression was analyzed by fluorescence activated cell sorting. Data are representative of two mice from two independent experiments.
Tumor-Bearing Mice Rapidly Generate High Titer Anti-EGFP Antibodies in Response to EGFP Induction.
We determined whether tumor-bearing mice were capable of generating an immune response that could detect an antigen induced to high levels of expression within the growing tumor. Because antibody responses against EGFP have been reported in the BALB/c strain, we used (BALB/c × C57BL/6)F1 (F1) mice that retained host reactivity against the EGFP antigen while remaining tolerant to the MC57G tumor (17, 18).
First, we determined whether F1 mice generated a cellular response to MC57 tumors expressing EGFP. We immunized mice with either EGFP-transgenic C57BL/6 splenocytes, MCME cells induced in vitro to express high levels of EGFP, or the parental MCM cells. Whereas EGFP-expressing splenocytes induced the generation of anti-EGFP CD8+ T cells in the peripheral blood, in vitro-induced MCME tumor cells and the parental MCM cell lines failed to induce a similar response (Fig. 5). In addition, we failed to detect IL-4 production, IFN-γ production, or cytolytic activity in the CD8+ or CD4+ T cells from mice immunized with in vitro-induced MCME cells. (Data not shown). However, F1 mice did generate high titer anti-EGFP antibodies against in vitro-induced MCME tumors (P < 0.001) by day 12 (Fig. 3B and Table 2). In contrast, uninduced MCME tumors (Fig. 3C) did not elicit detectable EGFP antibody titers because these titers did not differ from those of naive mice or of mice bearing the parental MCM tumors (Fig. 3A) for a similar amount of time.
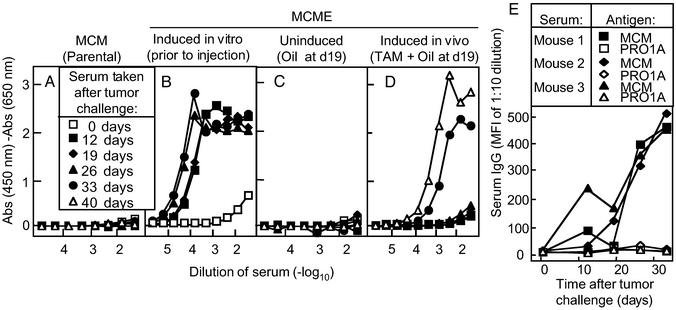
Figure 3.
EGFP induction in an established tumor elicits an anti-EGFP antibody response in the presence of a preexisting anti-tumor antibody response. (A–D) Mice were implanted with parental MCM tumor fragments (A), uninduced MCME tumor fragments (C and D), or in vitro-induced MCME tumor fragments that expressed high levels of EGFP (B). On day 19, mice bearing uninduced MCME tumors were treated with TAM + oil (D) or with oil alone (C) for 3 days. Total numbers of mice in each group are indicated in Table 2. (E) Mice were challenged with uninduced MCME tumors, and sera was collected on day 0, day 12, day 19, day 26, and day 33. Twelve days after tumor challenge, mice were treated with oil (Mouse 1) or oil + TAM (Mouse 2 and 3). At day 33, the anti-EGFP antibody titer of Mouse 1, Mouse 2, and Mouse 3 were 30, 7,290, and 2,430, respectively (summarized in Table 2). Sera at these times was also tested for anti-MC57 antibodies by fluorescence cytometry. Namely, MCM cells or PRO1A cells were incubated with a 1:10 dilution of serum samples, washed, and incubated with phycoerythrin-conjugated goat anti-mouse IgG secondary antibody. Because anti-EGFP antibodies were detected only in Mouse 2 and 3 at day 33, these results indicate that anti-tumor antibodies preceded the anti-EGFP antibody response.
Because mice generated antibodies against tumors expressing EGFP at the time of challenge, we determined whether mice bearing established tumors also generated anti-EGFP antibodies when EGFP was induced in vivo. When mice bearing uninduced MCME tumors for almost 3 wk were treated with TAM, we observed that these mice generated anti-EGFP antibodies within 2–3 wk (Fig. 3D). Three weeks after TAM treatment, eight tumor-bearing mice in five independent experiments possessed a serum anti-EGFP antibody titer of 2,970 ± 989, which was 27- to 243-fold greater than the anti-EGFP antibody serum titer of 30 in control treated mice (P < 0.001; Table 2). Thus, the tumor-bearing host rapidly generated antibodies against new tumor antigens.
Anti-EGFP Antibodies Undergo Class Switching and Arise in the Context of Preexisting Anti-Tumor Antibody Responses.
We characterized IgG class of these new anti-EGFP antibodies to elucidate how antibody responses may arise against new antigens. The predominant anti-EGFP antibody response was of the IgG1 subclass regardless of whether EGFP expression was induced after 3 wk of tumor growth or expressed at the time of tumor challenge (data not shown). Because antibodies can be generated to other tumor antigens, we determined whether specific anti-tumor antibodies existed before the EGFP antibody was detected. We incubated antigen-specific MCM cells or the nonspecific PRO1A cells with sera of mice bearing MCME tumors before or after TAM treatment. Indeed, mice generated specific IgG anti-tumor antibodies before the anti-EGFP antibody response was induced (Fig. 3E).
Preexisting, Tumor-Specific CD4+ T Cell Immunity Potentiates Anti-EGFP Antibody Responses.
Because both the anti-EGFP antibodies and the anti-tumor antibodies were of the IgG class, these observations suggested that CD4+ T cell help existed before and was likely required for the anti-EGFP antibody response. In fact, mice treated with anti-CD4 antibody failed to make anti-EGFP antibodies, suggesting that CD4+ T cells were required for an anti-EGFP antibody response (Fig. 4A). To determine whether MCME cells induced significant anti-EGFP CD4+ T cell responses, we examined whether immunity against induced MCME cells expressing high levels of EGFP conferred EGFP-specific and/or MC57 tumor-lineage-specific T cell immunity. We compared the DTH response to the induced MCME tumor, the parental MCM tumor, or the control PRO1A tumor in mice immunized with these cell lines (Fig. 4B). Immunity to the parental MCM cells was sufficient to induce responses to MCME cells expressing EGFP. Furthermore, mice immunized with induced MCME cells failed to elicit greater DTH responses to MCME cells expressing EGFP compared with the parental MCM cells, suggesting that the EGFP expressed by MCME cells did not induce significant CD4+ T cell-mediated anti-EGFP immunity.
Because we failed to detect EGFP-specific CD4+ T cell responses, we hypothesized that CD4+ T cells responding to other antigens on the cancer cell provided the T cell help necessary to generate anti-EGFP antibodies. We immunized mice with the uninduced MCME tumor or an unrelated tumor PRO1A. After 12 days, we challenged these immunized mice or naive mice with tumor fragments of in vitro-induced MCME cells expressing high levels of EGFP. Mice immunized with uninduced MCME tumors generated substantial anti-EGFP antibodies within 9–12 days after challenge with the induced EGFP tumor (Fig. 4C). In contrast, naive mice and mice immunized with the unrelated PRO1A tumor failed to generate anti-EGFP antibody responses to challenges with EGFP-expressing MCME tumors, indicating that existing tumor immunity was tumor-lineage specific. Because uninduced MCME tumors expressed low levels of EGFP, we assessed whether immunization with the MC57G tumor, which is the parental tumor of the MCME tumor, also potentiated an anti-EGFP antibody response (Fig. 4C). As with the uninduced MCME immunization, mice immunized against MC57G generated greater anti-EGFP antibody responses compared with mice immunized against PRO1A. Thus, existing tumor-associated CD4+ T cell help may potentiate antibody responses against new antigens even when these antigens fail to induce specific CD4+ T cell responses.
Discussion
We find that the tumor-bearing host can rapidly generate antibodies against new antigens that arise from genetic changes occurring within a growing tumor. Furthermore, our observations establish that the immune system can respond to new tumor antigens in the context of an existing anti-tumor immune response. We modeled the appearance of these antigenic variants by inducing genes with a novel Cre-loxP system. When antigen expression increased, the host rapidly generated high titer IgG antibody responses. Although the uninduced tumors expressed antigens at low levels, these tumors did not elicit detectable antibodies. Therefore, new antibody responses may recognize either newly expressed proteins or existing proteins that become overexpressed.
Preexisting CD4+ T cell responses likely potentiated antibody responses to the induced antigen, because CD4+ T cell responses were required but specific CD4+ T cell responses to the induced antigen were not detected. Previous reports have described this mechanism of intermolecular help in which distinct molecules within the same cell or complex contain the helper T cell and B cell epitopes (21–23). Thus, antibody responses may reflect genetic changes occurring within a growing tumor without the need for these antigens to induce new cellular immune responses that may be difficult to generate in a tumor-bearing host. In any case, cancer patients possess tumor-specific CD4+ T cell responses (24) that may directly or indirectly induce an antibody response to a new tumor antigen.
To study antibody responses to new antigens, we used a TAM-regulated site-specific recombination method to induce antigens in established tumors. Ligand-regulated recombination has induced multiple genes in vitro and in transgenic mice (14, 25). Cre recombinase activity does not require energy or cofactors (26), and, therefore, Cre recombinase can efficiently recombine DNA even in a hypoxic tumor environment. Because Cre recombination is essentially irreversible, a short-term TAM treatment permanently induces gene expression, with minimal effects on the tumor-bearing host. In contrast, ligand-regulated promoters reversibly induce genes to levels proportional to the concentration of the ligand in the surrounding environment (27). Because ligand-regulated promoters transiently control gene expression, the level of gene expression in vitro most likely does not approximate the level of gene expression in vivo. Therefore, ligand-regulated recombination provides an effective tool to study the biology of established tumors.
Induction of new antigens in a growing tumor results in new antibody responses despite the down-regulation of cellular immune responses that can be observed in the tumor-bearing host. The tumor-bearing state can induce regulatory T cells, suppressive cytokines, and defects in the signal transduction pathways of lymphocytes (for review see ref. 1). Furthermore, existing antibody responses in a tumor-bearing host may cause antigenic competition (28, 29) thereby suppressing antibody responses against new antigens arising in growing tumors. Whereas CD8+ T cell responses are frequently inhibited in tumor-bearing hosts, our evidence is consistent with previous observations that antibody responses against tumors or other antigens were not suppressed (12, 30). Furthermore, our data support the idea that CD4+ T cells induced at earlier stages of tumor growth help the generation of antibody responses that occur later. In contrast, antigens present at earlier stages of tumor development may elicit both cellular and humoral responses (31). Although it remains unclear whether a similar robust antibody response would develop against less immunogenic tumor-associated self antigens, such antigens when up-regulated in cancers can generate detectable antibody responses (31). In addition, new mutations occurring during tumor progression may cause strongly immunogenic mutant antigens. Whereas passively transferred antibodies can have significant therapeutic effects (for review see ref. 1), the effect of endogenous antibodies on tumor growth remains unclear because endogenous antibodies may prevent CD8+ T cell responses and may promote tumor growth (32, 33). In any case, a patient's sera can identify new targets for cancer therapy (7).
By analyzing the humoral immune response of cancer patients, serological analysis of recombinant cDNA expression (SEREX) has revolutionized the ability to identify the molecular and genetic nature of the target of anti-tumor antibodies (3, 34). Some anti-tumor antibodies recognize important oncogenic proteins and/or can be correlated with certain malignancies (35–37). Because genetic alterations guide tumor progression, we propose that new antibody responses arising in a tumor-bearing host may reflect and identify genetic changes important for cancer progression. Although cancer progression may alter immune responses against tumors, antibody responses against tumor antigens can increase with increasing tumor burden (12, 30) and, thereby, increase the likelihood of detecting a new antibody response. Therefore, serial analysis of a patient's serum may complement direct genetic analysis by providing probes to identify the genetic origin of these changes and by providing a noninvasive means for monitoring these changes that occur during tumor progression. By knowing when and what molecular changes occur during tumor growth, we may better implement therapies to combat the progressing malignancy.
Supplementary Material
Acknowledgments
We thank Dr. D. A. Rowley for exceptional advice and G. Beck-Engeser for technical assistance. This work was supported by National Institutes of Health (NIH) Grants RO1-CA22677, RO1-CA37516, and PO1-CA97296 and by University of Chicago Cancer Research Center Grant CA-14599. M.T.S. is a recipient of NIH Growth and Development Training Grant HD 07009.
Abbreviations
- EGFP
enhanced GFP
- DTH
delayed type hypersensitivity reaction
- TAM
tamoxifen
- 4-OHTAM
4-hydroxytamoxifen
- MCM
MC57G cancer cell clone transfected with pAN-MerCreMer
- MCME
MC57G cancer cell clone transfected with pAN-MerCreMer and iEGFP
- iEGFP
inducible EGFP vector
- MFI
mean fluorescence intensity
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schreiber H. In: Tumor Immunology: Fundamental Immunology. 5th Ed. Paul W, editor. Philadelphia: Lippincott-Raven; 2003. , in press. [Google Scholar]
- 2.Monach P A, Meredith S C, Siegel C T, Schreiber H. Immunity. 1995;2:45–59. doi: 10.1016/1074-7613(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 3.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon T, Cerottini J C, Van den Eynde B, van der Bruggen P, Van Pel A. Annu Rev Immunol. 1994;12:337–365. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 5.Stoler D L, Chen N, Basik M, Kahlenberg M S, Rodriguez-Bigas M A, Petrelli N J, Anderson G R. Proc Natl Acad Sci USA. 1999;96:15121–15126. doi: 10.1073/pnas.96.26.15121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber K, Wu T H, Kast W M, Schreiber H. Clin Cancer Res. 2001;7:871s–875s. [PubMed] [Google Scholar]
- 7.Scanlan M J, Welt S, Gordon C M, Chen Y T, Gure A O, Stockert E, Jungbluth A A, Ritter G, Jager D, Jager E, et al. Cancer Res. 2002;62:4041–4047. [PubMed] [Google Scholar]
- 8.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 9.Nowell P C. Semin Cancer Biol. 2002;12:261–266. doi: 10.1016/s1044-579x(02)00012-3. [DOI] [PubMed] [Google Scholar]
- 10.Ward P L, Koeppen H K, Hurteau T, Rowley D A, Schreiber H. Cancer Res. 1990;50:3851–3858. [PubMed] [Google Scholar]
- 11.Beck-Engeser G, Monach P, Mumberg D, Yang F, Wanderling S, Schreiber K, Espinosa R, Le Beau M, Meredith S, Schreiber H. J Exp Med. 2001;194:285–299. doi: 10.1084/jem.194.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen C A, Urban J L, Van Waes C, Rowley D A, Schreiber H. J Exp Med. 1985;162:1665–1682. doi: 10.1084/jem.162.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizoguchi H, O'Shea J J, Longo D L, Loeffler C M, McVicar D W, Ochoa A C. Science. 1992;258:1795–1798. doi: 10.1126/science.1465616. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Riesterer C, Ayrall A M, Sablitzky F, Littlewood T D, Reth M. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verrou C, Zhang Y, Zurn C, Schamel W W, Reth M. Biol Chem. 1999;380:1435–1438. doi: 10.1515/BC.1999.184. [DOI] [PubMed] [Google Scholar]
- 16.Siegel C T, Schreiber K, Meredith S C, Beck-Engeser G B, Lancki D W, Lazarski C A, Fu Y X, Rowley D A, Schreiber H. J Exp Med. 2000;191:1945–1956. doi: 10.1084/jem.191.11.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D M, Fisher T L, Wei C, Frelinger J G, Lord E M. Immunology. 2001;102:486–497. doi: 10.1046/j.1365-2567.2001.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stripecke R, Carmen Villacres M, Skelton D, Satake N, Halene S, Kohn D. Gene Ther. 1999;6:1305–1312. doi: 10.1038/sj.gt.3300951. [DOI] [PubMed] [Google Scholar]
- 19.Angrand P O, Woodroofe C P, Buchholz F, Stewart A F. Nucleic Acids Res. 1998;26:3263–3269. doi: 10.1093/nar/26.13.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson S P, Langan-Fahey S M, Jordan V C. Eur J Cancer Clin Oncol. 1989;25:1769–1776. doi: 10.1016/0277-5379(89)90347-7. [DOI] [PubMed] [Google Scholar]
- 21.Lake P, Mitchison N A. Cold Spring Harbor Symp Quant Biol. 1977;41:589–595. doi: 10.1101/sqb.1977.041.01.068. [DOI] [PubMed] [Google Scholar]
- 22.Scherle P A, Gerhard W. J Exp Med. 1986;164:1114–1128. doi: 10.1084/jem.164.4.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roosnek E, Lanzavecchia A. J Exp Med. 1991;173:487–489. doi: 10.1084/jem.173.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardoll D, Topalian S. Curr Opin Immunol. 1998;10:588–594. doi: 10.1016/s0952-7915(98)80228-8. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Indra A K, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, Chambon P. Nature. 2000;407:633–636. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- 26.Stark W M, Boocock M R, Sherratt D J. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- 27.Blau H M, Rossi F M. Proc Natl Acad Sci USA. 1999;96:797–799. doi: 10.1073/pnas.96.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albright J F, Omer T F, Deitchman J W. Science. 1970;167:196–198. doi: 10.1126/science.167.3915.196. [DOI] [PubMed] [Google Scholar]
- 29.Waterston R H. Science. 1970;170:1108–1110. doi: 10.1126/science.170.3962.1108. [DOI] [PubMed] [Google Scholar]
- 30.Yagello M, Lespinats G, Fridman W H. Int J Cancer. 1978;22:136–141. doi: 10.1002/ijc.2910220206. [DOI] [PubMed] [Google Scholar]
- 31.Nishikawa H, Tanida K, Ikeda H, Sakakura M, Miyahara Y, Aota T, Mukai K, Watanabe M, Kuribayashi K, Old L J, Shiku H. Proc Natl Acad Sci USA. 2001;98:14571–14576. doi: 10.1073/pnas.251547298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monach P A, Schreiber H, Rowley D A. Transplantation. 1993;55:1356–1361. [PubMed] [Google Scholar]
- 33.Qin Z, Richter G, Schuler T, Ibe S, Cao X, Blankenstein T. Nat Med. 1998;4:627–630. doi: 10.1038/nm0598-627. [DOI] [PubMed] [Google Scholar]
- 34.Old L J, Chen Y T. J Exp Med. 1998;187:1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu T M, Kuriyama M, Johnson E, Papsidero L D, Killian C S, Murphy G P, Wang M C. Ann NY Acad Sci. 1983;417:383–389. doi: 10.1111/j.1749-6632.1983.tb32880.x. [DOI] [PubMed] [Google Scholar]
- 36.Sahin U, Tureci O, Pfreundschuh M. Curr Opin Immunol. 1997;9:709–716. doi: 10.1016/s0952-7915(97)80053-2. [DOI] [PubMed] [Google Scholar]
- 37.Naora H, Montz F J, Chai C Y, Roden R B. Proc Natl Acad Sci USA. 2001;98:15209–15214. doi: 10.1073/pnas.011503998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.