Abstract
Myxococcus xanthus is a Gram-negative bacterium with a complex life cycle that includes vegetative swarming and fruiting-body formation. Social (S)-motility (coordinated movement of large cell groups) requires both type IV pili and fibrils (extracellular matrix material consisting of polysaccharides and protein). Little is known about the role of this extracellular matrix, or fibril material, in pilus-dependent motility. In this study, mutants lacking fibril material and, therefore, S-motility were found to be hyperpiliated. We demonstrated that addition of fibril material resulted in pilus retraction and rescued this phenotype. The fibril material was further examined to determine the component(s) that were responsible for triggering pilus retraction. Protein-free fibril material was found to be highly active in correcting hyperpiliation. However, the amine sugars present in hydrolyzed fibril material, e.g., glucosamine and N-acetylglucosamine (GlcNAc) had no effect on fibril− mutants, but, interestingly, cause hyperpiliation in wild-type cells. In contrast, chitin, a natural GlcNAc polymer, was found to restore pilus retraction in hyperpiliated mutants, indicating that a polysaccharide containing amine sugars is likely required for pilus retraction. These data suggest that the interaction of type IV pili with amine-containing polysaccharides on cell and slime-trail surfaces may trigger pilus retraction, resulting in S-motility and slime-trailing behaviors.
Keywords: gliding motility‖type IV pilus‖microbial development‖biofilm
Myxococcus xanthus is a Gram-negative gliding bacterium that exhibits social (S)-motility, a behavior of hundreds to thousands of cells moving together coordinately on a solid surface without the aid of flagella (1). S-motility mutants are usually defective in aggregation, fruiting-body formation, and cellular agglutination (2, 3), indicating its importance for these cellular events of M. xanthus. Extensive genetic and behavioral analyses revealed that S-motility requires at least three major cellular components: type IV pili (TFP), extracellular fibril material, and lipopolysaccharide (LPS) O-antigen (4–10).
The polarly localized TFP of M. xanthus were initially discovered by Kaiser in 1979 (11), and were later shown to be associated with S-motility (5, 12, 13). Nonpiliated cells (generated by genetic mutations or mechanical depiliation) are defective in aggregation, S-motility, and cellular agglutination. Genetic analyses of the nonpiliated mutants led to the discovery of many M. xanthus pil genes, which are highly homologous with the pil genes found in Pseudomonas aeruginosa and Neisseria gonorrhoeae (14, 15). These findings suggested that S-motility of M. xanthus and twitching motility of Pseudomonas and Neisseria employ similar mechanisms. For a long time, however, the exact role of TFP in cellular motility remained a mystery. Recent studies on TFP of M. xanthus, N. gonorrhoeae, and P. aeruginosa by using very different experimental approaches provided clear evidence that social gliding or twitching motility is generated through retraction of pilus filaments, a mechanism originally proposed by Bradley in 1972 (16). A tethering assay developed by Sun et al. (17) for M. xanthus cells allowed observation of tethered M. xanthus cells being drawn closer to the tethering surface in a pilus-dependent manner. Merz et al. (18) used laser tweezers to demonstrate that N. gonorrhoeae cells were able to pull TFP antibody-coated beads by means of the retraction of TFP. Direct observation of pilus retraction in P. aeruginosa was recently achieved by Skerker and Berg (19) by using fluorescent labeling of pili with a Cy3 dye and visualization by means of total internal reflection microscopy. These findings strongly indicate that social-gliding motility of M. xanthus and twitching motility of Pseudomonas and Neisseria are closely related. In this study, we propose to call this mode of cellular translocation “TFP-dependent motility.”
S-motility in M. xanthus requires not only TFP but also extracellular fibril material. This extracellular fibril material was initially identified as cell-surface appendages, since fixed M. xanthus cells contain peritrichous, filamentous structures 10–30 nm in diameter (called fibrils) when viewed with electron microscopy (7, 20, 21). Later studies revealed that these fibrils likely form a capsule over the entire cell body and are likely to be a component of the extracellular matrix composed of protein and carbohydrate moieties (22). Interestingly, like pil mutants, all mutants lacking this extracellular fibril material are defective in aggregation, S-motility, and cellular agglutination (6, 7, 23). Genetic analyses indicated that biogenesis of extracellular fibril material appears to involve a DnaK homolog (encoded by sglK), a set of chemotaxis homologues (encoded by the dif genes), and some uncharacterized dsp genes (7, 23–25). Only recently, some mutations resulting in lack of fibril material and S-motility have been localized to genes associated with carbohydrate transport and biosynthesis (A.L., Y.L., K. Cho, D.Z., and W.S., unpublished data).
Extracellular fibril material is composed of approximately equal amounts of protein and carbohydrate (22). Fibril protein A (FibA), a zinc-metalloprotease homologue and major protein in fibril material, appears not to be required for known functions of fibril material (26, 27). HPLC analysis of fibril carbohydrates showed that they are composed of monosaccharides such as galactose, glucose, glucosamine, rhamnose, and xylose (22). However, little is known about the structure of extracellular fibril material or its role in M. xanthus S-motility on a molecular level.
LPS O-antigen is another major component of the extracellular matrix of M. xanthus (10) that may play an important role in TFP-dependent functions. M. xanthus possesses the typical LPS of a Gram-negative bacterium, consisting of lipid A, core oligosaccharide, and O-antigen (28). LPS O-antigen mutants of M. xanthus are severely defective in gliding motility (especially in S motility), but resemble the wild-type phenotype in fibril-material synthesis and cellular agglutination (10). The exact function of LPS O-antigen in S-motility still needs to be determined.
Both TFP and extracellular matrix are required for S-motility, as demonstrated by the above genetic and behavioral analyses. The role of TFP in S-motility is now well understood through recent advances (17–19) made in uncovering the mechanism of TFP-dependent motility. On the other hand, little is known about the function of extracellular matrix in S-motility. TFP are suggested to enable bacteria to move on solid surfaces by means of the extension of pilus filaments and attachment to the surface at their distal tips followed by retraction. The details of the mechanism, however, especially how TFP attach their distal tips to a solid matrix and how retraction is triggered, remain to be elucidated. This study reports that the carbohydrate portion of extracellular fibril material of M. xanthus can provide an anchor for TFP that induces their retraction, suggesting that the extracellular matrix is involved in regulating TFP-dependent cellular processes.
Materials and Methods
Bacterial Strains and Growth Conditions.
The M. xanthus strains used in this study are listed in Table 1. M. xanthus cells were grown in CYE medium (10 g/liter casitone/5 g/liter yeast extract/8 mM MgSO4 in 10 mM Mops buffer, pH 7.6; ref. 29) at 32°C on a rotary shaker at 225 rpm. Strain SW509 was constructed by transducing an Mx4 lysate of SW501 into DK10409 and selecting for kanamycin resistance (Kmr).
Table 1.
Bacterial strains
| Strain | Relevant genotype | Ref. or source |
|---|---|---|
| DK1622 | Wild type | 11 |
| DK10407 | Δ pilA∷Tcr | Dale Kaiser's lab |
| SW504 | Δ difA | 37 |
| SW501 | Δ difE∷Kmr | 37 |
| SW301 | sglK | 25 |
| DSP1689 | dsp-1689 | Larry Shimkets' lab |
| SW509 | pilT∷Tn5Kmr/Δ difE | This study |
| SW600 | frzE∷Tn5TcΩ234 | 38 |
| SW601 | frzD∷Tn5TcΩ224 | 38 |
| SW602 | Δ difA frzE∷Tn5TcΩ234 | 38 |
| SW603 | Δ difA frzD∷Tn5TcΩ224 | 38 |
| HK1324 | Δ wzt wzm wbgA | 10 |
Detection of Pilin Protein with Western Blotting.
M. xanthus whole-cell pilin (encoded by pilA) was examined by Western blotting using anti-PilA antibody as reported by Wu and Kaiser (14). Cell-surface pilin was isolated and assayed as described (14).
Detection of Cell-Surface Pili with Transmission Electron Microscopy (TEM).
Cell-surface pili were examined by using negative staining and TEM as described (11), with the following modifications: The cells were pelleted and resuspended to 5 × 109 cells per ml in TPM buffer (10 mM Tris, pH 7.6/1 mM KH2PO4/8 mM MgSO4). One drop of cell suspension was added onto the carbon-coated microscope grid and incubated for 5 min at room temperature. This droplet was washed off by addition of several drops of the negative-staining solution (1% uranyl acetate/30 μg/ml bacitracin). The staining solution was then carefully removed with filter paper and the grid was air dried. Samples were examined with a JEOL-100CX electron microscope and micrographs were taken at an original magnification of ×10,000 or ×1,900.
Isolation and Treatment of Fibril Material.
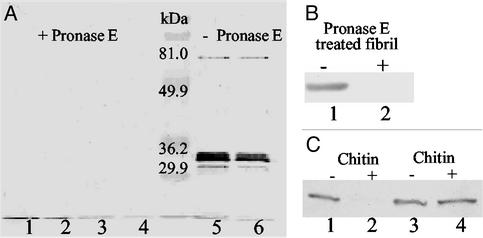
Extracellular fibril material was isolated and quantified as described (30), with the following modifications to generate protein-free fibril material: To remove proteins, cell lysates were treated with 1 mg/ml Pronase E (Sigma) at 37°C for 180 min as described by Sambrook and Russell (31). Final pellets obtained through centrifugation during fibril preparation were washed four times with 0.1% SDS to inactivate residual Pronase E. Inactivation was ensured by the inability of treated fibril material to digest pilin sheared off from fibril− mutant SW504. The removal of proteins from fibril material by Pronase E was confirmed by Western blotting using mAb 2105 (21).
Mixing Assay.
Fibril+ cells, isolated fibril material, monosaccharides, or the polysaccharide chitin (Sigma), and granular cellulose (Sigma) were mixed with 5 × 109 fibril− mutant cells and incubated at 32°C for 30 min. Chitin and cellulose were suspended to 1 mg/ml in cohesion buffer [10 mM Mops buffer (pH 6.8)/1 mM CaCl2/1 mM MgCl2]. Cell-surface pilin/pili were then analyzed with immunoblotting, or TEM, as described above.
Pilin Precipitation Assay.
Cell-surface pili/pilin were sheared off 109 SW504 cells by vortexing as described by Wu and Kaiser (14). The isolated pili/pilin were then incubated with a chitin or cellulose suspension (final concentration 0.2 mg/ml for both) or fibril material (0.2 mg/ml carbohydrate) at 32°C for 30 min. The mixtures were pelleted by centrifugation at 6,000 × g for 5 min. Because of the viscous nature of isolated fibril material, the spin was performed at 10,000 × g for 10 min for incubation with fibril material. The supernatants were discarded, and the were pellets resuspended in 80 μl of Mops buffer (pH 6.8) and boiled with protein-loading dye for SDS/PAGE.
Agglutination Assay.
The cohesion of M. xanthus cells was measured with an agglutination assay described by Chang and Dworkin (30).
Results
Fibril− Mutants Contain an Unusually High Amount of Pilin Protein on the Cell Surface.
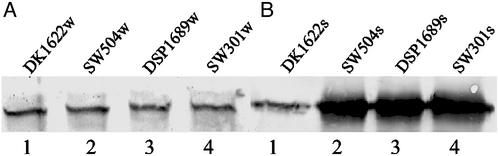
Previous work (3, 6, 23, 25) showed that sglK, dif, and dsp mutants were defective in the production of extracellular fibril material and S-motility. To explore the role of extracellular fibril material in TFP-dependent motility, cellular and cell-surface PilA (pilin protein) of vegetative wild-type or mutants deficient in fibril material were assayed by using immunoblotting with anti-PilA antibody. All mutants lacking extracellular fibril material tested, including SW504 (ΔdifA), SW501 (difE), DSP1689 (dsp), and SW301 (sglK), still produce PilA at levels comparable to wild-type DK1622 (Fig. 1A). The amount of cell-surface pilin, in contrast, was significantly elevated in the fibril− mutants (Fig. 1B). These data show that, whereas the fibril material does not affect production of PilA, it is involved in controlling the cellular localization of pilin.
Figure 1.
Detection of pilin protein in wild-type and fibril− mutants by Western blotting analysis. (A) Whole-cell pilin. Lanes 1–4, whole-cell lysates from 5 × 107 DK1622, SW504, DSP1689, and SW301 cells, were separated by SDS/15% PAGE, electroblotted, and probed with a polyclonal antibody against PilA. The apparent molecular mass of PilA is ≈25 kDa. (B) Cell-surface pilin. Lanes 1–4, pilin was sheared off from 109 DK1622, SW504, DSP1689, and SW301 cells and was immunoblotted as in A. Shown are representative data for triplicate experiments.
Fibril− Mutants Are Hyperpiliated.
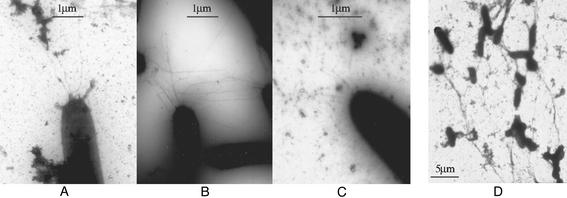
To further investigate the above finding, negative staining and TEM were used to directly visualize pili on the cell surface. More than 100 wild-type and fibril− mutant cells were examined. Under our experimental conditions, ≈10% of the wild-type cells were found to contain an average of 3–4 TFP at the cell poles, and most of the TFP observed in wild-type cells were ≈3–5 μm in length (Fig. 2A). Under the same conditions, however, all fibril− mutants examined were hyperpiliated (Fig. 2B), with >90% of the fibril− mutant cells containing 9–10 TFP at their cell poles. Additionally, the majority of TFP observed in fibril− mutants were longer than 5 μm, with some extending to >20 μm. Consistent with the above immunoblotting results, the high amount of pilin protein found on the cell surface of fibril− mutants is, therefore, likely the result of hyperpiliation.
Figure 2.
TEM micrographs of cell-surface pili. Cells were negatively stained and viewed with a JEOL-100CX electron microscope. (A) DK1622. (×10,000.) (B) SW504. (×10,000.) (C) SW504 after 1-h incubation with fibril material isolated from DK1622. (×4,000.) (D) DK1622 after 1-h incubation with 50 mM GlcNAc. (×1,900.) For each strain, >100 cells were examined. Shown are representative images.
Hyperpiliation of Fibril− Mutants Can Be Rescued Through Mixing with Wild-Type Cells.
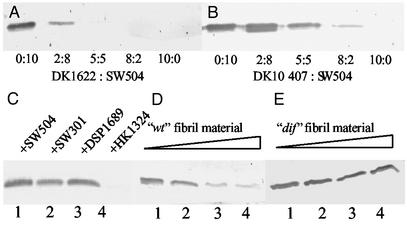
The above findings demonstrate that the lack of extracellular fibril material results in a considerable increase in both the number of surface pili and filament length for the majority of cells observed. To test the possibility of whether the addition of cells with wild-type extracellular matrix could rescue this phenotype, the hyperpiliated fibril− mutant SW504 was mixed with wild-type DK1622 cells at the various ratios indicated (Fig. 3). The addition of DK1622 to SW504 cells caused a striking reduction in cell-surface pilin (Fig. 3A). To exclude the possibility of interference from cell-surface pili of the wild-type cells, the PilA−, fibril+, and LPS+ strain DK10407 was used for mixing with SW504 as well. As shown in Fig. 3B, DK10407 was also able to rescue the hyperpiliation, although to a lesser extent as compared with wild type. This result is consistent with the finding that DK10407 has reduced level of fibril material on the cell surface (H.S. and W.S., unpublished data). A time course showed that the reduction of cell-surface pilin usually occurred at ≈5–10 min after mixing (data not shown).
Figure 3.
Effects of fibril+ cells and isolated fibril material on overpiliation of SW504. (A and B) SW504 cells (4 × 108) were mixed with different amounts of DK1622 (A) or DK10407 (B) cells at the indicated ratio and the surface pilin of the mixture was immunoblotted. (C) Surface pilin from the following 1:1 mixtures (2 × 108 cells for each): SW504/SW504 (lane 1), SW504/SW301 (lane 2), SW504/DSP1689 (lane 3), and SW504/HK1324 (lane 4). (D and E) Surface pilin from 4 × 108 SW504 cells after incubating SW504 with fibril material isolated from DK1622 (D) or SW504 (E). Respectively, 0, 0.015, 0.15, and 1.5 μg/μl fibril material from DK1622 (lanes 1–4 in D) or from SW504 (lanes 1–4 in E) was used for incubation. Data shown are representative for triplicate experiments.
These results clearly demonstrate that the extracellular matrix provided by wild type or the pilA mutant strain is responsible for the reduction of cell-surface pilin in the fibril− mutant SW504. Because fibril material and LPS O-antigen are two major components of the extracellular matrix known to be required for S-motility, we examined whether different mutants lacking fibril material or LPS O-antigen can complement each other and reduce hyperpiliation in fibril− mutants. The various mutants lacking fibril material (dsp, dif, and sglK) could not rescue the hyperpiliation phenotype of SW504, but the LPS O-antigen mutant HK1324 was able to do so (Fig. 3C). Therefore, the extracellular fibril material is most likely the factor responsible for the reversion of hyperpiliation of fibril− mutants, whereas LPS O-antigen does not seem to contribute to this bioactivity.
Hyperpiliation of Fibril− Mutants Is Rescued by Addition of Isolated Fibril Material.
To obtain direct evidence that extracellular fibril material per se is responsible for the correction of hyperpiliation observed with the fibril− mutants, fibril material was isolated from DK1622 according to a procedure described by Chang and Dworkin (30). Isolated fibril material was added to fibril− mutants (e.g., SW504) and incubated for 30 min. Cell-surface pilin protein and pilus filaments of the fibril− mutants were then examined with Western blotting and TEM, respectively. SW504 treated with isolated fibril material exhibited a significant reduction of cell-surface pilin (Fig. 3D), suggesting that the extracellular fibril material indeed regulates piliation. Consistent with the Western blotting data, TEM analysis revealed that fibril− mutants treated with isolated fibril material had significantly fewer pili protruding from their cell poles (Fig. 2C), compared with untreated mutant cells (Fig. 2B). As a control, various fibril− mutants were subjected to the same isolation procedure for fibril material, and the extract was then used to treat the fibril− mutants. This isolated extracellular material had no effect on the hyperpiliation of fibril− mutants (results for SW504 are shown in Fig. 3E as an example). These data demonstrate that extracellular fibril material is both sufficient and necessary for the rescue of hyperpiliation in mutants lacking this surface feature.
Reduced Cell-Surface Pilin and Pili Are Due to Fibril-Induced TFP Retraction.
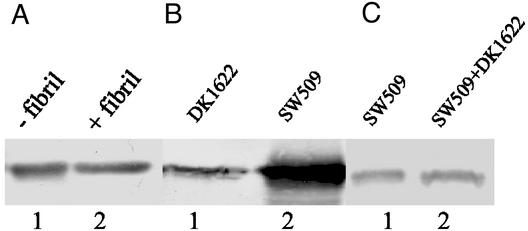
The above results strongly indicate that extracellular fibril material triggers the reduction of cell-surface pilin and pili, but how this reduction is achieved remains unclear. One possibility could be that cell-surface pilin is degraded by proteases (such as FibA) present in fibril material (26). Alternatively, fibril material may induce TPF retraction. To address the first possibility, isolated fibril material was added to cell-surface pilin extracted from SW504 and immunoblotted with anti-PilA antibody to test for possible degradation. Fig. 4A shows that pilin protein was not degraded by isolated fibril material, thus rendering this possibility unlikely. To examine the second hypothesis, a pilT difE double mutant (SW509) was constructed by transducing a Mx4 lysate of SW501 (difE∷Kmr) into DK10409 (pilT) and selecting for kanamycin resistance. PilT has been shown to be required for TFP retraction because the pilT mutants contain nonfunctional/paralyzed TFP (32). SW509, like other fibril− mutants, has the hyperpiliation phenotype (Fig. 4B), showing that the PilT mutation does not affect TFP extrusion. The mixing experiment of SW509 and DK1622 (Fig. 4C) demonstrated that contact with wild-type cells failed to rescue the overpiliation phenotype in this mutant, presumably because of the inability of the pili to retract. These results provide further evidence that fibril-induced pilus retraction, rather than pilin degradation, is the mechanism for the fibril-mediated cell-surface pilin reduction.
Figure 4.
Rescue of overpiliation in fibril− mutants by isolated fibril material. Cell-surface pilin was assayed by Western blotting analysis as described earlier. (A) Surface pilin from 4 × 108 SW504 before (lane 1) or after (lane 2) incubating extracted pilin with 20 μl of isolated fibril material (1.5 μg/μl) for 1 h at 37°C. (B) Surface pilin from 109 DK1622 (lane 1) and SW509 cells (lane 2). (C) Surface pilin from 2 × 108 SW509 cells alone (lane 1) or from the SW509/DK1622 mixture (1:1; lane 2). Shown are representative data for triplicate experiments.
The Carbohydrate Portion of the Fibril Material Is Responsible for Inducing TFP Retraction.
Previous biochemical analyses revealed that extracellular fibril material is composed of approximately equal amounts of protein and carbohydrate (22). To further investigate which component of fibril material is responsible for inducing TFP retraction, the fibril material isolation protocol was modified to obtain protein-free fibril material (see Materials and Methods). This modified procedure effectively removed the protein component (Fig. 5A). The addition of this protein-free fibril material to the fibril− mutant (SW504) triggered pilus retraction (Fig. 5B) as efficiently as the untreated fibril material, indicating a minimal role for the protein component in this bioactivity.
Figure 5.
Effect of carbohydrate on TFP retraction. (A) The protease treatment of fibril material was monitored by immunoblotting analysis using mAb 2105 (anti-FibA antibody). Lanes 1–4, various concentrations of DK1622 fibril material (8, 4, 2, and 1 mg/ml, respectively) were treated with 1 mg/ml Pronase E at 37°C for 180 min; lanes 5 and 6: 2 and 1 mg/ml untreated DK1622 fibril material. In each lane, 20 μl samples were loaded. (B) Surface pilin from 4 × 108 SW504 cells before (lane 1) or after (lane 2) incubating SW504 with protein-free fibrils (1.5 mg/ml) as described in Materials and Methods (lane 2). PilA was probed with anti-PilA antibody as for Fig. 3. (C) Lanes 1 and 2, surface pilin from 4 × 108 SW504 before (lane 1) or after (lane 2) incubating SW504 with 1 mg/ml chitin suspension (30 min at 32°C); lanes 3 and 4, pilin isolated from 4 × 108 SW504 before (lane 3) or after incubating isolated pilin with 1 mg/ml chitin suspension (1 h at 37°C). Shown are representative data for triplicate experiments.
Specific Monosaccharides in Extracellular Fibril Material Play Key Roles in Mediating TFP Retraction.
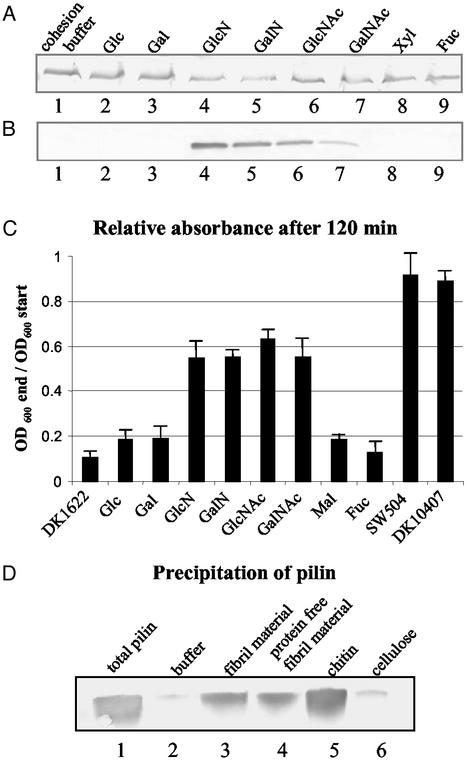
HPLC analysis of fibril material carbohydrates showed that they are composed of monosaccharides such as galactose, glucose, glucosamine (GlcN), rhamnose, and xylose (22), among which GlcN was reported to inhibit cellular agglutination of DK1622. In this study, monosaccharides found in fibril material along with other sugar moieties abundant in the bacterial cell wall, e.g., GlcNAc and those with similar structure such as galactosamine (GalN) and N-acetylgalactosamine (GalNAc) were tested for their effect on TFP. None of the monosaccharides examined was able to reduce hyperpiliation of the fibril− mutants (Fig. 6A), excluding the possibility that the individual monosaccharides alone can trigger pilus retraction. Interestingly, the modified sugars carrying amine substitutions, including GlcN, GalN, GlcNAc, and GalNAc, were able to cause hyperpiliation of the wild type (Figs. 6B and 2D). A plausible explanation for this phenomenon is that the amine-substituted monosaccharides block the interaction between fibril material and TFP in wild type by competitively binding to the fibril recognition site(s) on pilus, thus preventing wild-type TFP from retracting. In support of this explanation, these four monosaccharides were also found to inhibit cellular agglutination of wild-type cells (Fig. 6C), a feature that requires the presence of both pili and the extracellular fibril material. Additionally, preincubation of SW504 with these four amino sugars, but not the other monosaccharides, abolishes the rescue effect of wild-type cells on the overpiliation of this mutant (data not shown). This result also provides further evidence for our previous conclusion that retraction, rather than pilin degradation, accounts for the reduction in cell-surface pilin observed for the mixing experiments described earlier.
Figure 6.
Effect of monosaccharides on TFP retraction. Cell-surface pilin was assayed as described for Fig. 3. (A) SW504 (4 × 108 cells) was incubated at 32°C for 150 min with cohesion buffer (lane1); 100 mM glucose (Glc), galactose (Gal), GlcN, GalN, GlcNAc, GalNAc, xylose (Xyl) (lanes 2–8), or fucose (Fuc) (as negative control, lane 9) and surface pilin was immunoblotted. All sugar solutions had been adjusted to a pH of 7 and cell viability was tested after the incubation, showing no noticeable difference when compared with the cohesion buffer control. (B) DK1622 (4 × 108 cells) was incubated at 32°C for 150 min with 100 mM designated sugars and cell-surface pilin was immunoblotted. Lanes 1–9, same as in A. Data presented in A and B are representative for triplicate experiments. (C) Cohesion inhibition of wild-type DK1622 on the addition of different sugars at a final concentration of 50 mM. Mal, maltose; other abbreviations are the same as in A. SW504 and DK10407 were used as negative controls. OD600 was taken every 10 min for 120 min. The OD600 of each sample after 120 min (OD600 end), relative to its OD600 at the beginning of the assay (OD600 start), is shown in the graph. Data represent the mean ± SD of three experiments. (D) Cell-surface pilin was sheared off from 5 × 108 SW504 cells and the total pilin amount as prepared by MgCl2 precipitations shown in lane 1. Same amounts of isolated pilin were also precipitated with cohesion buffer (lane 2), fibril material (0.2 mg/ml carbohydrate, lane 3), Pronase-treated fibril material (0.2 mg/ml carbohydrate, lane 4), chitin suspension (0.2 mg/ml, lane 5, and cellulose suspension (0.2 mg/ml, lane 6).
Chitin, a GlcNAc Polymer, Induces TFP Retraction.
Although GalN, GlcN, GlcNAc, and GalNAc appear to play a key role in mediating the interaction with TFP, these individual monosaccharides could not induce TFP retraction in fibril− mutants (Fig. 6A). This phenomenon can be explained by the requirement for certain anchoring resistance naturally provided by the extracellular matrix to generate pulling forces involved in pilus retraction. To validate this hypothesis, chitin, a natural polymer of GlcNAc, was tested for its effect on TFP retraction. As shown in Fig. 5C (lanes 1 and 2), chitin was indeed able to induce TFP retraction of fibril− mutants, similar to what we observed above for addition of extracellular fibril material. The presence of pili-degrading agents in chitin was ruled out by addition of chitin to pilin isolated from SW504 cell surfaces (Fig. 5C, lanes 3 and 4). As control, cellulose, a glucose polymer with the same β1–4 linkage as chitin was used. Interestingly, cellulose failed to rescue the hyperpiliation phenotype in fibril− mutants under the same conditions (data not shown), strongly suggesting the presence of specific TFP-recognition moieties in chitin that are absent in cellulose (i.e., acetylated amino groups).
Isolated Pili/Pilin Directly Bind to Fibril Material and Chitin.
To test whether pilin can directly bind to fibril material or chitin, pili/pilin were sheared off SW504 cell surface, mixed with fibril material or chitin, and precipitated by a centrifugation as described in Materials and Methods. Cellulose and Mops buffer served as controls. Our data clearly demonstrated that pilin protein can be effectively precipitated by fibril material and chitin, but not by cellulose (Fig. 6D), which strongly suggests direct binding of pilin protein to fibril material and chitin. This observation is consistent with the hyperpiliation rescue data detailed above (chitin, but not cellulose, can rescue the hyperpiliation of SW504).
Discussion
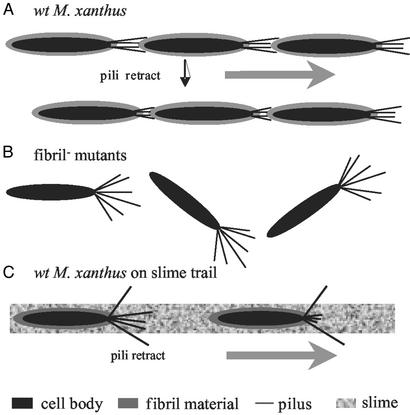
Both TFP and extracellular matrix are required for S-motility and cellular agglutination (4–8). Despite the increasing knowledge on TFP of M. xanthus and other bacteria on a molecular level (4, 5, 17), little is known about the role of the extracellular matrix in these cellular events. By analyzing fibril− mutants that are defective in S-motility and cellular agglutination, we provide evidence that the extracellular matrix, in particular, the extracellular fibril material, is able to interact with TFP and induce TFP retraction. In addition to the data presented here, we were able to block the rescue effect of fibril material on SW504 overpiliation with an anti-PilA antibody reacting with native PilA protein (Y.L. and W.S., unpublished data). This finding further supports the pilus-retraction model presented in this study. The observation that fibril material triggers pilus retraction would explain why the extracellular matrix is an essential part of the TFP-dependent motility, as illustrated by the following model (Fig. 7A). TFP “grab onto” specific sugar moieties (e.g., GlcN and GlcNAc) in the extracellular fibril material of an adjacent bacterium. This contact triggers TFP retraction, hence providing the pulling force for TFP-dependent motility (S-motility in the case of M. xanthus). This model explains why the lack of fibril material results in defective S-motility as well as the hyperpiliation, because in fibril− mutants, TFP are still able to extrude, but fail to retract, in the absence of fibril material (Fig. 7B).
Figure 7.
Model for the pili–fibril material interaction. (A) The interaction between TFP and fibril material on the surface of wild-type cells allows TFP retraction and S-motility. (B) The absence of fibril material in fibril− mutants abolishes fibril–TFP interaction, resulting in their overpiliation phenotype and defects in S-motility. (C) The interaction between TFP and fibril material present in slime trails guides M. xanthus cells along these trails.
Another unique feature of the M. xanthus gliding motility is its ability to follow slime trails laid by other M. xanthus cells (33). A recent study by Bowden (34) shows that the natural slime trails contain fibril material along with LPS and pili. Our work suggests that the fibril material shed by other M. xanthus cells would enable M. xanthus cells to find and follow these fibril material trails. As illustrated in Fig. 7C, M. xanthus extrudes its TFP randomly at the leading pole; pili that “hit” and attach to some specific carbohydrate moiety in the fibril material retract and pull the bacterium in the direction of the leading pole. Thus, the specific interaction between TFP and fibril material can guide M. xanthus cells along the slime trails. In support of this model, preliminary data from our lab showed that wild-type M. xanthus cells preferentially follow a fibril trail laid on agar surface (H.S. and W.S., unpublished data).
How extracellular fibril material and TFP interact is a key question that will provide insight into the mechanism of TFP-dependent motility. Our data demonstrate that the carbohydrate moiety of fibril material, in particular, the sugar residues GlcN and GlcNAc, as well as residues with similar structure (e.g., GalN and GalNAc), are recognized by a yet unidentified binding site on M. xanthus TFP. Although only GlcN and GalN were previously reported to be present in fibril material (22), the finding that all amine sugars tested have similar activity may indicate that only the amine group, but not a specific monosaccharide, is relevant for the interaction. Another possibility would be that GlcNAc and GalNAc are present in fibril material but have not yet been revealed by earlier methods. Without further in-depth chemical examination of fibril material, this remains an open question.
Attachment of TFP to fibril material triggers retraction by an unknown mechanism. One possibility is that this binding causes physical restrain through pilus filaments, which induces a tension-sensitive apparatus resulting in TFP retraction, in which case not only a TFP-recognition site, but also an “anchor” are required for retraction to take place, or TFP pilin (or other proteins associated with TFP) may undergo conformational changes upon binding to these exposed carbohydrate residues in fibril material, which in turn initiate retraction. In this scenario, binding per se would be sufficient to trigger the retraction. Our finding that the polymeric (chitin), but not the monomeric, form of GlcNAc can induce pili retraction (Figs. 5C and 6A) favors the physical restrain/tension-sensitive model. However, further studies are needed to clearly distinguish these possibilities. Detailed investigation is also needed to identify the natural pilus-recognition site(s) in fibril material. Furthermore, these findings raised more interesting questions regarding this cellular process, e.g., how is retraction terminated? How is pili extrusion regulated? How do these regulations relate to S-motility on a molecular level? Answers to these questions will shed new light on the detailed mechanism for M. xanthus social-gliding motility.
The requirement of extracellular matrix for TFP-dependent cellular processes is not limited to M. xanthus. It has been reported that in P. aeruginosa not only TFP but also exopolysaccharides are required for biofilm formation (35). Based on our observations for M. xanthus, we propose that the exopolysaccharide-induced TFP retraction is a key event that may also play a role in P. aeruginosa biofilm formation. In other studies, it was shown that P. aeruginosa pili bind specifically to the carbohydrate sequence βGalNAc(1–4)βGal found in glycosphingolipids asialo-GM1 and asialo-GM2, which are present on eukaryotic cell surfaces (36). While it is known that P. aeruginosa TFP bind to glycosylation sites on a eukaryotic cell surface, TFP retraction may aid in the adherence to the host cell, thus facilitating infection. Pathogens such as Neisseria and Escherichia coli may similarly employ pilus retraction for adherence and invasion. Because TFP-mediated processes are important steps in bacterial pathogenesis, an in-depth understanding of the underlying mechanism for the interactions between TFP and extracellular matrix will provide insights into TFP-mediated bacterial infections.
Acknowledgments
We thank Dale Kaiser and Larry Shimkets for providing antibodies and bacterial strains, and Melissa Sondej for helpful discussion and careful review of the manuscript. This work was supported by National Institutes of Health Grant GM54666 (to W.S.).
Abbreviations
- TFP
type VI pili
- S-motility
social motility
- LPS
lipopolysaccharide
- TEM
transmission electron microscopy
- GlcN
glucosamine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dworkin M. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 3.MacNeil S D, Clara F, Hartzell P L. Mol Microbiol. 1994;14:785–795. doi: 10.1111/j.1365-2958.1994.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu S S. Ph.D. thesis. Stanford, CA: Stanford University; 1997. [Google Scholar]
- 5.Wu S S, Kaiser D. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 6.Shimkets L J. J Bacteriol. 1986;166:842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnold J W, Shimkets L J. J Bacteriol. 1988;170:5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dworkin M. BioEssays. 1999;21:590–595. doi: 10.1002/(SICI)1521-1878(199907)21:7<590::AID-BIES7>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Spormann A M. Microbiol Mol Biol Rev. 1999;63:621–641. doi: 10.1128/mmbr.63.3.621-641.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowden M G, Kaplan H B. Mol Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser D. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbluh A, Eisenbach M. J Bacteriol. 1992;174:5406–5413. doi: 10.1128/jb.174.16.5406-5413.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser D, Crosby C. Cell Motil. 1983;3:227–245. [Google Scholar]
- 14.Wu S S, Kaiser D. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall D, Kaiser D. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 16.Bradley D E. J Gen Microbiol. 1972;72:303–319. doi: 10.1099/00221287-72-2-303. [DOI] [PubMed] [Google Scholar]
- 17.Sun H, Zusman D, Shi W. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 18.Merz A J, So M, Sheetz M P. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 19.Skerker J M, Berg H C. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnold J W, Shimkets L J. J Bacteriol. 1988;170:5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behmlander R M, Dworkin M. J Bacteriol. 1991;173:7810–7821. doi: 10.1128/jb.173.24.7810-7820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behmlander R M, Dworkin M. J Bacteriol. 1994;176:6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Ma X, Tong L, Kaplan H B, Shimkets L J, Shi W. J Bacteriol. 2000;182:5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weimer R M, Creighton C, Stassinopoulos A, Youderian P, Hartzell P L. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Geng Y, Shi W. J Bacteriol. 1998;180:218–224. doi: 10.1128/jb.180.2.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behmlander R M, Dworkin M. J Bacteriol. 1994;176:6304–6311. doi: 10.1128/jb.176.20.6304-6311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearns D B, Bonner P J, Smith D R, Shimkets L J. J Bacteriol. 2002;184:1678–1684. doi: 10.1128/JB.184.6.1678-1684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink J M, Zissler J F. J Bacteriol. 1989;171:2028–2032. doi: 10.1128/jb.171.4.2028-2032.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campos J M, Geisselsoder J, Zusman D R. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 30.Chang B Y, Dworkin M. J Bacteriol. 1994;176:7190–7196. doi: 10.1128/jb.176.23.7190-7196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 32.Wu S S, Wu J, Kaiser D. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 33.Burchard R P. J Bacteriol. 1982;152:495–501. doi: 10.1128/jb.152.1.495-501.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden M G. Ph.D. thesis. Houston: University of Texas; 1999. [Google Scholar]
- 35.Davies D G. In: Microbial Extracellular Polymeric Substances. Flemming H C, editor. Berlin: Springer; 1999. pp. 93–117. [Google Scholar]
- 36.Sheth H B, Lee K K, Wong W Y, Srivastava G, Hindsgaul O, Hodges R S, Paranchych W, Irvin R T. Mol Microbiol. 1994;11:715–723. doi: 10.1111/j.1365-2958.1994.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Geng Y, Xu D, Kaplan H B, Shi W. Mol Microbiol. 1998;30:1123–1130. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- 38.Shi W, Yang Z, Sun H, Lancero H, Tong L. FEMS Microbiol Lett. 2000;192:211–215. doi: 10.1111/j.1574-6968.2000.tb09384.x. [DOI] [PubMed] [Google Scholar]