Abstract
The chemical synthesis of peptides and small proteins is a powerful complementary strategy to recombinant protein overexpression and is widely used in structural biology, immunology, protein engineering, and biomedical research. Despite considerable improvements in the fidelity of peptide chain assembly, side-chain protection, and postsynthesis analysis, a limiting factor in accessing polypeptides containing greater than 50 residues remains the time taken for chain assembly. The ultimate goal of this work is to establish highly efficient chemical procedures that achieve chain-assembly rates of approximately 10–15 residues per hour, thus underpinning the rapid chemical synthesis of long polypeptides and proteins, including cytokines, growth factors, protein domains, and small enzymes. Here we report Boc chemistry that employs O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU)/dimethyl sulfoxide in situ neutralization as the coupling agent and incorporates a protected amino acid residue every 5 min to produce peptides of good quality. This rapid coupling chemistry was successfully demonstrated by synthesizing several small to medium peptides, including the “difficult” C-terminal sequence of HIV-1 proteinase (residues 81–99); fragment 65–74 of the acyl carrier protein; conotoxin PnIA(A10L), a potent neuronal nicotinic receptor antagonist; and the pro-inflammatory chemotactic protein CP10, an 88-residue protein, by means of native chemical ligation. The benefits of this approach include enhanced ability to identify and characterize “difficult couplings,” rapid access to peptides for biological and structure–activity studies, and accelerated synthesis of tailored large peptide segments (<50 residues) for use in chemoselective ligation methods.
Advances in solid-phase peptide synthesis (SPPS) (1–3) over the past decade have encouraged increased efforts toward the total chemical synthesis of large complex peptides and small proteins (4–7). More recently, the value of merging chemical and recombinant technologies was beautifully demonstrated by the “expressed protein ligation” strategy developed at Rockefeller University (8). Despite these successes, target molecules containing 50 to 200 residues still represent a significant challenge. The rate-determining step in long stepwise SPPS remains slow chain assembly, in which incorporation of each amino acid usually requires a 20- to 120-min cycle, depending on the choice of chemistry and the synthetic protocol followed. In addition, chain assembly times are further increased with “difficult” synthetic sequences, for which double or triple coupling is often necessary to achieve acceptable acylation yields (9). Consequently, the ability to assemble peptides faster is highly desirable and has several benefits, including (i) the early identification of difficult synthetic sections in the target sequence; (ii) the rapid synthesis of small complex peptides; (iii) the rapid synthesis of protein mutants to establish more comprehensive structure–activity-relationship (SAR) studies in shorter times; and (iv) the rapid assembly of peptide segments for use in chemoselective ligation methods to construct small proteins. Therefore, we have endeavored to accelerate the process of SPPS, with the ultimate aim being the efficient synthesis of long peptides and protein domains at a chain-assembly rate of 10–15 residues per hour (or 100–200 residues per day over several peptide segments). To realize this goal it is necessary to develop a strategy for rapid protein synthesis that reduces the coupling cycle times and addresses “difficult” synthetic sequences.
The recent introduction of chemoselective ligation strategies has created an exciting new avenue for the construction of large peptides and small proteins (5, 7, 10–12). This strategy involves the chemical ligation of two or more 40- to 50-residue unprotected peptide segments to form the target protein. In native chemical ligation, an unprotected peptide with a C-terminal α-thioester is reacted with a second unprotected peptide containing an N-terminal cysteine residue in aqueous solution around pH 7 to form a native peptide bond at the ligation site (5). Currently, chemical ligation is the most promising route for protein synthesis, since it takes advantage of the ability to synthesize and purify to homogeneity peptide segments approximately 50 amino acids in length that have incorporated chemoselective functionality. Alternative approaches to protein synthesis, such as classical stepwise chain assembly, are not suited to the synthesis of peptides containing more than approximately 70 residues (13). Such longer stepwise syntheses are impeded by the accumulation of side-products arising from incomplete acylation and cleavage-related side reactions which make purification of the target peptide from a large family of closely related side-products both time consuming and difficult. As a result, most protein chemical synthesis strategies now adopted employ chemoselective ligation chemistry with its clear benefit of providing homogeneous target molecules in acceptable yields. However, the challenge firmly remains to deliver these tailored peptide segments on a daily basis.
Although carefully implemented stepwise SPPS protocols are effective in producing up to 50-residue peptides, few have been developed with rapid protein synthesis as the goal. Typical SPPS protocols have standard coupling times that are long and allow kinetically slower side reactions to become competitive (14–16). For example, in the early 1990s, standard protocols used by the Peptide Synthesis Research Committee (PSRC) of the Association of Biomolecular Resource Facilities (ABRF) in SPPS studies were based on activation with symmetric anhydrides or O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) (17) with a cycle time of 2 hr (18, 19). Other peptide laboratories commonly employed dicyclohexylcarbodiimide (20) (DCC) or diisopropylcarbodiimide (21) (DIC)/1-hydroxybenzotriazole (HOBt) chemistry (22), or benzotriazole-1-yl-oxytris(dimethylamino)phosphonium hexafluorophosphate (BOP) (23) chemistry, which typically incorporates a single amino acid residue every 30–120 min (18, 24, 25). In recent years there has been a gradual decrease in coupling cycle times recommended by manufacturers of automated peptide synthesizers (26). In our earlier efforts to improve the efficiency of SPPS we reported HBTU in situ neutralization tert-butoxycarbonyl (Boc) and 9-fluorenylmethoxycarbonyl (Fmoc) chemical approaches, which incorporate 3 amino acid residues per hour (27, 28). Even though these protocols are approximately 6 times faster then most current chemistries, they still do not fulfil our requirements.
The rate of amino acid acylation either in solution or on solid support is heavily dependent on the properties of the coupling reagent. Mass transfer rates have shown that diffusion of activated Boc-amino acids through a swollen 1% divinylbenzene-polystyrene resin matrix is at least 10 times faster than coupling rates and that diffusion is not the limiting factor (29, 30). Coupling reactions follow good second-order reaction rates and are generally 99% complete within 10–100 sec on cross-linked divinylbenzene-polystyrene resin, depending on the method of activation (29–31). That is, coupling times are primarily determined by the acylation reactivity of activated Nα-protected amino acids. Therefore, it is feasible that the coupling may be achieved efficiently within 1–2 min when a highly activated acylation species is selected. Recently, a new generation of coupling reagents that have superior acylation kinetics, reduce racemization, and minimize side reactions has been reported (32, 33). O-(7-Azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HATU) (34) and acid fluorides (35–37) are members of this current generation of highly efficient coupling reagents suitable for the synthesis of peptides or proteins containing “difficult” sequences, β-branched and/or N-methylated amino acids. While HATU chemistry was initially introduced and predominantly examined for the assembly of short peptides by Fmoc chemical strategies, the full potential of this coupling chemistry in Boc-based SPPS has not yet been thoroughly investigated and provides the basis of our rapid synthetic strategy.
We thus investigated the feasibility of developing a new coupling protocol for the rapid, efficient, and reliable synthesis of peptides by using HATU/Boc in situ neutralization chemistry. The initial goal was to incorporate each amino acid at the rate of one every 5 min by using robust chemistry that would provide, after cleavage, crude peptide of good quality. Thus we chose to develop chemistry that was functional and fast rather than fully optimizing each chemical step in the chain assembly. Such a strategy would also deliver important information within a working day such as the identification of “difficult or slow” acylations. This protocol was evaluated by the synthesis of the Ala-10 → Leu (A10L) mutant of PnIA conotoxin from Conus pennaceus, which blocks the nicotinic acetylcholine receptor (nAChR); the “difficult” 65–74 fragment of the acyl carrier protein (ACP)(38–40); the “difficult” C-terminal sequence of human immunodeficiency virus (HIV)-1 proteinase (PR)(81–99) (27, 41); and a 40-residue domain of chemotactic protein 10 kDa (CP10) (42), which was ligated with CP10(41–88) to construct the full-length protein. In this study, we found that the efficiency and reliability of this chemistry especially at “difficult” coupling sites was improved by use of dimethyl sulfoxide (DMSO) as the coupling solvent instead of N,N-dimethylformamide (DMF). This work establishes robust coupling chemistry for the rapid synthesis of peptides and peptide ligation segments at a chain-assembly rate of approximately 12 residues per hour.
EXPERIMENTAL PROCEDURES
Materials.
Nα-Boc-l-amino acids and reagents used during chain assembly were peptide synthesis grade purchased from Auspep (Melbourne, Australia) and Novabiochem (San Diego, CA). Nα-Boc-(l)-amino acid-phenylacetamidomethyl (Pam)-resin and 4-methylbenzylhydrylamine (MBHA) resin were purchased from Applied Biosystems (Foster City, CA) and Peptide Institute (Osaka, Japan), respectively. Anhydrous DMSO, p-cresol, p-thiocresol, and resorcinol were purchased from Aldrich (Sydney, Australia). HBTU was purchased from Richelieu Biotechnologies (Quebec, Canada) and HATU was prepared as previously described (32, 34). Screw-cap glass peptide synthesis reaction vessels (27) (4 ml) with sintered glass filter frit were obtained from Embell Scientific Glassware (Queensland, Australia). Anhydrous HF was purchased from Matheson Gas (BOC Gases, Melbourne, Australia).
Solid-Phase Peptide Synthesis.
Chain assembly of peptides ACP(65–74), PnIA(A10L), PR(81–99), and CP10(1–40)-αCOSR (SR indicates S-mercaptopropionoyl-Ala) were performed on 0.1 mmol of Boc-Gly-Pam resin (0.71 mmol/g), MBHA (0.66 mmol/g), Boc-Phe-Pam resin (0.72 mmol/g), and Boc-Gly-SR-Pam resin (0.753 mmol/g), respectively. The following amino acid side-chain protection was used: Boc-Asn(β-xanthyl)-OH, Boc-Arg(4-toluenesulfonyl)-OH, Boc-Asp(O-cyclohexyl)-OH, Boc-Cys(4-methylbenzyl)-OH, Boc-Gln(γ-xanthyl)-OH, Boc-Glu(O-cyclohexyl)-OH, Boc-His(Nπ-benzyloxymethyl)-OH, Boc-His(Nim-dinitrophenyl)-OH, Boc-Lys-(Nɛ-2-chlorobenzyloxycarbonyl)-OH, Boc-Ser(O-benzyl)-OH, and Boc-Thr(O-benzyl)-OH. The assembly of peptides consisted of a 4- to 5-min cycle for each residue at ambient temperature as follows: (i) a 5-s vacuum-assisted TFA flow wash followed by a 55-s neat TFA Boc deprotection step; (ii) a 45-s vacuum-assisted DMF flow wash (typically 7–10 reaction vessel volumes) to give the N-terminal ammonium⋅TFA salt; (iii) a 1- to 2-min in situ neutralization coupling step with vigorous shaking using 0.4 mmol of Boc-amino acid pre-activated for approximately 3 min† with 0.39 mmol of 0.5–0.625 M HATU in DMF or DMSO solution and 0.52 mmol of diisopropylethylamine (DIEA) (reaction volume approximately 900 μl); and (iv) a 40-s vacuum-assisted DMF flow wash. After each coupling step a resin sample was taken and the acylation efficiency was determined by the quantitative ninhydrin reaction after assembly (43). The average yields of chain assembly of ACP(65–74), PnIA(A10L), and PR(81–99) were 99.2%, 99.4%, and 97.9%, respectively. Prior to HF cleavage the N-terminal Boc group was removed with neat TFA, and the resin was washed with dichloromethane and dried under a nitrogen atmosphere. Approximately 200 mg of the PR(81–99), PnIA(A10L), or [Glu40Gly]CP10(1–40)-αCOSR peptide-resins was treated with 10 ml of HF/p-cresol (9:1, vol/vol), HF/p-cresol/p-thiocresol (18:1:1, vol/vol), or HF/p-cresol/resorcinol (18:1:1, vol/vol/wt) at 0°C for 1.5 h, respectively. After evaporation of the HF, the crude peptide was precipitated and washed with cold anhydrous diethyl ether (twice with 10 ml), dissolved in 50% aqueous acetonitrile, and lyophilized after aqueous dilution. The crude lyophilized peptide was reconstituted in 50% acetonitrile and then analyzed by reversed-phase (RP)-HPLC, electrospray mass spectrometry (ES-MS), and liquid chromatography–mass spectrometry (LC-MS).
HPLC.
Analytical reversed-phase HPLC was performed with a Waters 600E solvent delivery system. Data were collected by using a 484 absorbance detector (Applied Biosystems) at 214 nm. Chromatographic separations were achieved with a 1%/min linear gradient of buffer B in A (A = 0.1% TFA in H2O; B = 90% CH3CN/10% H2O/0.09% TFA) over 80 min at a flow rate of 1 ml/min and 8 ml/min using Vydac C18 analytical (5 μm, 0.46 cm × 25 cm) and preparative C18 (10 μm, 2.2 cm × 25 cm) columns, respectively.
ES-MS and LC-MS.
Mass spectra were acquired on a PE-Sciex API-III triple quadrupole mass spectrometer equipped with an Ionspray atmospheric pressure ionization source. Samples (typically 10 μl) were injected into a moving solvent (30 μl/min; 1:1 CH3CN/0.05% TFA in H2O) coupled directly to the ionization source by a fused silica capillary interface (50 μm i.d. × 50 cm length). Sample droplets were ionized at a positive potential of 5 kV and entered the analyzer through an interface plate and subsequently through an orifice (100–120 μm in diameter) at a potential of 80–100 V. Full scan mass spectra were acquired over the mass range of 700 to 2000 Da with a scan step size of 0.2 Da. Molecular masses were derived from the observed m/z values by using MacSpec 3.3 software (PE-Sciex, Toronto). Theoretical monoisotopic and average masses were calculated by using the MacBiospec program (PE-Sciex, Toronto). LC-MS runs were carried out on a 140B Applied Biosystems dual syringe pump solvent delivery system and a Vydac reversed-phase C18 (5 μm, 300 Å, 0.21 × 25 cm) column using a 35–65% B linear gradient over 40 min at a flow rate of 150 μl/min. Samples were loaded directly on the column and the eluent was directly connected to the mass spectrometer with a 30-cm, 75-μm i.d. fused silica capillary. The application of Turbo Ionspray (5 liters of N2 per min at 500°C) allowed the introduction of the total eluent without splitting and loss in sensitivity. Acquisition parameters were as described above.
Oxidation of PnIA(A10L).
Air oxidation was carried out by dissolving 10 mg of the lyophilized crude PnIA(A10L) cleavage material in 45 ml of 1:1 0.1 M NH4HCO3/isopropyl alcohol (pH 8.25) with vigorous stirring at room temperature for 1 h. Prior to purification the solution was acidified to pH 3 with TFA and analyzed by analytical C4 HPLC using a linear gradient of 0–80% B at 1%/min while monitoring by UV absorbance at 214 nm and ES-MS. Oxidized PnIA(A10L) was then purified by semipreparative HPLC using the same chromatographic conditions; the yield was 27%.
Native Chemical Ligation.
The ligation of the two unprotected synthetic peptide fragments was carried out as previously described (5, 44). Briefly, 4% (vol/vol) phenylmercaptan was added to a solution of CP10(1–40)-αCOSR (6 mg/ml) and CP10(41–88) (6 mg/ml) in degassed 6 M guanidine⋅HCl/100 mM tris(hydroxymethyl)aminomethane buffered to pH 8.2. The ligation mixture was incubated at 37°C for 8 h, and the reaction was monitored until completion by RP-HPLC. Chromatographic separations were achieved by using a 35–65% B linear gradient at 0.75%/min on a C18 HPLC column. HPLC fractions containing the ligated polypeptide chain of CP10(1–88) were identified by ES-MS and lyophilized; the yield was 61%.
RESULTS AND DISCUSSION
Synthetic Strategy.
The initial step in the development of this rapid SPPS was the selection of Boc chemistry. This strategy has the advantage of clean, reliable, and rapid acidolytic Nα-Boc deprotection compared with its Fmoc counterpart, which is prone to kinetically slower, sequence-dependent, and/or incomplete Nα-Fmoc deprotection (45–47). This phenomenon may prove to be an intrinsic limitation for the acceleration of Fmoc SPPS. Furthermore, the suitability of Boc chemistry for the total chemical synthesis of peptide and proteins, especially when using chemoselective ligation methods (4–6, 10–12, 48–51), was a crucial factor in its selection. By comparison the alternative Fmoc approach is limited and at the present time is not as compatible (52) with the synthesis of ligation domains, which generally employs base-sensitive thioester chemistry.
HATU coupling chemistry was selected in preference to other efficient benzotriazole-based activators such as HBTU or benzotriazole-1-yloxytripyrrolidinophosphonium hexafluorophosphate (pyBOP) (53) because it leads to superior acylation rates, reduces racemization, and has higher solubility in the common coupling solvent DMF. The use of acid fluorides was considered an attractive alternative, but the lack of commercially available derivatized amino acids was a severe drawback notwithstanding the recent introduction of in situ fluoride activation using tetramethylfluoroformamidium hexafluorophosphate (TFFH) (33). We chose to perform the syntheses by using batch methodology in preference to the continuous-flow approach with solvent and reactant recycling. The batch system allows for rapid solvent drainage, fast solvent washes, thorough mixing and solvation of resin, and of critical importance, for the high reactant concentrations to be maintained through the coupling phase (27). To maximize the benefit of this method in longer syntheses, the molar concentration of HATU-activated Boc-amino acids was increased to 0.625 M. Although further increases in HATU molarity are possible (32, 34), higher molar concentrations of HATU-activated Boc-amino acids were found to result in poor resin coverage when a 3-fold excess of Boc-amino acids was used. If necessary, this limitation may be overcome simply by increasing the molar excess of Boc-amino acids.
Streamlining HATU/Boc Chain Assembly.
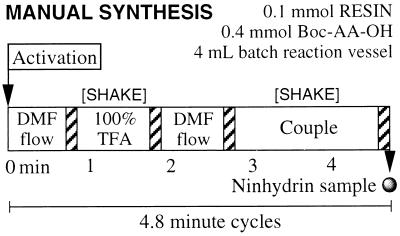
The synthesis of polypeptides on a solid support consists of a repetitive cycle of deprotection, draining, washing, and coupling (Fig. 1). For that reason, one of the aims of the protocols was to streamline SPPS by performing the following short steps for each cycle. (i) A 1-min neat TFA treatment at ambient temperature was sufficient for the quantitative removal of Nα-Boc groups (27). (ii) A 45-s DMF flow wash in a fast flowing reaction vessel removes excess TFA, leaving the N-terminal amine group as the ammonium salt with a trifluoracetate counterion. (iii) In situ neutralization and coupling with a 3-fold excess of Boc-amino acid using 0.625 M HATU in the presence of 5 equivalents of DIEA for 1–2 min. Importantly, although many laboratories still separate neutralization and coupling steps, in situ neutralization has been shown to give higher acylation yields in shorter times (27, 46). In this protocol all Boc-amino acids were activated approximately 3 min prior to use†. (iv) To complete the cycle another 45-s DMF flow wash was necessary for the removal of solution-phase reaction components. The total cycle took less than 5 min as outlined in Fig. 1 and was repeated until the desired peptide sequence was assembled. After chain assembly, cleavage from the solid support and side-chain deprotection was performed by treating the resin-bound peptides with anhydrous HF in the presence of aromatic scavengers at 0°C for 1.5 h (54, 55).
Figure 1.
Manual HATU/Boc in situ neutralization synthetic protocol. The hatched regions indicate a drainage step where excess solvent and/or reactant components are removed from the peptide-resin by filtration.
Evaluation of HATU/Boc Chain Assembly.
We initially prepared three peptides ranging in length from 10 to 20 amino acids. These included the benchmark “difficult” sequence PR(81–99) (27, 41), the A10L mutant of PnIA (a nanomolar nicotinic acetylcholine antagonist containing two disulfide bonds), and the “difficult” segment of the acyl carrier protein, ACP(65–74) (38–40). Later, a 40-residue ligation domain of 10-kDa chemotactic protein (CP10) (42) served to demonstrate the applicability of this protocol for rapid protein synthesis using native ligation. Importantly, chemical synthesis has proven to be an effective and reliable method for the preparation of CP10, as it required 2 years before an adequate recombinant expression system was developed. The evaluation of shorter sequences reflected the empirical observation that the maximum occurrence of “difficult” couplings is approximately 5–15 residues from the resin (9) and subsequent chain assembly tends to proceed less problematically.
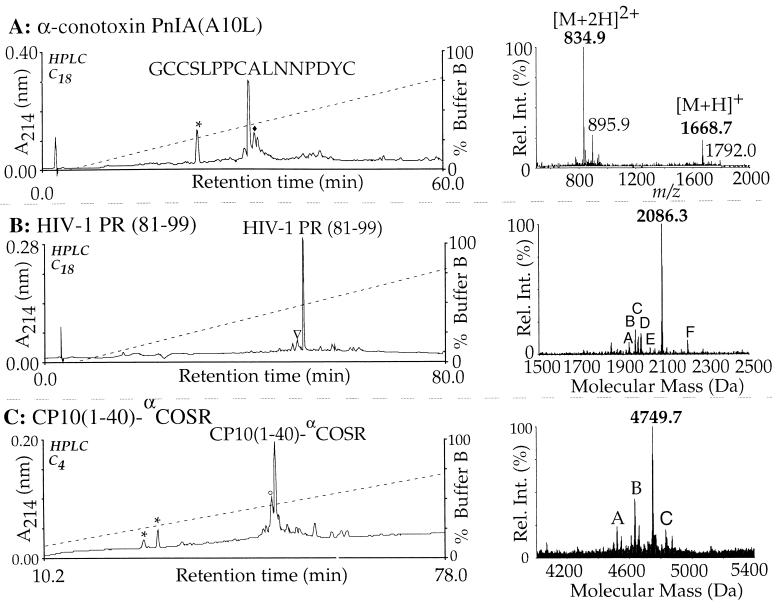
All peptides were synthesized manually by using the single-couple cycles described above. The assembly times for PnIA(A10L), ACP(65–74), PR(81–99), and CP10(1–40) sequences were 67 min, 57 min, 2.1 h, and 4.4 h, respectively. In all syntheses the major component in the resulting crude cleavage product was the target peptide, as indicated by RP-HPLC and ES-MS analysis (Fig. 2). No effort was made to optimize chain-assembly times, and the coupling efficiencies and crude peptide analyses reflect the ease of the chain assembly under these conditions.
Figure 2.
Rapid syntheses of PnIA(A10L), PR(81–99), and [Glu40Gly]-CP10(1–40)-αCOSR assembled in 67 min, 2.1 h, and 4.4 h, respectively. Peaks labeled with ∗ indicate residual p-cresol or resorcinol from the HF cleavage. (A) HPLC and ES-MS analysis of the crude PnIA(A10L) reaction products. The major HPLC peak has the correct mass for PnIA(A10L). The HPLC peak labeled with ♦ refers to the PnIA(A10L) peptide with an oxidized Cys(p-MeBzl) group. (B) HPLC and ES-MS analysis of the crude PR(81–99) synthesis. The HPLC peak labeled with ▿ indicates the des-Arg deletion product. In the deconvoluted ES-MS spectrum (Right) peaks A, B, C, D, E, and F correspond to the presence of the des-Arg, des-Gln, des-Asn/Ile/Leu, des-Thr, and des-Gly deletion side-products and the correct peptide with an oxidized Cys(p-MeBzl) group. (C) HPLC and ES-MS analysis of the crude [Glu40Gly]CP10(1–40)-αCOSR synthesis. The HPLC peak labeled with ○ indicates the des-Leu/Ile deletion side-product. In the deconvoluted ES-MS spectrum (Right) peaks A, B, and C correspond to a double des-Ile/Leu deletion, single des-Ile/Leu deletion, and [Glu40Gly]CP10(1–40)-αCOSR containing residual glutamyl O-cyclohexyl ester protection, respectively.
Rapid Chemical Synthesis of the PnIA(A10L) Conotoxin.
As a part of ongoing structure–activity investigations on a nanomolar nicotinic acetylcholine receptor antagonist from Conus pennaceus venom, the mutant PnIA(A10L) conotoxin, GCCSLPPCALNNPDYC, was chemically synthesized. Previously (J. Gehrmann, personal communication) the native sequence was found to be “difficult” in our laboratory when HBTU/Boc in situ neutralization protocols were used (27) and thus was a good starting point for our evaluation. As the sequence is relatively short, the coupling cycle (Fig. 1) was altered by halving the coupling time to 1 min, giving a total cycle time of approximately 4 min. After chain assembly, cleavage from the resin and side-chain deprotection were carried out with HF/p-cresol (9:1) at 0°C for 1.5 h. The average coupling yield for the chain assembly of PnIA(A10L) was estimated to be 99.4% by using the quantitative ninhydrin assay (43). Apart from minor deletion impurities, HPLC and LC-MS analysis confirmed the major product (Fig. 2A) as reduced PnIA(A10L). A HF-resistant‡ minor product was also observed as a consequence of partial oxidation of the Cys(p-MeBzl) protecting group. Oxidation of this crude material was carried out smoothly to yield PnIA(A10L) in sufficient quantities for comprehensive bioassay and crystallization studies. Interestingly, the A10L point mutation was found to enhance potency for the neuronal nicotinic response (B. Livett, personal communication).
Rapid Chemical Synthesis of the “Difficult” Sequence PR(81–99).
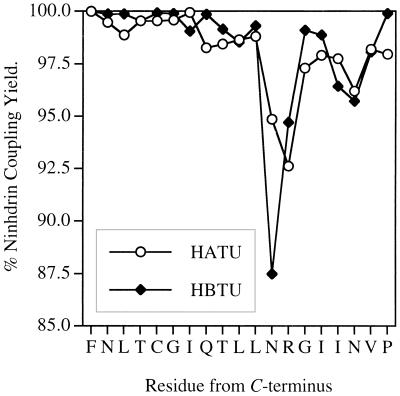
The chemical synthesis of PR(81–99), PVNIIGRNLLTQIGCTLNF, forms a rigorous test for any SPPS protocol (27, 41), as it is difficult to assemble efficiently by using either Boc or Fmoc chemistry. The protected peptide was assembled with the 5-min cycle (Fig. 1) using 0.5 M HATU and resulted in an average coupling yield of 97.9%. After HF cleavage, RP-HPLC and ES-MS analysis of the crude product (Fig. 2B) showed that the major component was the desired PR(81–99). The only notable side-product is approximately 5% des-Arg-87-PR(81–99). Arg-87 is notoriously difficult to couple in this sequence and requires double coupling followed by acetylation. To compare the efficiency of rapid HATU/Boc in situ neutralization with respect to existing HBTU/Boc in situ neutralization chemistry, the coupling profiles of PR(81–99) with both synthetic strategies are shown Fig. 3. The HBTU/Boc assembly was carried out using 10-min couplings of HBTU-activated Boc-amino acids without recoupling or acetylation and required approximately 7 h to complete. Whereas the HBTU and rapid HATU coupling profiles were generally similar, HBTU coupling of Asn-88 was considerably slower than with HATU. Moreover, in both syntheses the last eight residues displayed relatively slow coupling rates. This observation led us to improve the yields of rapid HATU chemistry at difficult coupling sites by incorporating DMSO as discussed below.
Figure 3.
Comparison of the coupling yields for synthesis of PR(81–99). ♦, HBTU/Boc in situ neutralization chemistry (27), consisting of 10-min coupling times, 22-min cycles, and an average manual synthesis time of ≈7 h. ○, HATU/Boc in situ neutralization chemistry, consisting of 2-min coupling times, 5 min cycles, and a synthesis time of 2.1 h.
Overcoming Difficult Synthetic Sequences with DMSO.
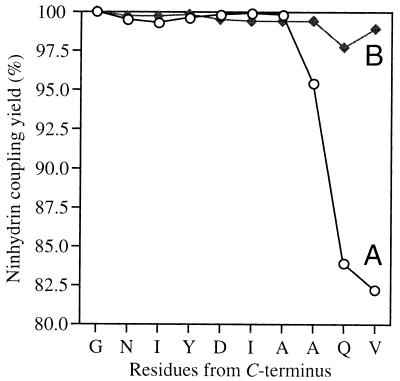
To further improve the efficiency and reliability of HATU/Boc in situ neutralization chemistry, especially when confronted with difficult sequences, the use of DMSO as a coupling solvent was investigated. DMSO has been reported to delay or eliminate the onset of interchain aggregation and also improve acylation yields (56, 57). We chose to examine any beneficial effect of coupling in DMSO with the synthesis of the well known difficult sequence ACP(65–74), VQAAIDYING. This decapeptide was an appropriate test case because it undergoes strong chain aggregation during the deprotection and coupling of the ninth and tenth residues, Gln and Val, respectively (9, 38, 40). A comparison of the 1-min coupling yields based on the quantitative ninhydrin assay (44) for assembly of ACP(65–74) employing HATU/Boc chemistry using DMF and DMSO as the coupling solvent is shown in Fig. 4. One-minute coupling times were used to emphasize the effect of the two solvents on amino acid acylation rates (coupling yields). When DMF was used as the sole coupling solvent the chain assembly proceeded well until the Gln and Val residues, where the 1-min HATU coupling yields decreased to 83% and 82%, respectively. However, when DMSO was used as the sole coupling solvent the coupling yields increased to 97.6% and 98.8% for Gln and Val, respectively. Even though these coupling yields in DMSO are significantly higher than those in DMF, they can be further improved (data not shown) by doubling the coupling time to the recommended 2 min, while still giving a total cycle time of less than 5 min. The beneficial effect of DMSO may arise from improved solvation of both cross-linked polymers and growing peptide chains, which enables greater penetration of activated amino acids throughout the resin matrix. Consequently, at difficult coupling sites where chain association is problematic it appears likely that coupling yields can be significantly increased by replacing the usual coupling solvent, DMF, with DMSO.§
Figure 4.
Comparison of the coupling yields (43) for the chain assembly of ACP(65–74) using 1-min HATU/Boc in situ neutralization coupling cycles in DMF (curve A, ○) and DMSO (curve B, ♦).
Rapid Chemical Synthesis of CP10.
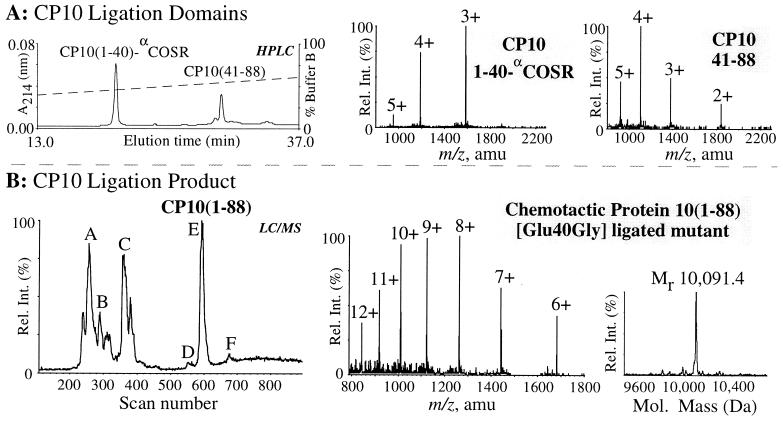
Because the rapid chain assembly results for the small PnIA(A10L) and PR(81–99) peptides were encouraging, the next stage was the synthesis of a larger polypeptide, [Glu40Gly]CP10, PSELEKALSNLIDVYHNYSNIQGNHHALYKNDFKKMVTTGCPQFVQNINIENLFRELDINSDNAINFEEFLAMVIKVGVASHKDSHKE. The Glu-40 → Gly mutant of CP10 was selected both to probe the role of the Glu residue, which is conserved in most S100 calcium-binding proteins (58), and as a convenient “native ligation” site adjacent to the sole Cys residue in CP10. As a result, the N-terminal [Glu40Gly]CP10(1–40)-αCOSR ligation domain was manually assembled by using the 5-min rapid HATU/Boc coupling cycle (Fig. 1) commencing with Boc-Gly-SR-Pam resin in approximately 4.4 h. The three His residues in the peptide were protected at the π-imidazole nitrogen atom with a benzyloxymethyl (Bom) group instead of the usually preferred dinitrophenyl (DNP) derivative, as it is conveniently removed during HF cleavage in the presence of resorcinol. It should be noted that DNP removal prior to HF cleavage results in the premature cleavage of peptide thioesters from the resin (6).
The major component in the crude cleaved material was found to be target peptide as confirmed by RP-HPLC and ES-MS analysis (Fig. 2C). As expected, the crude material contained more minor deletion side-products in comparison to the other significantly shorter syntheses. For instance, the peak labeled with ○ in the RP-HPLC trace in Fig. 2C had a mass corresponding to a des-Ile/Leu side-product, while other side-products with longer retention times were mainly due to incomplete side-chain deprotection of the [Glu40Gly]CP10(1–40)-αCOSR polypeptide. After cleavage, [Glu40Gly]CP10(1–40)-αCOSR was readily purified in good yield by preparative RP-HPLC. The purified [Glu40Gly]CP10(1–40)-αCOSR was then used in the native ligation with CP10(41–88) containing an N-terminal Cys residue to construct the CP10 mutant protein (Fig. 5). The ligation reaction was initiated by addition of 4% phenylmercaptan and after 8 h gave [Glu40Gly]CP10 in a yield of 61%.
Figure 5.
Total synthesis of [Glu40Gly]CP10 by native chemical ligation. (A) Analytical RP-HPLC of the synthetic peptide fragments [Glu40Gly]CP10(1–40)-αCOSR and CP10(41–88) dissolved at 6 mg/ml in 0.1 M Tris⋅HCl buffer (pH 8.2) containing 6 M guanidine⋅HCl at t = 0. (Center and Right) Electrospray mass spectra show the m/z ratios of the [Glu40Gly]CP10(1–40)-αCOSR peptide (3rd to 5th ionized states) with an observed molecular mass of 4749.8 ± 0.5 Da (calc. 4750.2 Da, average isotope composition), and the CP10(41–88) peptide (2nd to 5th ionized states) with an observed molecular mass of 5518.6 ± 0.9 Da (calc. 5518.2 Da, average isotope composition). (B) LC-MS analysis of the ligation reaction after t = 3 h. Peaks A, B, and C represent unreacted [Glu40Gly]CP10(1–40)-αCOSR, [Glu40Gly]CP10(1–40) as the free C-terminal acid, and [Glu40Gly]CP10(1–40)-αCOS-C6H5, respectively. Peaks D and F had a mass indicating the presence of CP10(1–88) esterified through the Cys-41 by CP10(1–40) (14,665 Da), and the CP10(1–88)[Cys-41-thiophenol] mixed disulfide (10,199 Da), respectively. ES-MS of purified [Glu40Gly]CP10 (6th to 12th ionized states), peak E, had an observed molecular mass of 10,091.4 ± 1.1 Da; (calc. 10,091.3 Da, average isotope composition).
Implications for Protein Synthesis.
The advent of chemoselective ligation technology has opened new routes to the chemical synthesis of proteins where the fundamental prerequisite is the capacity to synthesize homogenous peptide segments containing approximately 40–50 residues. As the existing chemistries employed in most laboratories lead to long chain-assembly times for medium-length peptide segments, the routine total chemical synthesis of proteins has remained in the hands of only a few practitioners. However, with the integration of recombinant and chemical technologies (8) the power and applicability of chemical synthesis is now more widespread and attractive to biomedical researchers. It is now possible to introduce sequences of unnatural amino acids, post-translational modifications, and biophysical probes into proteins of any size (8).
In this work we have addressed the need for accelerated SPPS by developing fast and reliable coupling chemistry with accompanying short cycle times. Using robust HATU/Boc in situ neutralization coupling chemistry with a 5-min cycle time (Fig. 1), we manually synthesized four challenging polypeptides of various sizes from 10 to 40 residues with good quality. Implementation of this chemistry will now enable fast access to medium-length peptides for protein ligation and will also provide rapid and precise identification of difficult or slow coupling sites within each sequence. The difficult regions thus identified could be addressed in subsequent rapid syntheses by optimizing the reaction conditions at such sites by a variety of methods, including the substitution of DMSO for DMF as the coupling solvent, variation of the amino acid activation chemistry, and/or increasing the concentration or coupling time of the activated amino acid. Implementation of this rapid coupling strategy together with improvements in SPPS, such as more reliable side-chain protection, the development of specialized and more reliable resins (59), and improved coupling agents should allow us to target longer peptides and peptide segments (50–100 residues). The construction of combinatorial libraries and routine peptide synthesis will also benefit from the faster cycle times described here. Faster acylation and deprotection chemistry should help in a number of ways by (i) minimizing the risk of side reactions becoming kinetically competitive; (ii) preventing racemization; and (iii) minimizing the premature cleavage of some “permanent” side-chain protecting groups.
Finally, to maximize the advantage of rapid HATU/Boc in situ neutralization chemistry, we had hoped to adapt the protocols for machine-assisted synthesis. However, none of the currently available commercial instruments, such as the Applied Biosystems ABI 433 and ABI 430 synthesizers (28), meet the requirements for automation of this chemistry. Issues such as slow solvent transfer times, software constraints, instrument reliability, and the inability to perform syntheses at the microscale level limit the effectiveness of the current machines for conversion to rapid HATU/Boc chemistry. Such compatibility issues should be considered in the design and development of the next generation of solid-phase polypeptide and combinatorial library synthesizers.
Acknowledgments
We gratefully acknowledge Prof. Carolyn Geczy and Prof. Bruce Livett for performing the bioassays. We also thank Dianne Alewood for kindly providing CP10(41–88) for ligation trials. L.P.M. was supported by an Australian Postgraduate Research Award scholarship from the Australian Government.
ABBREVIATIONS
- ACP
acyl carrier protein
- Boc
tert-butoxycarbonyl
- Bzl
benzyl
- CP10
chemotactic protein 10 kDa
- DIEA
diisopropylethylamine
- DMF
N,N-dimethylformamide
- DMSO
dimethyl sulfoxide
- ES-MS
electrospray mass spectrometry
- Fmoc
9-fluorenylmethoxycarbonyl
- HATU
O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- HBTU
O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- LC-MS
liquid chromatography–mass spectrometry
- Pam
phenylacetamidomethyl
- PnIA
Conus pennaceus conotoxin
- PR
HIV-1 proteinase
- pyBOP
benzotriazole-1-yloxytripyrrolidinophosphonium hexafluorophosphate
- RP-HPLC
reversed-phase high-performance liquid chromatography
- SPPS
solid-phase peptide synthesis
- SR
S-mercaptopropionoyl-Ala
- TFA
trifluoroacetic acid
Footnotes
All 20 naturally encoded amino acids have been successfully incorporated by using this method. Racemization of Boc-amino acids after HATU pre-activation was not evident, although excessive cysteine pre-activation should be avoided (60).
This oxidative side-product is caused by air oxidation, which may be encountered during manual synthesis; however, it is largely avoided when carrying out machine-assisted syntheses because the resin and reactants are continually maintained under a nitrogen atmosphere.
The beneficial effect DMSO has been observed in many cases in our laboratory. Treatment of the resin-bound peptide Cys(MeBzl)-Met-Cys(MeBzl)-Met with DMSO in the presence of 5 equivalents of DIEA for 18 h resulted in the formation of a small amount of oxidative products (≪5%). On the basis of this amount, it appears that DMSO-induced oxidative side-products are formed at <0.005%/min. Further studies are needed to evaluate the effectiveness of N-methylpyrrolidine (NMP) (57) as a rapid coupling solvent for difficult sequences.
References
- 1.Merrifield R B. Fed Proc Amer Soc Exp Biol. 1962;21:412. (abstr.). [Google Scholar]
- 2.Merrifield R B. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 3.Fields, G. B., ed. (1997) Methods Enzymol.289. [DOI] [PubMed]
- 4.Dawson P E, Kent S B H. J Am Chem Soc. 1993;115:7263–7266. [Google Scholar]
- 5.Dawson P E, Muir T W, Clarklewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 6.Hackeng T M, Mounier C M, Bon C, Dawson P E, Griffin J H, Kent S B H. Proc Natl Acad Sci USA. 1997;94:7845–7850. doi: 10.1073/pnas.94.15.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C F, Tam J P. Proc Natl Acad Sci USA. 1994;91:6584–6588. doi: 10.1073/pnas.91.14.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muir T W, Sondhi D, Cole P A. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent S B H. Annu Rev Biochem. 1988;57:957–989. doi: 10.1146/annurev.bi.57.070188.004521. [DOI] [PubMed] [Google Scholar]
- 10.Englebretsen D R, Garnham B G, Bergman D A, Alewood P F. Tetrahedron Lett. 1995;36:8871–8874. [Google Scholar]
- 11.Liu C F, Rao C, Tam J P. J Am Chem Soc. 1996;118:307–312. [Google Scholar]
- 12.Baca M, Muir T W, Schnölzer M, Kent S B H. J Am Chem Soc. 1995;117:1881–1887. [Google Scholar]
- 13.Kent S B H, Alewood D, Alewood P F, Baca M, Jones A, Schnölzer M. In: Innovation and Perspectives in Solid Phase Synthesis: Peptides, Polypeptides and Oligonucleotides. Epton R, editor. Canterbury, U.K.: Intercept; 1992. pp. 1–22. [Google Scholar]
- 14.Baba T, Sugiyama H, Seto S. Chem Pharm Bull. 1973;21:207–209. [Google Scholar]
- 15.Tam J P, Riemen M W, Merrifield R B. Pept Res. 1988;1:6–18. [PubMed] [Google Scholar]
- 16.Kates S A, Albericio F. Lett Pept Sci. 1994;1:213–220. [Google Scholar]
- 17.Knorr R, Trzeciak A, Bannawarth W, Gillessen D. Tetrahedron Lett. 1989;30:1927–1930. [Google Scholar]
- 18.Fields G B, Carr S A, Marshak D R, Smith A J, Stults J T, Williams K R, Young J D. In: Techniques in Protein Chemistry IV. Angeletti R H, editor. San Diego: Academic; 1993. pp. 229–238. [Google Scholar]
- 19.Smith A J, Young J D, Carr S A, Marshak D R, Williams L C, Williams K R. In: Techniques in Protein Chemistry III. Angeletti R H, editor. Orlando, FL: Academic; 1992. pp. 219–229. [Google Scholar]
- 20.Sheenan J C, Hess G P. J Am Chem Soc. 1955;77:1067–1068. [Google Scholar]
- 21.Sarantakis D, Teichman J, Lien E L, Fenichel R L. Biochem Biophys Res Commun. 1976;73:336–342. doi: 10.1016/0006-291x(76)90712-9. [DOI] [PubMed] [Google Scholar]
- 22.König W, Geiger R. Chem Ber. 1970;103:2024–2034. doi: 10.1002/cber.19701030705. [DOI] [PubMed] [Google Scholar]
- 23.Castro, B., Dormoy, J. R., Evin, G. & Selve, C. (1975) Tetrahedron Lett. 1219–1222.
- 24.Briand J-P, Van Dorsselaer A, Raboy B, Muller S. Pept Res. 1989;2:381–388. [PubMed] [Google Scholar]
- 25.Gausephol H, Schnölzer M, Frank R W. In: Innovation and Perspectives in Solid Phase Synthesis: Peptides, Polypeptides and Oligonucleotides. Epton R, editor. Canterbury, U.K.: Intercept; 1992. pp. 387–389. [Google Scholar]
- 26.Applied Biosystems. FastMoc® Chemistry: HBTU Activation Chemistry in Peptide Synthesis on Model 430A. Foster City, CA: Applied Biosystems; 1990. [Google Scholar]
- 27.Schnölzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Pept Prot Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 28.Alewood P, Alewood D, Miranda L P, Love S, Meutermans W D F, Wilson D. Methods Enzymol. 1997;289:14–29. doi: 10.1016/s0076-6879(97)89041-6. [DOI] [PubMed] [Google Scholar]
- 29.Hetnarski B, Merrifield R B. In: Peptides, Chemistry and Biology. Marshall G R, editor. Leiden, the Netherlands: ESCOM; 1988. pp. 220–222. [Google Scholar]
- 30.Pickup S, Blum F D, Ford W T, Periyasamy M. J Am Chem Soc. 1986;108:3987–3990. [Google Scholar]
- 31.Erickson B W, Merrifield R B. In: The Proteins. Neurath H, Hill R L, editors. New York: Academic; 1975. pp. 361–368. [Google Scholar]
- 32.Carpino L. PCT International Patent. USA; 1990. 5,590,981. [Google Scholar]
- 33.Carpino L A, El-Faham A. J Am Chem Soc. 1995;117:5401–5402. [Google Scholar]
- 34.Carpino L A. J Am Chem Soc. 1993;115:4397–4398. [Google Scholar]
- 35.Carpino L A, Sadat-Aalee D, Chao H G, DeSelms R. J Am Chem Soc. 1990;112:9651–9652. [Google Scholar]
- 36.Bertho J-N, Loffet A, Pinel C, Reuther F, Sennyey G. Tetrahedron Lett. 1991;32:1303–1306. [Google Scholar]
- 37.Kaduk C, Wenschuh H, Beyermann M, Forner K, Carpino L A. Lett Pept Sci. 1995;2:285–288. [Google Scholar]
- 38.Hancock W S, Prescott D J, Vagelos P R, Marshall G R. J Org Chem. 1973;38:774–781. [Google Scholar]
- 39.Kent S B H, Merrifield R B. In: Peptides 1980. Proceedings of the 16th European Peptide Symposium. Brunkfeldt K, editor. Copenhagen: Scriptor; 1981. pp. 328–333. [Google Scholar]
- 40.Hyde C, Johnson T, Owen D, Quibell M, Sheppard R C. Int J Pept Prot Res. 1994;43:431–435. [PubMed] [Google Scholar]
- 41.Eggenweiler M, Clausen N, Fritz H, Zhang L, Bayer E. In: Peptides. Proceedings of 23rd European Peptide Symposium. Maia H L, editor. Braga, Portugal: ESCOM; 1994. pp. 275–276. [Google Scholar]
- 42.Lackman M, Cornish C J, Simpson R J, Moritz R L, Geczy C L. J Biol Chem. 1992;267:7499–7504. [PubMed] [Google Scholar]
- 43.Sarin V, Kent S B H, Tam J P, Merrifield R B. Anal Biochem. 1981;117:147–157. doi: 10.1016/0003-2697(81)90704-1. [DOI] [PubMed] [Google Scholar]
- 44.Dawson P E, Churchill M J, Ghadiri M R, Kent S B H. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]
- 45.Dryland, A. & Sheppard, R. C. (1986) J. Chem. Soc. Perkin Trans 1, 125–137.
- 46.Beyermann M, Bienert M. Tetrahedron Lett. 1992;33:3745–3748. [Google Scholar]
- 47.Larsen B D, Holm A. Int J Pept Prot Res. 1994;43:1–9. doi: 10.1111/j.1399-3011.1994.tb00368.x. [DOI] [PubMed] [Google Scholar]
- 48.Baca M, Kent S B H. Proc Natl Acad Sci USA. 1993;90:11638–11642. doi: 10.1073/pnas.90.24.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C F, Tam J P. J Am Chem Soc. 1994;116:4149–4153. [Google Scholar]
- 50.Miller D, Sathyanarayana B K, Toth M V, Marshall G R, Clawson L, Selk L, Schneider J, Kent S B H. Science. 1989;246:1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L S, Tam J P. J Am Chem Soc. 1997;119:2363–2370. [Google Scholar]
- 52.Futaki S, Sogawa K, Maruyama J, Asahara T, Niwa M. Tetrahedron Lett. 1997;38:6237–6240. [Google Scholar]
- 53.Coste J. Tetrahedron Lett. 1990;31:669–672. [Google Scholar]
- 54.Sakakibara S, Shimonishi Y. Bull Chem Soc Jpn. 1965;38:1412–1413. doi: 10.1246/bcsj.38.1412. [DOI] [PubMed] [Google Scholar]
- 55.Miranda L P, Jones A, Meutermans W D F, Alewood P F. J Am Chem Soc. 1998;120:1410–1420. [Google Scholar]
- 56.Hyde, C., Johnson, J. & Sheppard, R. C. (1992) J. Chem. Soc. Chem. Commun. 1573–1575.
- 57.Tam J P, Lu Y A. J Am Chem Soc. 1995;117:12058–12063. [Google Scholar]
- 58.Schäfer B W, Heizmann C W. Trends Biochem Sci. 1996;21:134–140. doi: 10.1016/s0968-0004(96)80167-8. [DOI] [PubMed] [Google Scholar]
- 59.Adams J H, Cook R M, Hudson D, Jammalamadaka V, Lyttle M H, Songster M F. J Org Chem. 1998;63:3706–3716. [Google Scholar]
- 60.Han Y, Albericio F, Barany G. J Org Chem. 1997;62:4307–4312. doi: 10.1021/jo9622744. [DOI] [PubMed] [Google Scholar]