Abstract
In the visual system, differential gene expression underlies development of the anterior–posterior and dorsal–ventral axes. Here we present the results of a microarray screen to identify genes differentially expressed in the developing retina. We assayed gene expression in nasal (anterior), temporal (posterior), dorsal, and ventral embryonic mouse retina. We used a statistical method to estimate gene expression between different retina regions. Genes were clustered according to their expression pattern and were ranked within each cluster. We identified groups of genes expressed in gradients or with restricted patterns of expression as verified by in situ hybridization. A common theme for the identified genes is the differential expression in the dorsal-ventral axis. By analyzing gene expression patterns, we provide insight into the molecular organization of the developing retina.
A fundamental aspect of nervous system development is the spatial and temporal regulation of gene expression that underlies cell fate specification and connectivity. In the visual system, generation of the anterior–posterior (A-P) and dorsal–ventral (D-V) axes underlies positional specificity within the retina. The best example is the formation of a topographic map on the retinal axon output target, the superior colliculus (SC). Axons from D and V retina project to V and D SC, respectively, while A (also defined as nasal, N) and P (also defined as temporal, T) retinal axons project to P and A SC, respectively (1–4). Map formation depends upon complementary gradients of positional labels in the retina and the SC (5), specifically retinal EphA receptors and collicular ephrin-A ligands. In chick, EphA3 is expressed in a high-T to low-N gradient in the retina (6), whereas ephrin-A2 and ephrin-A5 are expressed in corresponding P-A gradients across the target (6, 7). Current models suggest that retinocollicular mapping is independent of the absolute level of EphA receptor signaling in retinal ganglion cells (RGCs), but, rather, is dependent on relative differences between neighboring RGCs (8).
Other molecules are differentially expressed in the visual system. Two EphB receptors and A and B type ephrins are expressed in gradients in the retina (9–12). The early eye can also be subdivided by expression of retinoic acid-generating enzymes along the D-V axis (13–15). Transcription factors and signaling components are expressed in gradients in the retina during development (16–19). To gain a broader view of gene expression patterns in the retina, we applied a microarray-based approach to identify new genes with restricted patterns of expression during development. Our results provide insight into the organization of the retina structure as well as identify candidate genes with potential roles in positional identity.
Materials and Methods
Microarray Hybridizations.
Embryonic day 14.5 (E14.5) CD-1 mouse retinas were divided into N, T, D, and V portions for RNA isolation with TRIzol (Invitrogen) and RNeasy columns (Qiagen, Valencia, CA). RNA amplifications and microarray hybridizations were carried out according to published methods (20). Arrays were constructed with the RIKEN 19K cDNA mouse clone set (21).
Image Analysis.
A total of 19 hybridizations containing comparisons between different portions of the retina (DN, NT, TV, and VD; 11 slides) and comparisons between whole retina (W) and different portions (DW, NW, TW, and VW; 8 slides) were used. Images were processed by spot (22) with foreground seeds set to five pixels square. The data were normalized by the “print-tip group-scale” method followed by multiple-slide scale normalization (23). We used an ℳ𝒜-plot (24) to represent the (R, G) data where ℳ = log2 (R/G) and 𝒜 = log2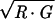 .
.
Data Analysis.
Linear models.
For every gene, we use a linear model to estimate each of the six contrasts [(NT), (DT), (VT), (DN), (VN), and (DV)]. We fit the linear model Y = X β̂ + ɛ where Y is a vector of log-ratios from the different slides, X is the design matrix, β is a vector of parameters (d(w), n(w), t(w), and v(w)), and ɛ is the error. Specifically, we fit the following linear model:
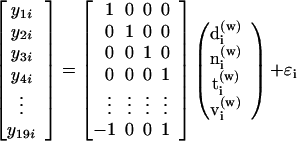 |
where y is the slide number and i is the gene number, by the iterated reweighted least squares procedure. We used the function rlm in the library MASS in the statistical software package r (25, 26). The six contrasts (designated retinal profiles) are calculated by linear combination of the vector of parameter estimates β̂ (e.g., NT = n(w) − t(w)). Retinal profiles can be simplified into four values, an in silico average representation, where each effect (d, n, t, and v) is compared with the average of all four regions (yielding d̃, ñ, t̃, and ṽ, respectively). For detailed design considerations, see ref. 27.
Two-stage clustering.
For each contrast, genes were selected based on the estimated contrast vs. average signal intensity plot where genes with average intensity >28 = 256 were considered. We selected genes based on four intensity bins in an attempt to take into account the change in variability as intensity increases. Genes were clustered by building a dendrogram based on a modified Mahalanobis distance and Ward agglomeration (28, 29). Considering all maximal disjoint clusters with heterogeneity score <0.3 provides nine groups. Genes within each group were ranked, based on their modified Mahalanobis distance Dx = (x)′(X′X)(x) where X is the design matrix. We carried out a second hierarchical clustering using the average profiles of the nine groups. A more detailed description of the statistical analysis and the complete data set are provided in Supporting Text and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org.
In Situ Hybridization.
First-strand cDNA from E14.5 retina was used as template for PCR with 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min. Primer sequences are as follows: neuroendocrine differentiation factor (NEDF) (TTCCTCTAATCCCTTGGCACCC and AACGAAGAAATCCAGGCGGC); Id3 (AACCCAGCCCTTTTCACTTACC and TAGTTCATCCCCACACTTGACCCC); e(y)2 (TTTCTGTGTGCTTAGGTGCCCGAG and CTCTGCCTTTTGGAGTGATTTCAG); μ-crystallin (ATGAGGCAAGCGGTGCTGTATGTG and TGACAATCACTACCATTCACTGGG); and cellular retinoic acid-binding protein 1 (CRABP1) (AGACACTTCTTGAGGGGGATGG and TGAGGGGAGGACACTACAACAATG). PCR products were cloned with the TA Cloning kit (Invitrogen) and used as template for in vitro transcription to produce antisense 35S-labeled probes. Probes were purified on S-200 microspin columns (Amersham Pharmacia) and were diluted to 8 × 108 cpm/ml in Wilkinson's hybridization buffer (30). Fixed or fresh-frozen CD-1 E14.5 heads were cut into 20-μm sections. Sections were fixed in 4% paraformaldehyde/1× PBS for 10 min, washed in PBS, and incubated with proteinase K (Sigma; 20 μg/ml in 50 mM Tris, pH 7.5/5 mM EDTA) for 4 min. Fresh-frozen sections were not treated with proteinase K. Sections were acetylated for 10 min, washed with PBS, and incubated with hybridization solution for 4 h. RNA probes were applied by “painting” with Parafilm. Slides were coverslipped and incubated in a humidified chamber overnight at 65°C. The next day, slides were washed in 0.2× SSC/10 mM DTT at 72°C for four times, each for 30 min (100 mM DTT in the first wash). Slides were cooled in RNase buffer (0.5 M NaCl/10 mM Tris, pH 7.5/5 mM EDTA), incubated with 5 μg/ml RNase A for 30 min at 37°C, washed twice for 30 min each with RNase buffer, four times for 15 min each with 0.2 × SSC at 72°C, and dehydrated through an ethanol series with 0.3 M NH4OAc. Slides were coated with NTB2 emulsion (Kodak) and were exposed for 10–14 days. Slides were developed with D19 developer (Kodak), rinsed with water, and fixed at 15°C. Slides were dehydrated through an ethanol series, incubated with xylenes, and mounted.
Results
Our main goal was to molecularly characterize the topography of RGC populations. We therefore chose to assay gene expression in E14.5 tissue. At this stage, there is a reduced complexity in the number of cell types, and the RGCs, which are the first cells to differentiate, are beginning to form connections with their targets. We tested whether our microarrays are able to detect the known differential expression of Eph receptors and their cognate ligands in our RNA samples. We found ephrin-A2 and ephrin-A5 to be differentially expressed 2.2- and 5.5-fold higher, respectively, in P relative to A SC (Fig. 1a). Furthermore, ephrin-A5 shows a higher differential in its expression as compared with ephrin-A2, in agreement with published results (31). In the retina, EphA5 and EphA6 are differentially expressed 2.6- and 2.8-fold higher, respectively, in T retina as compared with N retina (Fig. 1b). These values are in good agreement with published results for the equivalent EphA3 in chick (6).
Figure 1.
Differential expression of ephrins and Eph receptors by microarray analysis. (a) ℳ𝒜-plot of anterior SC RNA (G) vs. posterior SC RNA (R) from E14.5 embryos where ℳ = log2 (R/G) and 𝒜 = log2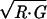 . Ephrin-A2 (red) and ephrin-A5 (blue) are highlighted. (b) ℳ𝒜-plot of N retina RNA (G) vs. T retina RNA (R) from E14.5 embryos. EphA5 (red) and EphA6 (blue) are highlighted.
. Ephrin-A2 (red) and ephrin-A5 (blue) are highlighted. (b) ℳ𝒜-plot of N retina RNA (G) vs. T retina RNA (R) from E14.5 embryos. EphA5 (red) and EphA6 (blue) are highlighted.
Rather than restricting our analysis to comparisons along a single axis in isolation (e.g., N vs. T or D vs. V), we decided to look at all pairwise comparisons. To this end, we prepared RNA from D, N, T, and V retina (Fig. 2a) and made several direct comparisons between the different regions (DN, NT, TV, and VD). We estimated the differential expression between all pairwise comparisons [denoted as (NT), (DT), (VT), (DN), (VN), and (DV)] by robust regression and used the resulting set of six values to generate a four-component in silico-average representation. This representation shows that EphA6 has relative higher expression in T and V retina as compared with N and D retina (Fig. 2b). As expected, the pattern of EphA6 expression with in situ hybridization matches the predicted expression profile (Fig. 2 c and d).
Figure 2.
Retina expression profile of EphA6. (a) Schematic of retina dissection protocol. Shading indicates the expected expression pattern of EphA6 and lines represent tissue taken for RNA purification. (b) The in silico-average representation for EphA6, where there is one component for each region measuring the relative expression between each region to the average expression across all regions. (c and d) Dark-field view of horizontal section (c) or coronal section (d) of E14.5 eye hybridized in situ with EphA6 probe. (Scale bar, 100 μm.)
Having established that we can identify known patterns of gene expression, we proceeded to identify expression patterns for all genes. We selected the 100 genes with the greatest differences (50 positive and 50 negative) in each of the six comparisons for further analysis (Fig. 3a), which resulted in a unique set of 362 genes. (We realize that these selection criteria include a substantial amount of noise, i.e., many genes display very small contrast measurements. However, we purposely chose to relax our selection criteria as the top 10% of genes with the highest differences encode hemoglobin subunits. This observation suggested that the largest differences we measured were because of dissection artifact and that the biologically interesting genes would be buried in this noise.) We then used hierarchical clustering as an organizational guide of the expression data. A two-step approach was used, based on the following considerations. A simple ranking of genes according to their overall variance would enrich for a single, dominant expression pattern. We reasoned that by separating the genes into groups first and then ranking them within a cluster, we could identify genes with different patterns. To this end, genes were divided into nine groups based on a modified Mahalanobis distance measurement and ranked within each cluster (Table 1). To identify potential similarities between these nine groups, we carried out a second clustering step using the average expression profile of each group (Fig. 3c). The corresponding dendrogram has three main branches (Fig. 3b). For each group, we generated a coefficient of variation (CV; standard deviation divided by the mean); a small CV value suggests that a particular cluster on average is less likely to be similar to random noise.
Figure 3.
Identification of genes differentially expressed in the retina by statistical analysis. (a) Representative scatter plots of estimated contrast versus average intensity. The y axis shows the estimated contrasts between two regions of the retina and the x axis shows the average intensity values for all data. Two of six plots are shown [(NT) and (DV)]. Genes with average intensity >256 (28) included for further analysis are red. Five genes validated by in situ hybridization (see Fig. 4) are highlighted: Id3 (green), μ-crystallin (purple), CRABP1 (aqua), e(y)2 (pink), and NEDF (blue). (b) Cluster dendrogram of the average expression profiles of the nine groups. (c) Visualization of the average expression profiles of the nine groups using a red/green diagram (41) where the order corresponds to the dendrogram in b. Red and green indicate increased or decreased relative expression, respectively. Black indicates equivalent expression, all relative to the average across all regions.
Table 1.
Identities and scores of select candidate genes from each group
| Group | CV | No. of genes | RIKEN ID | Name | Score |
|---|---|---|---|---|---|
| 1 | 0.11 | 4 | ZX00024M03 | μ-Crystallin | 3.07 |
| ZX00026L02 | Inhibitor of DNA binding 3 | 2.6 | |||
| 2 | 0.47 | 17 | ZX00030P22 | delta-like homolog | 4.21 |
| ZX00019A18 | Neuroendocrine differentiation factor | 3.79 | |||
| ZX00012B09 | Enhancer of yellow 2 | 2.73 | |||
| 3 | 0.45 | 29 | ZX00004M02 | Hemoglobin β, pseudogene bh3 | 4.66 |
| ZX00035H14 | Hemoglobin β-chain complex | 4.17 | |||
| 4 | 0.58 | 25 | ZA00002N13 | Glutamine synthetase | 2.22 |
| ZX00009N19 | Elongation factor 1-β | 2.06 | |||
| 5 | 1.0 | 25 | ZX00035J12 | ATPase-like vacuolar proton channel | 5.11 |
| ZX00049I17 | Ribosomal protein S4 | 3.63 | |||
| 6 | 0.62 | 42 | ZX00030P13 | Related to CG3450 gene product | 2.16 |
| ZX00032I10 | EST | 2.15 | |||
| ZX00029E09 | EST, similar to BPM1 antigen | 1.37 | |||
| 7 | 0.66 | 58 | ZX00034M16 | Pleiotrophin | 2.01 |
| ZX00004P13 | EST | 1.97 | |||
| ZX00024L05 | CRABP1 | 1.15 | |||
| 8 | 0.89 | 64 | ZX00035H16 | EST | 2.36 |
| ZX00035D15 | Template activating factor-1 | 2.28 | |||
| ZX00025D01 | Split hand/foot deleted gene 1 | 1.40 | |||
| 9 | 1.52 | 98 | ZX00022O24 | PKC substrate 80 K-H | 2.34 |
| ZX00016L23 | EST | 1.95 | |||
| ZA00003J01 | Cofilin 2 | 1.89 |
Branch I.
Branch I (groups 1, 4, 6, and 8) consists of genes with retinal expression profiles that are high in T and D relative to N and V retina. The average profile of group 1 (CV = 0.11) shows the highest degree of relative expression difference (Fig. 3c). This group contains two genes, μ-crystallin, a lens structural protein, and the inhibitor of DNA binding family 3 (Id3), a negative regulator of basic helix–loop–helix DNA-binding proteins (32), represented three times (Table 1). By in situ hybridization, Id3 is predominantly expressed in D and T retina (Fig. 4 a and b) as predicted (Fig. 4c). Furthermore, it is expressed in a high-T to low-N gradient in the ventricular zone (Fig. 4 a and b, asterisks). In addition to its role as a lens structural protein, μ-crystallin is hypothesized to be an enzyme involved in amino acid metabolism (33). In the developing retina, μ-crystallin is restricted to T and D retina (Fig. 4 d and e) as predicted (Fig. 4f). Unlike Id3, μ-crystallin is expressed in a patch that encompasses the ventricular zone as well as the RGC layer (Fig. 4 d and e, asterisks). The average profiles of group 4 (CV = 0.58), group 6 (CV = 0.62), and group 8 (CV = 0.89) have much smaller relative expression differences compared with group 1 (Fig. 3c). Furthermore, the CVs are much higher than for group 1, suggesting that these clusters are more similar to random noise. The split hand/foot-deleted gene 1 (group 8), glutamine synthetase (group 4), and an EST with weak similarity to the 230-kDa bullous pemphigoid antigen BPM1 (group 6), are restricted to the RGC layer but are not expressed in a graded fashion (data not shown). Given that the expression patterns for genes in these clusters (groups 4, 6, and 8) are more similar to random noise on average, it is not surprising that the genes tested by in situ hybridization do not match the microarray data prediction.
Figure 4.
In situ hybridization of predicted differentially expressed genes. (a–c) Id3. Dark-field view of horizontal section (a) or coronal section (b) of E14.5 eye hybridized in situ with Id3 probe. Note signal in the ventricular zone in a high-T to low-N and high-D to low-V gradient (compare areas marked by asterisks in a and b). A four-way expression prediction (in silico average) for Id3 is shown in c. (d–f) μ-Crystallin. μ-Crystallin probe labels D-T retina (asterisks in d and e) and the optic disk (arrows in d and e). A four-way prediction for μ-crystallin is shown in f. (g–i) CRABP1. A CRABP1 probe labels the RGC layer in a high-V to low-D gradient (compare areas marked by arrowheads in h). Note equal signal in N and T (compare areas marked by arrowheads in g). A four-way prediction for CRABP1 is shown in i. (j–l) e(y)2. e(y)2 probe labels the RGC layer in a high-D to low-V gradient (compare areas marked by arrowheads in k). A four-way prediction for e(y)2 is shown in l. (m–o) NEDF. NEDF is expressed in the RGC layer in a high-D to low-V gradient (compare areas marked by arrowheads in n). A four-way prediction for NEDF is shown in o. (Scale bar, 100 μm.)
Branch II.
Branch II (groups 2 and 3) consists of genes with expression profiles that are high in D relative to V retina and, to a lesser degree, high in N relative to T retina (Fig. 3c). Group 3 (CV = 0.45) is composed almost entirely of genes encoding hemoglobin subunits (Table 1); this cluster may represent differential vascularization of the retina tissue. From group 2 (CV = 0.47), the enhancer of yellow 2 gene, e(y)2, encodes a putative transcription factor (34) and is predicted to be expressed in D retina (Fig. 4l). Indeed, e(y)2 is expressed in the RGC layer in a high-D to low-V gradient (Fig. 4 j and k). NEDF (group 2) has a similar prediction (Fig. 4o) and is also expressed in a high-D to low-V gradient in differentiated RGCs (Fig. 4 m and n). Delta-like (dlk) homolog (group 2) is a member of the notch/delta/serrate family of transmembrane proteins (35). Its expression pattern is restricted to the optic disk, the exit point of the RGC axons (data not shown). Dlk does not match its expression profile; however, it is also highly expressed in the cartilage surrounding the eye (data not shown), which most likely contributes to its microarray profile.
Branch III.
Branch III (groups 5, 7, and 9) consists of genes with expression profiles that are high in V relative to D retina and show no difference in the N-T axis. Of the three groups, group 7 has the largest differential expression on average (Fig. 3c) and the lowest CV (0.66). CRABP1 has been shown previously to be expressed in a D-V asymmetrical pattern (36). Accordingly, CRABP1 (group 7) is predicted to be high in V retina and low in D retina (Fig. 4i), as is reflected in the in situ hybridization pattern (Fig. 4 g and h). However, pleiotrophin (group 7), an extracellular matrix protein, is selectively expressed in the optic disk and optic nerve (not shown). Cofilin 2 (group 9) is restricted to the RGC layer (not shown). NORPEG (Novel retinal pigment epithelial cell gene; group 5) encodes a retinoic acid-inducible gene (37) and is restricted to the epithelium (not shown). The CVs are very high for group 5 (1.0) and group 9 (1.52); thus, these clusters are more similar to random noise on average.
Discussion
A major challenge in the newly emerging field of functional genomics is to uncover meaningful expression differences in complex tissues. Our approach allowed the identification of several genes expressed in biologically interesting patterns in the developing retina. Moreover, all expression differences were on the order of 2-fold or less, which highlights the sensitivity of our approach. We identified several genes expressed in gradients, and such genes are hypothesized to underlie positional identity in the retina (5). The data also recapitulated known expression differences (EphA6 and CRABP1). Thus, our method presents a useful molecular screen to identify candidate genes that may play a role in positional identity in retina development.
By using the CV and the average expression profile of each cluster as a guide, we identified three groups of genes (groups 1, 2, and 7) that were validated by in situ hybridization. Group 1 (four genes, Id3 is represented three times) showed the highest degree of relative expression differences for the average profile (Fig. 3c) and had the lowest CV (0.11). By in situ hybridization, 100% of the genes were validated. In contrast, group 7 (58 genes) had the highest CV value (0.66) of these three groups and the validation rate for this group was much lower. Indeed, we tested 6 genes in the top 10 and the validation rate was 0%. The only positive we have identified thus far in this group is CRABP1 (a previously known gene), whose score is 1.19. Thus, although we anticipate that there are other true positives within this group, the high false-positive rate (>90%) confounds our analysis. Group 2 (17 genes) has an intermediate CV value (0.47) and an intermediate validation rate. We tested eight genes in this group and the validation rate by in situ hybridization was 25% (2 of 8). The validation rate is higher (40%) if one considers only those genes tested in the top 10 (2 of 5). This result is not too surprising as the difference in CV value is large for group 2 compared with group 1. Indeed, there is a precipitous increase in the CV between groups 2 and 7 compared with group 1. Thus, we anticipate that ≈25% of genes in group 2 and <10% of genes in group 7 are true positives, in addition to 100% of the genes in group 1.
The other groups had much larger CV values, and of the few genes that we tested, we did not find any true positives. We anticipate that the majority, if not all, of these genes are false positives. Most likely, the greatest source that leads to such a high false-positive rate is dissection artifact. Tissue dissection is ultimately contaminated with blood, cartilage, and other nonneuronal tissue. However, our approach distributed genes into separate categories, which enabled discarding those genes that are most likely noise. Indeed, our method is superior to simple rank-ordering of genes. By separating the genes into distinct groups and then ranking them according to their score, we identified several genes that would have been buried in the noise had we simply ranked all genes. For instance, e(y)2 is ranked 42nd of the total 362 genes; however, it is ranked 6th within group 2. Thus, our approach is useful for identifying genes with complex patterns of expression that would otherwise have been difficult to detect. Perhaps we could increase our true-positive rate by being more selective in our criteria, but we feel that the subsequent analysis is sufficient to identify those groups that are most likely false.
Examination of the identified genes illustrates several features of our analysis. We identified three main expression patterns represented by group 1 (high in T and D relative to N and V), group 2 (high in N and D relative to T and V), and group 7 (high in V relative to D). All of the genes we identified showed some degree of differential expression along the D-V axis, suggesting that this is the most prominent expression pattern at this point in development. Indeed, D-V aspects of the eye are distinct very early in development (E8). The developing eye-cup grows dorsally, and the choroidal fissure is formed on its V side. While the EphA receptors and ephrin-A ligands clearly play a role in N-T topographic map formation, much less is known about the positional labels that specify the D-V map. The patterns of differential expression along the D-V axis may underlie or otherwise reflect these morphogenetic changes.
Id3 and μ-crystallin have similar predicted expression patterns; however, in situ hybridization reveals a striking difference in expression between these two genes. Id3 is expressed in a gradient in the ventricular zone in both the D-V and N-T axes. Id proteins are negative regulators of basic helix–loop–helix DNA-binding proteins (32). Interestingly, Math5, a basic helix–loop–helix gene, is expressed during early stages of retinal neurogenesis (38). Similar to Id3, it is specifically expressed in the ventricular zone, whereas it is excluded from differentiated RGCs (38). It is tempting to speculate that Id3 and Math5 function together to specify positionally distinct RGCs. Alternatively, Id3 may reflect a gradient of differentiation within the ventricular zone. However, Id3 expression is maintained at E17.5 and postnatal day 0 in a similar pattern, supporting the gradient of positional information model. Further experiments are necessary to test this directly. μ-crystallin expression is highest in the medial cell layers of D-T retina. Crystallin proteins are structural components of the lens but their role in the retina is unknown. Expression in D retina could serve as marker for the asymmetric distribution of a particular cell type, for instance, the photoreceptor cells known to be spatially restricted (39).
The gene e(y)2 is homologous to a putative transcription factor in flies (34). It is expressed in a high-D to low-V gradient in the RGC layer and its pattern suggests a role in D-V topographic map formation, perhaps by directing the graded expression of targeting molecules such as EphB receptors and ephrin-B ligands, which are expressed in D-V gradients. NEDF is a recently characterized protein that can induce a neuritic-like morphology in culture (40). It has an expression pattern similar to that of e(y)2, suggesting that it may also play a role in D-V topographic map formation. These genes are excellent candidates to test functionally by transgenic approaches and in vitro assays.
Intriguingly, several of the genes identified in our microarray screen in the retina also show differential expression in the olfactory epithelium, suggesting a common mechanism between these systems. For example, CRABP1 is excluded from the D portion of the olfactory epithelium (not shown). It will be exciting to explore this concept in more detail by comparing expression data from other topographic mapping systems. Finally, it will be interesting to combine the temporal program of gene expression with our spatial analysis during development. We predict that as the retina develops, some patterns may become stronger while others become weaker. New patterns may also emerge as more cell types are formed. For instance, we predict that the Id3 pattern would disappear as the ventricular zone of undifferentiated cells is lost. Such a spatiotemporal map of gene expression could then be combined with expression profiles in the target, the SC. By adding more profiles to a compendium of expression profiles, it should be possible to build a precise spatiotemporal map of gene expression that underlies development of the visual system, from axon to target.
Supplementary Material
Acknowledgments
We thank J. DeRisi for use of his microarrayer; S. Choksi, A. Finn, D. Lin, P. Luu, V. Peng, L. Simirenko, D. Speca, M. Szpara, and T. Alioto, who contributed to the microarray fabrication; F. Wang for the Eph and ephrin constructs; H.-J. Cheng for help with dissections; L. Brunet for help with in situ hybridizations; and members of the Ngai and Tessier-Lavigne laboratories, especially F. Wang, H.-J. Cheng, and K. Mitchell, for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health (to J.N. and T.P.S.) and by funds from the Department of Molecular Cell Biology and Helen Wills Neuroscience Institute, University of California, Berkeley. Funding for generation of the 19K RIKEN cDNA set was supported in part by the Special Coordination Funds for Promoting Science and Technology and a Research Grant for the RIKEN Project from the Science and Technology Agency of the Japanese government (to Y.H.). M.T.-L. is an Investigator of the Howard Hughes Medical Institute. E.D. was supported by fellowships from the Helen Hay Whitney Foundation and the National Institutes of Health. T.F. was supported by a predoctoral fellowship from the National Science Foundation.
Abbreviations
- A
anterior
- P
posterior
- D
dorsal
- N
nasal
- T
temporal
- V
ventral
- SC
superior colliculus
- RGC
retinal ganglion cell
- CV
coefficient of variation
- CRABP1
cellular retinoic acid-binding protein 1
- NEDF
neuroendocrine differentiation factor
- En
embryonic day n
References
- 1.Tessier-Lavigne M. Cell. 1995;82:345–348. doi: 10.1016/0092-8674(95)90421-2. [DOI] [PubMed] [Google Scholar]
- 2.O'Leary D D, Yates P A, McLaughlin T. Cell. 1999;96:255–269. doi: 10.1016/s0092-8674(00)80565-6. [DOI] [PubMed] [Google Scholar]
- 3.Flanagan J G, Vanderhaeghen P. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 4.Drescher U, Bonhoeffer F, Muller B K. Curr Opin Neurobiol. 1997;7:75–80. doi: 10.1016/s0959-4388(97)80123-7. [DOI] [PubMed] [Google Scholar]
- 5.Sperry R W. Proc Natl Acad Sci USA. 1963;50:703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H J, Nakamoto M, Bergemann A D, Flanagan J G. Cell. 1995;82:371–381. doi: 10.1016/0092-8674(95)90426-3. [DOI] [PubMed] [Google Scholar]
- 7.Drescher U, Kremoser C, Handwerker C, Loschinger J, Noda M, Bonhoeffer F. Cell. 1995;82:359–370. doi: 10.1016/0092-8674(95)90425-5. [DOI] [PubMed] [Google Scholar]
- 8.Brown A, Yates P A, Burrola P, Ortuno D, Vaidya A, Jessell T M, Pfaff S L, O'Leary D D, Lemke G. Cell. 2000;102:77–88. doi: 10.1016/s0092-8674(00)00012-x. [DOI] [PubMed] [Google Scholar]
- 9.Holash J A, Pasquale E B. Dev Biol. 1995;172:683–693. doi: 10.1006/dbio.1995.8039. [DOI] [PubMed] [Google Scholar]
- 10.Braisted J E, McLaughlin T, Wang H U, Friedman G C, Anderson D J, O'Leary D, D. Dev Biol. 1997;191:14–28. doi: 10.1006/dbio.1997.8706. [DOI] [PubMed] [Google Scholar]
- 11.Connor R J, Menzel P, Pasquale E B. Dev Biol. 1998;193:21–35. doi: 10.1006/dbio.1997.8786. [DOI] [PubMed] [Google Scholar]
- 12.Marcus R C, Gale N W, Morrison M E, Mason C A, Yancopoulos G D. Dev Biol. 1996;180:786–789. doi: 10.1006/dbio.1996.0347. [DOI] [PubMed] [Google Scholar]
- 13.McCaffery P, Lee M O, Wagner M A, Sladek N E, Drager U C. Development (Cambridge, UK) 1992;115:371–382. doi: 10.1242/dev.115.2.371. [DOI] [PubMed] [Google Scholar]
- 14.Marsh-Armstrong N, McCaffery P, Gilbert W, Dowling J E, Drager U C. Proc Natl Acad Sci USA. 1994;91:7286–7290. doi: 10.1073/pnas.91.15.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godbout R, Packer M, Poppema S, Dabbagh L. Dev Dyn. 1996;205:319–331. doi: 10.1002/(SICI)1097-0177(199603)205:3<319::AID-AJA11>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Yuasa J, Hirano S, Yamagata M, Noda M. Nature. 1996;382:632–635. doi: 10.1038/382632a0. [DOI] [PubMed] [Google Scholar]
- 17.Schulte D, Furukawa T, Peters M A, Kozak C A, Cepko C L. Neuron. 1999;24:541–553. doi: 10.1016/s0896-6273(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 18.Koshiba-Takeuchi K, Takeuchi J K, Matsumoto K, Momose T, Uno K, Hoepker V, Ogura K, Takahashi N, Nakamura H, Yasuda K, Ogura T. Science. 2000;287:134–137. doi: 10.1126/science.287.5450.134. [DOI] [PubMed] [Google Scholar]
- 19.Sakuta H, Suzuki R, Takahashi H, Kato A, Shintani T, Iemura S, Yamamoto T S, Ueno N, Noda M. Science. 2001;293:111–115. doi: 10.1126/science.1058379. [DOI] [PubMed] [Google Scholar]
- 20.Diaz E, Ge Y, Yang Y H, Loh K C, Serafini T A, Okazaki Y, Hayashizaki Y, Speed T P, Ngai J, Scheiffele P. Neuron. 2002;36:417–434. doi: 10.1016/s0896-6273(02)01016-4. [DOI] [PubMed] [Google Scholar]
- 21.Miki R, Kadota K, Bono H, Mizuno Y, Tomaru Y, Carninci P, Itoh M, Shibata K, Kawai J, Konno H, et al. Proc Natl Acad Sci USA. 2001;98:2199–2204. doi: 10.1073/pnas.041605498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y H, Buckley M J, Dudoit S, Speed T P. J Comput Graph Stat. 2002;11:109–136. [Google Scholar]
- 23.Yang Y H, Dudoit S, Luu P, Lin D M, Peng V, Ngai J, Speed T P. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudoit S, Yang Y H, Callow M J, Speed T P. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- 25.Ihaka R, Gentleman R. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 26.Venables W N, Ripley B D. Modern Applied Statistics with S-PLUS. New York: Springer; 1999. [Google Scholar]
- 27.Yang Y H, Speed T P. Nat Rev Genet. 2002;3:579–588. doi: 10.1038/nrg863. [DOI] [PubMed] [Google Scholar]
- 28.Mardia K V, Kent J T, Bibby J M. Multivariate Analysis. London: Academic; 1979. [Google Scholar]
- 29.Hartigan J A. Clustering Algorithms. New York: Wiley; 1975. [Google Scholar]
- 30.Wilkinson D G. In Situ Hybridization. Oxford: Oxford Univ. Press; 1998. [Google Scholar]
- 31.Frisen J, Yates P A, McLaughlin T, Friedman G C, O'Leary D D, Barbacid M. Neuron. 1998;20:235–243. doi: 10.1016/s0896-6273(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 32.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 33.Segovia L, Horwitz J, Gasser R, Wistow G. Mol Vis. 1997;3:3–9. [PubMed] [Google Scholar]
- 34.Georgieva S, Nabirochkina E, Dilworth F J, Eickhoff H, Becker P, Tora L, Georgiev P, Soldatov A. Mol Cell Biol. 2001;21:5223–5231. doi: 10.1128/MCB.21.15.5223-5231.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laborda J, Sausville E A, Hoffman T, Notario V. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- 36.McCaffrery P, Posch K C, Napoli J L, Gudas L, Drager U C. Dev Biol. 1993;158:390–399. doi: 10.1006/dbio.1993.1197. [DOI] [PubMed] [Google Scholar]
- 37.Kutty R K, Kutty G, Samuel W, Duncan T, Bridges C C, El-Sherbeeny A, Nagineni C N, Smith S B, Wiggert B. J Biol Chem. 2001;276:2831–2840. doi: 10.1074/jbc.M007421200. [DOI] [PubMed] [Google Scholar]
- 38.Brown N L, Kanekar S, Vetter M L, Tucker P K, Gemza D L, Glaser T. Development (Cambridge, UK) 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- 39.Szel A, Rohlich P, Caffe A R, van Veen T. Microsc Res Tech. 1996;35:445–462. doi: 10.1002/(SICI)1097-0029(19961215)35:6<445::AID-JEMT4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 40.Wilson E M, Oh Y, Hwa V, Rosenfeld R G. J Clin Endocrinol Metab. 2001;86:4504–4511. doi: 10.1210/jcem.86.9.7845. [DOI] [PubMed] [Google Scholar]
- 41.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






