Abstract
The spontaneous activity of pacemaker cells in the sino-atrial node (SAN) controls the heart rhythm and rate under physiological conditions. Pacemaker activity in SAN cells is due to the presence of the diastolic depolarization, a slow depolarization phase that drives the membrane voltage from the end of an action potential to the threshold of a new action potential. SAN cells express a wide array of ionic channels, but we have limited knowledge about their functional role in pacemaker activity and we still do not know which channels play a prominent role in the generation of the diastolic depolarization. It is thus important to provide genetic evidence linking the activity of genes coding for ionic channels to specific alterations of pacemaker activity of SAN cells. Here, we show that target inactivation of the gene coding for α1D (Cav1.3) Ca2+ channels in the mouse not only significantly slows pacemaker activity but also promotes spontaneous arrhythmia in SAN pacemaker cells. These alterations of pacemaker activity are linked to abolition of the major component of the L-type current (ICa,L) activating at negative voltages. Pharmacological analysis of ICa,L demonstrates that Cav1.3 gene inactivation specifically abolishes ICa,L in the voltage range corresponding to the diastolic depolarization. Taken together, our data demonstrate that Cav1.3 channels play a major role in the generation of cardiac pacemaker activity by contributing to diastolic depolarization in SAN pacemaker cells.
Automaticity in sino-atrial node (SAN) pacemaker cells is due to the slow diastolic depolarization phase (1). Different ionic currents are believed to be involved in the generation of pacemaker activity, but their functional importance in the generation of the diastolic depolarization is still controversial. One possibility is that the diastolic depolarization is generated by the interaction of the decaying fast component of the delayed rectifier (IKr) and at least one of four different voltage-dependent inward currents; the hyperpolarization-activated current If, the sustained inward current Ist, the low-voltage activated T-type (ICa,T), and high-voltage-activated L-type Ca2+ current (ICa,L) (refs. 2–5; for review, see ref. 6). Particularly, ICa,L has been proposed to contribute to the diastolic depolarization (7). Indeed, pharmacological inhibition of ICa,L significantly slows pacemaker activity of isolated rabbit SAN cells in vitro (8). Also, the dihydropyridine Ca2+ channel antagonist nicardipine induces bradycardia in anesthetized mice in vivo (9).
L-type Ca2+ channels are multisubunit complexes formed by different isoforms of the pore forming α1 subunit named α1S (Cav1.1), α1C (Cav1.2), α1D (Cav1.3), and α1F (Cav1.4) (10). It has recently been reported that homozygous mice in which the gene coding for Cav1.3 Ca2+ channel has been inactivated (Cav1.3−/−) show prominent SAN dysfunction. Indeed, both bradycardia and sino-atrial arrhythmia have been observed in Cav1.3−/− mouse strains in vivo (11, 12). These observations suggest that Cav1.3 gene inactivation affects pacemaker activity of SAN cells. The recent possibility to isolate in vitro mouse SAN cells gives the opportunity to study pacemaker activity and ionic channels in SAN cells from genetically modified mice (5). We have thus studied pacemaker activity and ICa,L in SAN cells obtained from a Cav1.3−/− mouse strain (11).
Materials and Methods
RT-PCR Analysis and Southern Blotting.
For RT-PCR analysis, total RNAs from SAN, the right atrium, the left ventricle, retina, and skeletal muscle from C57BL/6J adult mice were extracted by using the SNAP Kit (Invitrogen). RT-PCR was performed with total RNA by using random primers and Superscript II RNase H reverse transcriptase (Invitrogen). After reverse transcription, the cDNAs for Cav1.3, Cav1.2, Cav1.4, and Cav1.1 were amplified by using the following set of primers (from 5′ to 3′): Cav1.3 (GenBank accession no. XM_138953) sense, GCA AAC TAT GCA AGA GGC ACC AG (position 43), antisense, GGG AGA GAG ATC CTA CAG GTG G (position 217, expected size 174 bp); Cav1.2 (accession no. NM_009781) sense GGA TGA GTG AAG AAG CCG AG (position 6682), antisense, AGG CCC TTC GAC CTA GAG AG (position 6855, 173 bp); Cav1.4 (accession no. AF192497) sense, CGA ACC CAG TCC TTT TAC CA (position 5376), antisense, TTG CCA AGG TAT CCT TCA CC (position 5672, 296 bp); and Cav1.1 (accession no. NM_014193) sense, AGA AAG CGC AGG AAG ATG TC (position 2157), antisense, GGG TAG GGG TCT TTC ACC TC (position 2326, 169 bp). PCR was performed in a final volume of 25 μl containing 2 μl of reverse transcription reaction, 10 pmol of each primer, 2.5 mM each dNTP (Pharmacia), 1.5 mM MgCl2 and 1 unit of Taq polymerase (Sigma) in an MJ Research (Cambridge, MA) Thermal Cycler with the following cycling protocol: after 2 min at 94°C, 35 cycles (94°C, 25 s; 53°C, 30 s; 72°C, 25 s) of PCR were performed followed by a final elongation period of 7 min at 72°C. To investigate the presence and size of the amplified fragments, 10 μl of PCR products were separated and visualized in an ethidium bromide-stained agarose gel (2%) by electrophoresis. Specific primers for Cav1.1 and Cav1.4 subunits detected corresponding fragments from mRNA of skeletal muscle and retina (data not shown). Cav1.3-specific PCR primers amplified a fragment from exon 2 of Cav1.3. Gene targeting in Cav1.3 mice involved introduction of a neomycin cassette into exon 2, thereby introducing multiple stop codons in all reading frames (11). Therefore, the used primer pair amplifies a fragment >1,800 bp in Cav1.3−/− mice that cannot be detected under our experimental conditions. For human tissue samples, small pieces of the atrioventricular node tissue, the right atrium, and ventricle were obtained from two cardiac graft recipients (aged 44–56 yr). The protocol was approved by the Ethical Committee of the Arnaud de Villeneuve University Hospital (Montpellier, France). The human SAN region could not be tested in this set of experiments because the cardiac grafting procedure required intact SAN of both donor and recipient. cDNAs for Cav1.3 and Cav1.2 were amplified by using the following set of primers (from 5′ to 3′): Cav1.3 (accession no. NM_000720) sense, GGA TTC TTG GAG GAC GAT GA (position 5843), antisense, TGC ATT AGA TGC AGA GGC AG (position 6042, 199 bp); and Cav1.2 (accession no. NM_000719) sense AGA TGA ACC ATG ACA CGG AGG CC (position 5784), antisense, AGG CCC TTC GAC CTA GTG AG (position 5957, 173 bp). The following cycling protocol was used: 3 min at 94°C, 35 cycles (94°C, 45 s; 57°C, 45 s; 72°C, 25 s). Elongation period lasted 10 min at 72°C. Specific PCR products were detected by Southern probing using internal oligonucleotides.
SAN Cell Preparation and Electrophysiological Recordings.
SAN tissue samples and pacemaker cells were isolated from age- and sex-matched wild-type or Cav1.3−/− C57BL/6J adult mice donors of either sex as described (Fig. 1Aa; ref. 5). The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (NIH publication no. 85-23, revised 1996), and European directives (86/609/CEE).
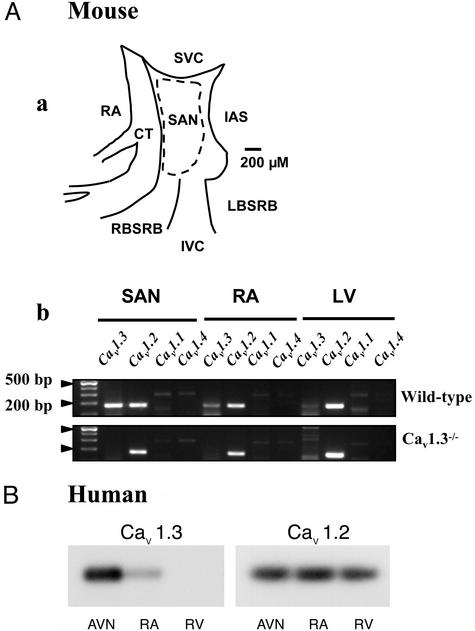
Figure 1.
(Aa) Experimental landmarks of the mouse SAN used in this study. RA, right atrium; CT, crista terminalis; SVC, superior vena cava; IVC, inferior vena cava; RBSRB, right branch of the sino-atrial ring bundle; LBSRB, left branch of the sino-atrial ring bundle; IAS, interatrial septum. The dotted line indicates the cutting edge used for obtaining SAN tissue samples for RT-PCR and pacemaker cells isolation. (Ab) RT-PCR analysis of Ca2+ channel Cav1 subunits, showing that the Cav1.3 mRNA is detected in both SAN and RA, but not in the left ventricle (LV) of wild-type mice. The Cav1.2 subunit is detected in SAN as well as in the other cardiac chambers. In two independent experiments, no mRNA expression is observed for the Cav1.1 and Cav1.4 subunit. (B) Southern blot analysis of Cav1.3 and Cav1.2 expression in the human heart. Contrary to the Cav1.2 subunit, which showed uniform expression in all cardiac tissues tested, including the right ventricle (RV), mRNA corresponding to Cav1.3 was found only in the atrioventricular node (AVN) and the RA.
Pacemaker activity of SAN cells was recorded at 35°C. Ionic currents in SAN cells were recorded at 26°C. The whole-cell patch-clamp technique (13) was used to record ionic currents and pacemaker activity. Recording of Ca2+ currents in SAN cells was performed in extracellular 4 mM Ca2+ as described (5). The voltage-dependence of ICa,L and ICa,T activation was determined from current-to-voltage (I–V) curves that were fitted by employing the Boltzmann relation: I = Gmax(V − Vrev)/{1 + exp[(V0.5,act − V)/s]}, where Vrev is the extrapolated reversal potential, V is the membrane potential, I is the peak current, Gmax is the cell maximum conductance, V0.5,act is the voltage for half current activation, and s is the slope factor of the Boltzmann term. Steady-state inactivation curves were fitted according to the Boltzmann relation: I/Imax = 1 − 1/{1 + exp[(V0.5,inact − V)/si]}, where V0.5,inact is the voltage for half-current inactivation and si is the slope factor of the Boltzmann term. Cellular automaticity was recorded under perforated-patch conditions by using Amphotericin and in extracellular Tyrode solution containing (mM): NaCl, 140; KCl, 5.4; CaCl2, 1.8; MgCl2, 1; Hepes-NaOH, 5; and d-glucose, 5.5; (adjusted to pH = 7.4 with NaOH). The composition of the pipette solution was (mM): KCl, 130; NaCl, 10; ATP-Na+ salt, 2; creatine phosphate, 6.6; GTP-Mg2+, 0.1; CaCl2, 0.04 (pCa = 7); Hepes-KOH, 10; (adjusted to pH = 7.2 with KOH). Amphotericin (Sigma) was added to the pipette solution at a final concentration of 200 μg/ml. All voltage values were corrected for the appropriate liquid junction potential.
Statistical Analysis.
Data are presented as mean ± SEM. Statistical significance was assessed by unpaired Student's t test. A value of P < 0.05 was considered statistically significant. Analysis was performed by employing origin software (ver. 6.0, Microcal Software, Northampton, MA).
Results
Expression of Cav1.3 mRNA in the Mouse and the Human Heart.
RT-PCR analysis readily detected a 180-bp Cav1.3-specific mRNA fragment in tissue samples obtained from the central SAN and the right atrium of wild-type mice (Fig. 1Ab). This fragment was almost completely absent in the left ventricle. No evidence for intact Cav1.3 transcripts was obtained in the SAN of Cav1.3−/− mice. Cav1.2 mRNA was detected in SAN of both wild-type and Cav1.3−/− mice. Cav1.2 mRNA was also found in the atrium and ventricle of both mouse strains. No mRNA coding for Cav1.1 and Cav1.4 subunits could be detected in these cardiac tissues. Because the Cav1.3 subunit showed differential expression in the mouse cardiac chambers, we also tested the expression of Cav1.3 and Cav1.2 mRNA in human cardiac tissue samples that were obtained from two end-stage failing hearts undergoing cardiac grafting (Fig. 1B). Cav1.3 expression was found in the atrioventricular node and, similarly to what found in the mouse, in the right atrium, but was almost absent in the right ventricle. In contrast, Cav1.2 mRNA showed uniform expression in the cardiac chambers and in the atrioventricular node.
Ca2+ Currents in Cav1.3−/− Pacemaker Cells.
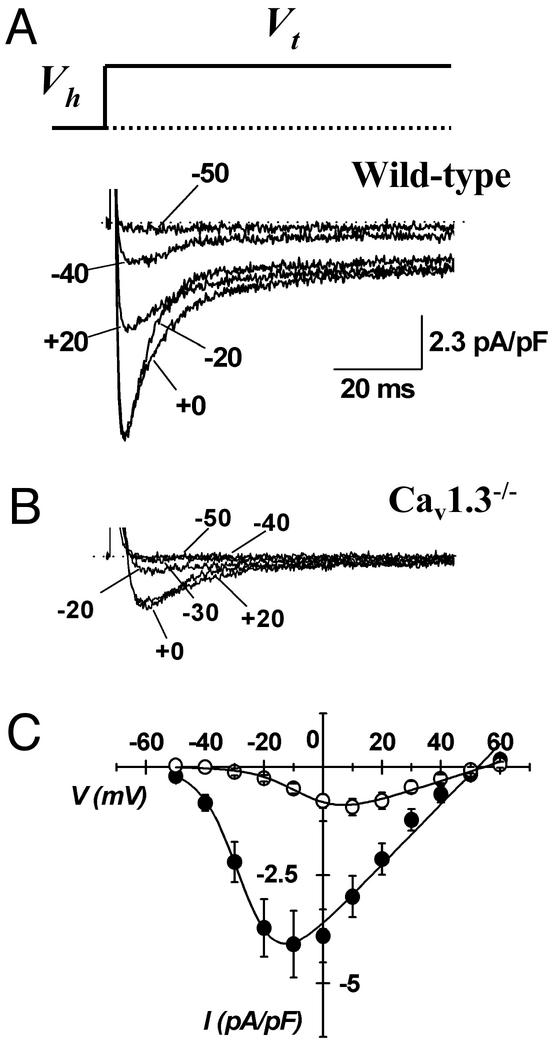
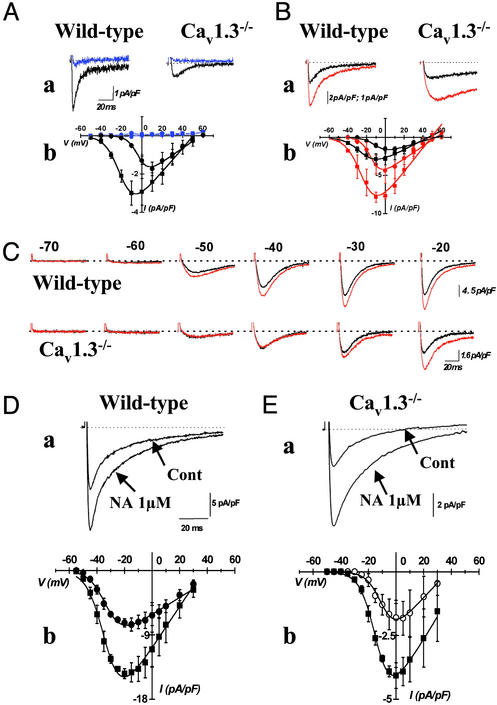
We next investigated the properties of ICa,L in SAN cells from wild-type and Cav1.3−/− mice (Fig. 2). In SAN cells from wild-type mice, ICa,L was activated from about −50 mV and peaked at −10 mV (Fig. 2A). In contrast, in SAN cells from Cav1.3−/− mice, ICa,L was undetectable negative to −20 mV, and peaked at +10 mV (Fig. 2B). All wild-type SAN cells showed ICa,L. In contrast, on 23 SAN cells isolated from 10 different Cav1.3−/− mice, 5 showed no detectable ICa,L. On average, ICa,L density was reduced by 69% in the Cav1.3−/− cell population from 3.32 ± 0.62 pA/pF (n = 20; wild-type) to 1.05 ± 0.21 pA/pF (n = 23, Cav1.3−/−) (P < 0.001, Fig. 2C). There was no difference in membrane capacitance between SAN cells from wild-type and Cav1.3−/− mice, indicating that gene inactivation did not affect cell size and morphology (32 ± 1.8 pF, n = 20, in wild-type and 30 ± 3 pF, n = 23, in Cav1.3−/− mice; P > 0.005). ICa,L current-to-voltage (I–V) relations showed that, in SAN cells from Cav1.3−/− mice, current activation was shifted to more positive voltages by 22 mV, when compared with wild-type SAN cells. Indeed, fitting I–V curves yielded parameter values for V0.5act of −3 ± 1 mV (n = 15) in Cav1.3−/−, and −25 ± 1 mV (n = 16), in wild-type SAN cells; (P < 0.0001, Fig. 2C). Application of 0.2 μM of isradipine similarly blocked ICa,L by 91 ± 6% (n = 4) in wild-type and by 98 ± 2% in Cav1.3−/− SAN cells (n = 4, P > 0.05, Fig. 3A). In contrast, application of 0.05 μM of isradipine, blocked ICa,L by 26 ± 8% (n = 4) in SAN cells from wild-type and by 72 ± 7% in Cav1.3−/− cells (n = 3, P < 0.05, data not shown), indicating that Cav1.3 gene inactivation abolished an ICa,L component that is less sensitive to dihydropyridines than ICa,L expressed in Cav1.3−/− cells. One micromolar of the Ca2+ channel activator (−)BayK8644 (BayK) increased ICa,L amplitude at the peak of the I–V relation by 263 ± 127% at −10 mV (n = 3) in wild-type and by 248 ± 82% at 0 mV in Cav1.3−/− SAN cells (n = 6, P > 0.05, Fig. 3B). BayK shifted ICa,L activation only in Cav1.3−/− cells (P > 0.05). Indeed, mean respective V0.5act values for ICa,L in cells from wild-type and Cav1.3−/− mice were −23 ± 1 and −8 ± 1 mV in control conditions, and −23 ± 2 and −14 ± 3 mV in the presence of BayK in wild-type and Cav1.3−/− cells. As a consequence, BayK application did not shift the apparent ICa,L threshold for activation. Indeed, in wild-type cells, BayK increased average ICa,L from 0.2 to 0.4 pA/pF at −50 mV, from 0.7 to 1.8 pA/pF at −40 mV and from 1.6 to 3.8 pA/pF at −30 mV. No measurable ICa,L was observed in Cav1.3−/− cells at a test potential of −50 and −40 mV even in the presence of BayK. Rather, detectable ICa,L was observed in Cav1.3−/− cells at −30 mV only in the presence of BayK (0.52 pA/pF). Mouse SAN cells also expressed ICa,T together with ICa,L. Because Cav1.3 gene inactivation significantly reduces ICa,L and because ICa,L and ICa,T may have partially overlapping voltage dependencies, we exploited the selective sensitivity of ICa,L to BayK to confirm the shift of the ICa,L threshold in SAN cells from Cav1.3−/− mice (Fig. 3C). From a holding potential of −80 mV, depolarizing steps activated both ICa,T and ICa,L. In SAN cells from wild-type mice, application of BayK increased the total ICa elicited from a test potential of −50 mV. In contrast, in Cav1.3−/− SAN cells, no BayK effect was evident for test potentials negative to −30 mV (Fig. 3C). This observation demonstrates that the positive shift on ICa,L activation observed in SAN cells from Cav1.3−/− mice was not a consequence of the reduced current density. Instead, it must reflect abolition of an ICa,L component activating from more negative voltages in the diastolic depolarization range. Application of 1 μM of the β-adrenergic receptor agonist noradrenaline stimulated ICa,L in SAN cells from both wild-type and Cav1.3−/− mice (Fig. 3 D and E). Indeed, noradrenaline enhanced ICa,L at the peak of I–V curve by 108 ± 32% (n = 5) in cells from wild-type and 104 ± 39% (n = 4) in cells from Cav1.3−/− mice (P > 0.05). Also, noradrenaline similarly shifted the V0.5act for ICa,L from −34 ± 2 to −40 ± 2 mV in cells from wild-type and from −9 ± 2 to −14 ± 2 mV in cells from Cav1.3−/− mice (P > 0.05, Fig. 3 Db and Eb). As a consequence, in the same group of cells tested, noradrenaline augmented ICa,L in the diastolic depolarization range from 0.02 ± 0.02 to 0.2 ± 0.08 pA/pF at −55 mV, from 0.33 ± 0.14 to 1.6 ± 0.4 pA/pF at −50 mV, from 1.5 ± 0.62 to 4 ± 1.5 pA/pF at −45 mV, and from 3.4 ± 1.2 to 10 ± 2 pA/pF at −40 mV. Again, even in the presence of noradrenaline, no measurable ICa,L was found in cells from Cav1.3−/− mice in this range of voltages, thus indicating that Cav1.3 gene inactivation prevents ICa,L activation in the diastolic depolarization even under β-adrenergic stimulation (Fig. 3Eb).
Figure 2.
Samples traces of ICa,L in SAN cells from wild-type (A) and homozygous Cav1.3−/− (B) mice. Recordings have been obtained by applying depolarizing voltage steps at variable voltages (Vt) indicated by arrows. Steps lasted 80 ms. Dotted lines indicate the zero current level. Holding potential (Vh) was −60 mV. (C) Averaged I–V relations of ICa,L in pacemaker cells from wild-type (filled circles) and Cav1.3−/− (open circles) mice, obtained from 21 independent experiments at a given voltage. Errors bars represent the SEM.
Figure 3.
(A) ICa,L is blocked by application of 0.2 μM isradipine in SAN cells from both mouse strains. ICa,L has been recorded from a holding potential of −60 mV, and sample traces corresponding to a test potential of −10 mV in SAN cells from wild-type and +10 mV in cells from Cav1.3 mice are shown. Traces (Aa) and I–V relations recorded in the presence of isradipine are displayed in blue (Aa). (Ab) Averaged I–V relations of ICa,L in cells from wild-type (filled squares) and Cav1.3−/− (filled circles) mice. (B) Up-regulation of ICa,L by 1 μM BayK. Experimental protocol is the same as in A. Sample traces (Ba) and I–V curves (Bb) obtained in the presence of BayK are displayed in red. Circles and squares as in A. (C) Application of BayK reveals threshold for activation of ICa,L in wild-type and Cav1.3−/− SAN cells. Stepping from a holding potential of −80 mV to the test potentials as indicated activate both ICa,T and ICa,L. Dotted lines indicate the zero current level. Traces shown are representative of five wild-type and four Cav1.3−/− cells recorded during three independent experiments. (D and E) Stimulation of ICa,L by 1 μM noradrenaline (NA) in SAN cells from wild-type (D) and Cav1.3−/− (E) mice. The effect of NA on sample traces is shown in a. Same voltage protocol and test potential as in A, for both wild-type and Cav1.3−/− cells, except the step duration, which lasted 100 mS. The effect of NA on averaged I–V relations is shown in b. Filled and open circles represent experimental data obtained in cells from wild-type and Cav1.3−/− cells, respectively. Filled squares represent experimental points obtained in the presence of NA.
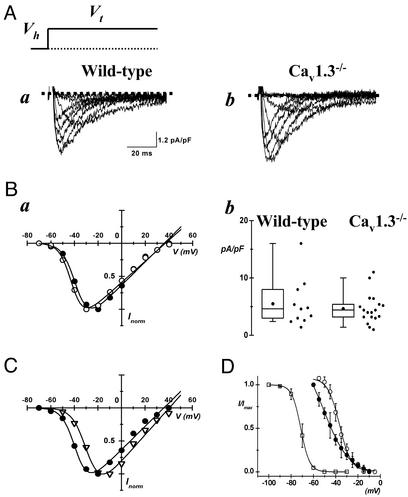
We also investigated whether inactivation of the Cav1.3 gene alters the functional expression of other ionic currents potentially involved in pacemaking. We observed no change on ICa,T (Fig. 4 A and B). The voltage dependence and the peak density of ICa,T were not significantly different in SAN cells from wild-type and Cav1.3−/− mice. Comparative values for the peak ICa,T density, the V0.5act and the slope factor s were 5.5 ± 1.2 pA/pF, −42 ± 2 mV and 3.7 ± 0.5 (n = 11) in SAN cells from wild-type and 4.6 ± 0.6 pA/pF, −42 ± 1 mV and 4.2 ± 0.6 (n = 18) in cells from Cav1.3−/− mice (P > 0.05, Fig. 4 Ba and Bb). Interestingly, the calculated V0.5act of the net ICa,L current component abolished by Cav1.3 gene inactivation was 12 mV more positive than ICa,T, (V0.5act = −28 mV, Fig. 4C), thus confirming that native ICa,T was the more negatively activated Ca2+ current in mouse SAN. Similarly to ICa,T, neither IKr or If densities showed significant change in SAN cells from wild-type and Cav1.3−/− mice. Respective values for deactivating IKr tails at −40 mV and steady-state If densities at −120 mV were 2.8 ± 0.3 pA/pF (n = 10) and 16.1 ± 2 pA/pF (n = 9) in cells from wild-type and 2.5 ± 0.4 pA/pF (n = 9) and 17.5 ± 3.6 pA/pF in cells from Cav1.3−/− mice (n = 9, P > 0.05 for each current tested, data not shown). These observations indicate that ICa,T, IKr, and If did not compensate for the lack of Cav1.3 channels in SAN cells. The positive shift of ICa,L voltage range for activation in SAN cells from Cav1.3−/− mice also allowed precise determination of the steady-state voltage dependence of inactivation for ICa,T, which was almost completely inactivated when the membrane potential was set to −60 mV (Fig. 4D). Comparison of the steady-state inactivation of ICa,L in the voltage range between −60 and −5 mV (Fig. 4D) showed that ICa,L in SAN cells from wild-type mice was more sensitive to inactivation than ICa,L of Cav1.3−/− cells. Indeed, the voltage for half-inactivation (V0.5inact) in cells from wild-type mice was −45 ± 2 mV (n = 7), about 10 mV more negative than the V0.5inact measured in Cav1.3−/− cells (−36 ± 2 mV, n = 5; P < 0.05).
Figure 4.
(A) Sample traces recorded from wild-type (Aa) and Cav1.3−/− (Ab) mice. Records of ICa,T are shown after subtraction of ICa,L measured from a Vh of −60 mV from total ICa recorded from a Vh of −80 mV (see Materials and Methods). (Ba) Superimposition of corresponding I–V relations from cells shown in A (filled circles, wild type; open circles, Cav1.3−/−). (Bb) Box histogram showing ICa,T peak densities in SAN cells from wild-type and Cav1.3−/− mice. Filled circles in boxes represent the mean density. Corresponding 25th and 75th percentiles define the box. The 5th and the 95th percentiles define whiskers. Experimental data points are shown on the right of the corresponding box. (C) Superimposition of ICa,T measured in wild-type cells (filled circles) and the difference ICa,L obtained by subtracting the averaged ICa,L measured in Cav1.3−/− cells from the total ICa,L of wild-type cells (open triangles). (D) Steady-state inactivation curves for ICa,T in SAN cells from Cav1.3−/− (open squares) and for ICa,L in wild-type (filled circles) and Cav1.3−/− (open circles) mice. Preconditioning steps were applied for 5 s at voltages indicated in the abscissa. ICa,T and ICa,L were then evoked at a test potential of −30 mV and 0 mV, respectively. Test pulses lasted 80 ms. ICa,L inactivation was investigated positive to −60 mV to avoid possible interfering ICa,T. Fitting inactivation curve yielded parameter values of −71 ± 2 mV and −3.4 ± 0.06 for V0.5inact and si, respectively, (n = 3) for ICa,T, −45 ± 2 mV and 10 ± 0.2 for wild-type ICa,L, and −36 ± 2 mV and 4.6 ± 0.4 for ICa,L measured in Cav1.3−/− cells.
Pacemaker Activity in SAN Cells of Cav1.3−/− Mice.
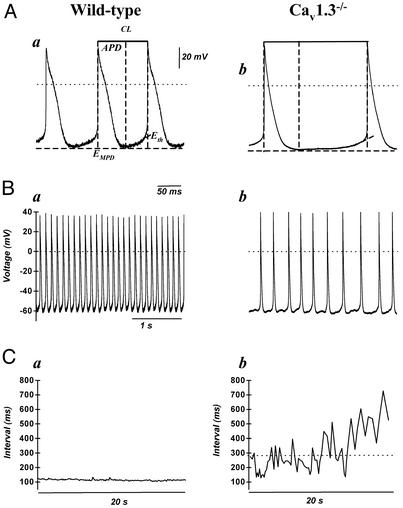
The prominent functional effect on ICa,L of Cav1.3 gene inactivation prompted us to investigate how the lack of the low-voltage activated ICa,L component affected pacemaking in SAN cells. Pacemaker activity in cells from wild-type mice was characterized by fast and regular pacing (Fig. 5 Aa and Ba). In contrast, slower and irregular pacing was found in all cells isolated from Cav1.3−/− mice (Fig. 5 Ab and Bb). Cellular arrhythmia was particularly evident in Cav1.3−/− cells during long intervals of recording as a substantial dispersion of consecutive measured cycle length intervals (compare in Fig. 5, Ca and Cb). Cells from Cav1.3−/− mice showed significantly longer cycle length, indicating that loss of Cav1.3 function induced negative chronotropism in SAN cells. On average, Cav1.3 gene inactivation prolonged the mean cycle length from 190 ± 22 ms (n = 6) in wild-type to 385 ± 84 ms (n = 5) in Cav1.3−/− SAN cells (P < 0.005, Fig. 5 Ba and Bb). Also, the action potential duration was significantly prolonged by Cav1.3−/− gene inactivation from 94 ± 10 ms in pacemaker cells from wild-type, to 151 ± 21 ms in Cav1.3−/− mice (P < 0.005). The maximum diastolic potential (EMDP) and the action potential threshold (Eth) were not significantly changed (EMDP, −59 ± 0.8 mV in wild-type and −54 ± 6 in Cav1.3−/− cells; Eth, −46 ± 5 mV and −56 ± 8 mV, respectively, Fig. 5A).
Figure 5.
(A) Representative sweeps of consecutive action potentials recorded in pacemaker cells from wild-type (Aa) and Cav1.3−/− (Ab) mice. The cycle length (CL) is defined as the time interval (in ms) between two consecutive action potentials. The action potential duration (APD) is defined as the interval between the maximum diastolic potential (EMDP) and the action potential threshold (Eth). (B) Cellular arrhythmia is evident as irregular cycle length duration in Cav1.3−/− cells (Bb) compared with wild-type cells (Ba). In A and B, dotted lines indicate the zero voltage level. Reference dotted lines used for calculations of the action potential parameters are also shown. (C) Examples of measurements of the cycle length show arrhythmia and negative chronotropism in Cav1.3−/− pacemaker cells. Wild-type cells display regular interval over 20-s-long recording periods. In contrast, the strong dispersion of measurements demonstrates erratic pacing rate in Cav1.3−/− cells (Cb). Furthermore, negative chronotropism is evident as an increased mean cycle length, as indicated by the dotted line.
Discussion
Our data constitute strong evidence that Cav1.3 Ca2+ channels play an important role in the generation of pacemaker activity. We show that Cav1.3 mRNA is expressed in the mouse SAN tissue and that Cav1.3 gene inactivation induces strong reduction of ICa,L in pacemaker cells and profoundly affects pacemaking.
The Cav1.2 subunit mRNA was also found in mouse SAN tissue (Fig. 1Ab). We thus propose that the Cav1.2 subunit underlies the residual ICa,L recorded in Cav1.3−/− pacemaker cells (see Fig. 2B). To this respect, our data are consistent with previous results showing that both Cav1.3 and Cav1.2 expression can be detected in the whole SAN region by in situ hybridization (14). However, the possibility that a currently unknown gene coding for ICa,L channels could also be involved in the generation of ICa,L in SAN cannot be excluded (15). Our data demonstrate that, also in SAN, Cav1.3 Ca2+ channels activate at more negative membrane voltages and show more negative steady-state inactivation with respect to Cav1.2 channels (Figs. 2 and 4). This finding is consistent with previous observations on recombinant Cav1.3 and Cav1.2 channels expressed in HEK293 cells (12, 16, 17). In our study, ICa,L activation significantly spans the diastolic depolarization voltage range in SAN cells of wild-type mice. Indeed, under voltage clamp conditions, the averaged peak ICa,L densities were 0.83 at −50 mV, 2.2 at −40 mV, and 3.7 pA/pF at −30 mV (Fig. 2C). Cav1.3 gene inactivation reduced ICa,L in the same voltage range by 100% at −50 and −40 mV and by 98% at −30 mV. In conclusion, our data demonstrate that Cav1.3 channels underlie a low-threshold ICa,L current component in mouse SAN, and that these channels are involved in the generation of pacemaker activity in the mouse by supplying inward current in the diastolic depolarization range. In our study, Cav1.3 gene inactivation induced negative chronotropism and cellular arrhythmia in SAN cells (Fig. 5).
Native Cav1.3 channels activated about 10 mV more positively than ICa,T (Fig. 4C). These channels are well available in the positive voltage range spanning the diastolic depolarization, thanks to their reduced sensitivity to steady-state voltage-dependent inactivation as compared with ICa,T, which showed very low availability at −60 mV (Fig. 4D).
The physiological role of Cav1.2 channels in pacemaking remains to be established. Due to the positive threshold for activation (Fig. 2C) and voltage-dependent inactivation (Fig. 4D) of ICa,L observed in Cav1.3−/− pacemaker cells, we suggest that Cav1.2 channels contribute to the generation of the upstroke phase of action potential, rather than to diastolic depolarization. It is thus tempting to suggest that the lack of expression of Cav1.3 channels in the ventricle may be reflective of the necessity to segregate the expression of ionic channels linked to pacemaker activity to the supraventricular stage and the conduction system (Fig. 1 A and B). Given that the lack of Cav1.3 channels did not stop pacemaker activity, our data also demonstrate that SAN pacemaking is the result of the activity of different ionic channels. To this respect, it is also possible that pacemaking in larger mammals such as humans could involve additional ionic channels that may not be expressed in the mouse. For example, the slow component of the delayed rectifier (IKs) has been reported to play a significant role in pacemaking in the pig heart (18). Understanding the ionic basis of pacemaking may yield important indications in the development of new drugs that specifically influence heart rate without affecting cardiac inotropism. Because Cav1.3 channels are not expressed in the human ventricle (Fig. 1B), our results suggest that Cav1.3 channels might constitute suitable candidate target for such drugs. This kind of chronotropic selectivity could be particularly important in the treatment of ischemic heart disease (19).
Acknowledgments
We thank Anne Cohen-Solal, Pierre Fontanaud, and Patrick Atger for their excellent technical assistance, and Jean-Charles Hoda for help with Cav1.3 primer. We are grateful to Dr. Wayne Giles and Robert Clark for helpful discussions and encouragement. We thank Dr. Philippe Lory for critical reading of the manuscript. We also thank three anonymous referees. This work was supported by the Fondation de France, the Action Concertée Incitative in Developmental Biology of the French Ministry for Education, the Austrian Science Fund (P14820), the Österreichische Nationalbank, and the European Community Grant Program (HPRN-CT-2000-00082). M.E.M. is the recipient of a postdoctoral fellowship by the Lefoulon-Delalande Foundation.
Abbreviations
- SAN
sino-atrial node
- ICa,L
L-type Ca2+ current
- ICa,T
T-type Ca2+ current
- BayK
Ca2+ channel activator (−)BayK8644
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Weidmann S. In: The Slow Inward Current and Cardiac Arrhythmias. Zipes D P, Bailey J C, Elharrar V, editors. The Hague, The Netherlands: Nijoff; 1980. pp. 3–9. [Google Scholar]
- 2.DiFrancesco D. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 3.Noma A. Jpn Heart J. 1996;37:673–682. doi: 10.1536/ihj.37.673. [DOI] [PubMed] [Google Scholar]
- 4.Hagiwara N, Irisawa H, Kameyama M. J Physiol (London) 1988;395:233–253. doi: 10.1113/jphysiol.1988.sp016916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangoni M E, Nargeot J. Cardiovasc Res. 2001;52:51–64. doi: 10.1016/s0008-6363(01)00370-4. [DOI] [PubMed] [Google Scholar]
- 6.Irisawa H, Brown H F, Giles W. Physiol Rev. 1993;73:197–227. doi: 10.1152/physrev.1993.73.1.197. [DOI] [PubMed] [Google Scholar]
- 7.Verheijck E E, van Ginneken A C, Wilders R, Bouman L N. Am J Physiol. 1999;276:H1064–H1077. doi: 10.1152/ajpheart.1999.276.3.H1064. [DOI] [PubMed] [Google Scholar]
- 8.Kodama I, Nikmaram M R, Boyett M R, Suzuki R, Honjo H, Owen J M. Am J Physiol. 1997;272:H2793–H2806. doi: 10.1152/ajpheart.1997.272.6.H2793. [DOI] [PubMed] [Google Scholar]
- 9.Lande G, Demolombe S, Bammert A, Moorman A F, Charpentier F, Escande D. Cardiovasc Res. 2001;50:328–334. doi: 10.1016/s0008-6363(01)00232-2. [DOI] [PubMed] [Google Scholar]
- 10.Striessnig J. Cell Physiol Biochem. 1999;9:242–269. doi: 10.1159/000016320. [DOI] [PubMed] [Google Scholar]
- 11.Platzer J, Engel J, Schrott-Fischer A, Stephan K, Bova S, Chen H, Zheng H, Striessnig J. Cell. 2000;102:89–97. doi: 10.1016/s0092-8674(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Xu Y, Song H, Rodriguez J, Tuteja D, Namkung Y, Shin H S, Chiamvimonvat N. Circ Res. 2002;90:981–987. doi: 10.1161/01.res.0000018003.14304.e2. [DOI] [PubMed] [Google Scholar]
- 13.Hamill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflügers Arch Eur J Physiol. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 14.Bohn G, Moosmang S, Conrad H, Ludwig A, Hofmann F, Klugbauer N. FEBS Lett. 2000;481:73–76. doi: 10.1016/s0014-5793(00)01979-7. [DOI] [PubMed] [Google Scholar]
- 15.Seisenberger C, Specht V, Welling A, Platzer J, Pfeifer A, Kuhbandner S, Striessnig J, Klugbauer N, Feil R, Hofmann F. J Biol Chem. 2000;275:39193–39199. doi: 10.1074/jbc.M006467200. [DOI] [PubMed] [Google Scholar]
- 16.Xu W, Lipscombe D. J Neurosci. 2001;21:5944–5951. doi: 10.1523/JNEUROSCI.21-16-05944.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koschak A, Reimer D, Huber I, Grabner M, Glossmann H, Engel J, Striessnig J. J Biol Chem. 2001;276:22100–22106. doi: 10.1074/jbc.M101469200. [DOI] [PubMed] [Google Scholar]
- 18.Ono K, Shibata S, Iijima T. J Physiol (London) 2000;524:51–62. doi: 10.1111/j.1469-7793.2000.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Striessnig J. Lancet. 2001;357:1294. doi: 10.1016/S0140-6736(00)04440-8. (lett.). [DOI] [PubMed] [Google Scholar]