Abstract
The shoot apical meristem of angiosperms shows a highly conserved cellular architecture in which a change of cell division orientation correlates with early events of leaf initiation. However, the causal role of this altered cellular parameter in leaf formation is debatable. We have used the dynamin-like protein phragmoplastin as a tool to modify the pattern of cell division within the apical meristem. Taking a microinduction approach, we show that local alteration in cell division orientation is not sufficient to induce morphogenesis in the meristem. Surprisingly, an altered cell division pattern did lead to an altered pattern of expression of genes implicated in aspects of leaf formation. Our data identify inducible expression of phragmoplastin as a tool to manipulate cell division pattern. Furthermore, they indicate that a mechanism exists by which cells in the meristem can respond at the level of gene expression to altered parameters of cell division. These data are discussed in the context of a model linking leaf morphogenesis and differentiation.
Cell division pattern within the apical meristem of angiosperms is highly conserved, leading to the classical definition of outer tunica layers (in which cell division is restricted to an anticlinal orientation) surrounding an inner corpus (in which cell division orientation is more random) (1). Despite this conservation of structure, the functional significance of the tunica/corpus organization remains unclear.
Although earlier works made detailed descriptions of apical meristems and used the number of tunica layers as a diagnostic for the phase of meristem growth and as an indicator of meristem evolution (2), the most compelling interpretation of cell division pattern in the meristem has come from the observed correlation with events of leaf initiation. Thus, numerous studies report the occurrence of nonanticlinal divisions in the tunica layers as indicative of the site of presumptive leaf formation (3). This altered pattern of cell division may also be accompanied by an increase in cell proliferation (4). However, whether the observed changes in cell division pattern are causally involved in leaf initiation remains debatable. For example, although mutants in which cell division pattern is disrupted can still generate morphologically normal leaves (suggesting that cell division orientation is not important during organogenesis) (5), it is possible that in such mutants local gradients of cell division, which are required for leaf initiation (6), still occur.
We recently developed a microinduction system that allows us to manipulate gene expression in intact apices to a spatial resolution of less than a meristem (7). Using this system, we were able to promote cell division locally within the meristem and show that no morphogenesis had occurred (8). In contrast, local expression of a gene encoding expansin (an effector of cell wall extensibility) led to morphogenesis and leaf formation (7). These data support the hypothesis that morphogenesis in the meristem occurs via a cell division-independent mechanism (9).
However, although our manipulations of the cell cycle led to a transient local increase in cell proliferation, the extra cells produced remained in their appropriate layers; i.e., the tunica/corpus organization was not obviously disrupted. Thus, the potential role of altered cell division orientation as a causal agent in leaf initiation was not directly addressed. We set out to test this hypothesis by using the microinduction approach to overexpress locally a gene product expected to disrupt the normal pattern of cell division in the meristem.
The orientation of cell division in plant cells is controlled by a complex series of events, the molecular basis of which is only now beginning to be unraveled (10). A key event is the formation of the phragmoplast, an agglomeration of vesicles and cytoskeletal elements that dictates the site of synthesis of the new cell plate within the parent cell. Phragmoplast formation and function depend on the appropriate activity of a number of proteins, including a dynamin-like protein termed phragmoplastin (11). Phragmoplastin was first identified in soybean and is involved in the delivery and fusion of vesicles within the phragmoplast. Overexpression of phragmoplastin leads to the disruption of cytokinesis in tobacco BY2 cells so that, although cell-plate formation occurs, the orientation of the new cell wall is oblique rather than perpendicular (12). Constitutive overexpression of phragmoplastin in transgenic tobacco leads to abnormal planes of cell division in embryos and roots and to premature termination of growth (13). These observations suggested to us that phragmoplastin could be used as a tool to disrupt cell division orientation in tobacco and that inducible expression of the gene might provide a means to overcome the lethality associated with constitutive overexpression.
In this article, we report on the generation of transgenic tobacco plants in which expression of a gene encoding soybean phragmoplastin is under chemical-inducible transcriptional regulation. Using a microinduction approach, we show that local expression of the phragmoplastin gene leads to local alteration of cell division pattern in the apical meristem. No change in meristem morphogenesis was observed, indicating that altered cell division orientation is not sufficient for leaf initiation. Surprisingly, however, the induction of altered cell division orientation or proliferation within the meristem led to an altered expression pattern of marker genes for meristem function. These data indicate that, in addition to specific gene products influencing cell proliferation in the meristem, a mechanism exists by which altered cell division can itself feed back into the expression of genes encoding developmental regulators. These data are discussed in the context of a mechanism linking morphogenesis and differentiation.
Materials and Methods
Plant Material and Transformation.
R7 Nicotiana tabacum seedlings (a gift from A. Jones, University of North Carolina, Chapel Hill) were transformed (7). Regenerants were grown in a greenhouse and F1 seeds were collected for analysis. For microinduction experiments, plants were grown on soil in a growth chamber (16 h light at 24°C/8 h dark at 20°C cycle) or on half-strength Murashige and Skoog (MS) medium (pH 5.6), 1% (wt/vol) agar (16 h light/8 h dark cycle at 24°C, 100 μmol⋅m−2⋅s−1). For RNA blot and protein analyses, seedlings were grown on MS medium with or without anhydrotetracycline (Ahtet) at the concentrations and for the times given in Results.
DNA Manipulation.
The SDL12 coding sequence (11) was inserted as a transcriptional fusion into the SmaI/SalI sites of the pBinHyg-Tx vector (14). The resultant clone, pBinHyg-Tx-SDL, was transformed into R7 tobacco plants. All DNA manipulations were by standard techniques (15).
Microinduction.
Microinductions were performed as described (7) by using Ahtet concentrations from 0.1 to 10 mg/ml. Controls were performed by using DMSO/lanolin/paraffin paste without Ahtet. After manipulation, apices were grown on half-strength Murashige and Skoog (MS) medium (pH 5.6), 1% (wt/vol) agar in a growth chamber (16 h light/8 h dark cycle at 24°C, 100 μmol⋅m−2⋅s−1).
RNA and Protein Analysis.
For blots, total RNA was extracted from 4-week-old seedlings by using RNeasy columns (Qiagen, Valencia, CA). Gel electrophoresis, blotting, and hybridization with a digoxigenin (DIG)-labeled riboprobe for SDL12 was according to the manufacturer's instructions (Roche Diagnostics). In situ hybridization was as described (7) by using DIG-labeled sense and antisense riboprobes for SDL12. Western blot analysis was by standard protocols with a horseradish peroxidase-linked second antibody and enhanced chemiluminescence visualization, as described by the manufacturer (Roche Diagnostics). Fifty micrograms of protein from leaf microsomal preparations (16) was loaded per lane.
Results
Generation of Transgenic Plants with Inducible Phragmoplastin Gene Expression.
A cDNA encoding a soybean phragmoplastin (SDL12) was cloned into the pBinHyg-Tx vector and then introduced by Agrobacterium-mediated transformation into R7 tobacco plants previously engineered to overexpress the tetracycline repressor protein. In the presence of the repressor protein, transcription of the SDL12 sequence is repressed until the addition of Ahtet. Seeds were collected from 10 independent transformants and each line was tested by using RT-PCR for the accumulation of SDL12 transcripts with or without prior induction with Ahtet. This process led to the identification of three lines showing accumulation of SDL12 transcripts after induction with undetectable expression under noninducing conditions (data not shown). Two of these lines (Tet∷SDL12-1 and Tet∷SDL12-2) were taken for further characterization by Northern and Western blot analysis, as shown in Fig. 1.
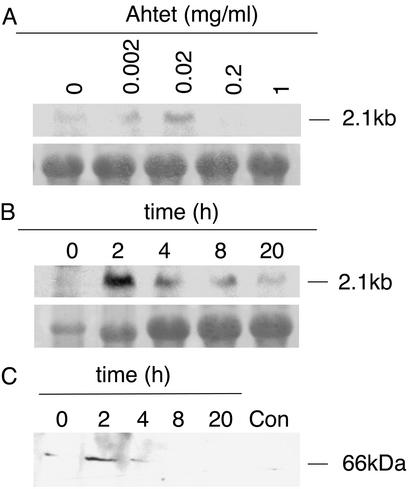
Figure 1.
Tetracycline-inducible phragmoplastin gene expression. (A) RNA blot analysis of total RNA (10 μg) from transgenic Tet∷SDL12-1 tobacco plants. Plants were treated for 2 h with the concentration of Ahtet indicated before RNA extraction. Blots were hybridized with a digoxigenin-labeled riboprobe for SDL12. Methylene blue staining of 25S rRNA is shown as a loading control. (B) Time course of SDL12 transcript accumulation by RNA blot analysis after treatment of seedlings with 0.02 mg/ml Ahtet for the times indicated. Hybridization was as in A with 10 μg of RNA per lane. (C) Analysis of phragmoplastin protein accumulation. Seedlings of Tet∷SDL12-1 plants were treated for the times indicated with 0.02 mg/ml Ahtet before protein extraction. Protein was also extracted from Tet∷SDL12-1 seedlings incubated for 20 h in buffer without Ahtet (Con). Blots (50 μg of protein per lane) were incubated with an antibody against soybean SDL before visualization of signal.
Treatment of tissue with Ahtet at concentrations of 0.002 or 0.02 mg/ml led to the accumulation of SDL12 transcripts after 2 h (Fig. 1A), higher Ahtet concentrations being supraoptimal. A low signal was occasionally observed in untreated controls, most probably reflecting hybridization with an endogenous phragmoplastin transcript (12), although the possibility of a low level of leaky expression of the SDL12 gene cannot be excluded. Time-course analysis of phragmoplastin transcript expression after Ahtet treatment indicated a rapid but transient accumulation of the SDL12 transcript, with a maximum level occurring after 2 h of induction (Fig. 1B). This transient accumulation of the SDL12 transcript was consistent with the time course of phragmoplastin protein accumulation observed in Western blots by using an antibody raised against the SDL protein (a gift from D. Verma, Ohio State University, Columbus). As shown in Fig. 1C, phragmoplastin protein levels rose to a high level at 2 h after Ahtet treatment and decreased rapidly so that they were barely detectable 8 h after induction. By 20 h after induction the detectable level of phragmoplastin protein was similar in induced and noninduced tissue. This low background signal probably reflects cross-reactivity of the antibody raised against the soybean protein with an endogenous tobacco phragmoplastin (12).
Local Induction of Phragmoplastin Gene Expression Leads to an Altered Pattern of Cell Division in the Shoot Apical Meristem.
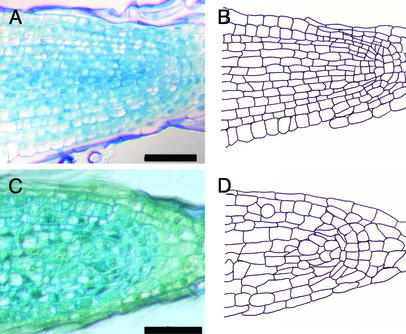
To investigate whether phragmoplastin expression could be used as a tool to manipulate cell division orientation in the shoot apical meristem, we performed a series of microinduction experiments in which Ahtet-impregnated lanolin was positioned onto the I2 position of meristems from Tet∷SDL12-1 plants. After localized induction, overall meristem morphology was unchanged compared with mock-treated controls or uninduced Tet∷SDL12-1 apices. Thus, as shown in Fig. 2, induced meristems of Tet∷SDL12-1 plants remained flattened domes similar to WT untreated controls (Fig. 2 A and B). However, histological analysis of the induced apices revealed an altered pattern of cell division. In untreated control tissue, cell division in the outer tunica layers was maintained in an anticlinal orientation at the I2 position. This layered organization is shown in more detail in Fig. 2C and as a cellular outline in Fig. 2E. After local induction of phragmoplastin gene expression, nonanticlinal cell divisions were observed in the tunica layers. This is shown in Fig. 2D and as a cellular outline in Fig. 2F. The differences in cellular organization between control meristems and tissue in which phragmoplastin expression has been induced is highlighted in Fig. 2 E and F by color coding of cells that share an anticlinal cell wall. In control meristems, this color coding highlights two outer cell layers (Fig. 2E), whereas in the induced meristem shown in Fig. 2F nonanticlinal division orientations are apparent in the outer layers.
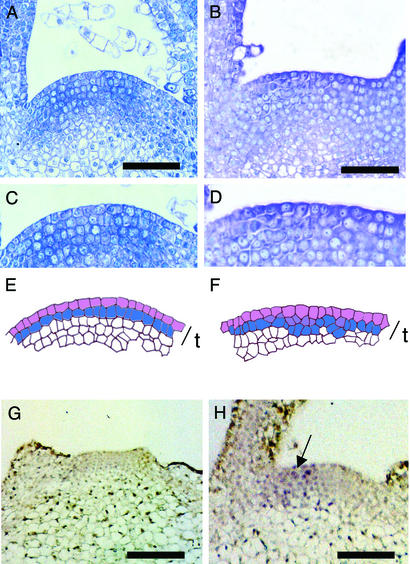
Figure 2.
Induction of phragmoplastin gene expression leads to altered cell division pattern in the apical meristem. (A) Longitudinal section through the meristem of a control apex. (B) Longitudinal section through the meristem of a Tet∷SDL12-1 plant in which microinduction has been performed. (C) Detail from the meristem shown in A. (D) Detail from the meristem shown in B. (E) Cellular outline of the meristem shown in C. The outer cells of the tunica (t) have been colored to highlight the layered cellular architecture. Cells connected by anticlinal cell walls are indicated by separate colors. (F) Cellular outline of the meristem shown in D. The outer cells of the tunica (t) have been colored to highlight the occurrence of nonanticlinal cell divisions at various positions within the tunica. Cells connected by anticlinal cell walls are indicated by separate colors. (G) In situ hybridization using a sense probe for SDL12 of a Tet∷SDL12-1 apex induced at the I2 position with Ahtet. No signal is visible. (H) In situ localization as in G except an antisense probe for SDL12 was used. Signal (blue/violet) is seen on one flank of the meristem (arrow). (Bars in A and B = 50 μm; bars in G and H = 100 μm.)
To confirm that the microinduction experiments performed led to a local induction of expression of the phragmoplastin gene, in situ hybridizations were performed by using an antisense probe for SDL12. Control hybridizations using an antisense probe on noninduced tissue or a sense probe on induced tissue did not reveal any signal above background (Fig. 2G). After microinduction of Tet∷SDL12-1 apices, SDL12 transcripts accumulated on one flank of the meristem (Fig. 2H).
Microinduction of Phragmoplastin Leads to an Altered Pattern of Marker Gene Expression in the Meristem.
Microinduction of SDL12 expression in the meristem did not lead to any obvious change in meristem function in terms of growth and morphogenesis. However, in situ hybridizations of such induced apices indicated an altered pattern of marker gene expression in response to the induction of phragmoplastin gene expression.
NTH15 encodes a homeobox transcription factor of the KNOX family (17). Functional expression and sequence analysis indicate that, of the tobacco KNOX genes described, it is most similar to STM from Arabidopsis. STM plays a key role in meristem initiation and maintenance and shows a distinct pattern of accumulation in which transcripts accumulate throughout the meristem but are excluded from the I1 site of presumptive leaf formation (18). In tobacco, NTH15 shows a similar pattern of expression (Fig. 3A and ref. 17). After local induction of phragmoplastin gene expression, a dramatic change in NTH15 mRNA expression pattern occurred, with a decrease in transcript level observed at the I2 site of Ahtet induction (Fig. 3B). To investigate whether this change in NTH15 transcript accumulation was specific to our manipulation of phragmoplastin gene expression, we performed similar in situ hybridizations on apices in which genes encoding other proteins had been locally induced with Ahtet. Our previous work showed that local overexpression of the cyclin Nt;CycA3;2 led to local cell proliferation in the meristem without morphogenesis, whereas local induction of the expansin Cs;Exp1 led to morphogenesis accompanied by appropriate cell division (7, 8). Local induction of Nt;CycA3;2 and Cs;Exp1 gene expression at the I2 position led to a decreased accumulation of NTH15 transcripts at this site, resulting in meristems in which NTH15 transcript accumulation was down-regulated both at the I1 and I2 positions (Fig. 3 C and D).
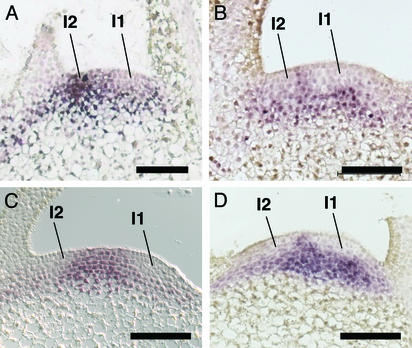
Figure 3.
Microinduction of phragmoplastin, cyclinA, and expansin gene expression leads to an altered pattern of NTH15 transcript accumulation. (A) Control apex hybridized with an antisense probe for NTH15. Transcripts do not accumulate at the presumptive site of leaf initiation, I1. (B) Longitudinal section of an apex from a Tet∷SDL12-1 plant in which microinduction had been performed 16 h previously at the I2 position, then hybridization had been performed as in A. NTH15 transcripts do not accumulate at the I1 or I2 position. (C) Conditions were as in B except that microinduction was performed on a Tet∷Nt;CycA3;2 apex. (D) Conditions were as in B except that microinduction was performed on a Tet∷Cs;Exp1 apex. (Bars = 100 μm.)
A paradigm has emerged in which a key function of STM-like KNOX genes is to repress the expression of PHANTASTICA-like myb transcription factors implicated in various aspects of early leaf development in a number of species (19, 20). Using a sequence in the GenBank database (accession no. AJ006181), we cloned a tobacco PHANTASTICA-like cDNA (NTPHAN) and used it as a probe in a series of in situ hybridizations of apices in which phragmoplastin gene expression had been locally induced in the meristem. Analysis of control apices indicated that the expression pattern of the NTPHAN gene in tobacco was similar to that described for similar genes in other species (20). Thus, transcripts accumulated within the meristem at the I1 site of presumptive leaf formation (Fig. 4A). Serial longitudinal sections through control apices confirmed a low to undetectable level of accumulation of NTPHAN transcript in the I2 region of control apices (Fig. 4B). After local induction of phragmoplastin gene expression at the I2 position, an accumulation of NTPHAN transcripts occurred at this site (Fig. 4C), equivalent to the region of NTH15 gene repression (Fig. 3B). This led to a high level of NTPHAN transcripts across the meristem, i.e., at both I1 and I2 positions, as revealed by in situ hybridizations of cross sections through induced Tet∷SDL12-1 meristems (Fig. 4 D and F). That NTPHAN transcript accumulation at the I2 position correlated with the position of induced phragmoplastin gene expression was indicated by the observation of an abnormal pattern of cell division at this site (Fig. 4E).
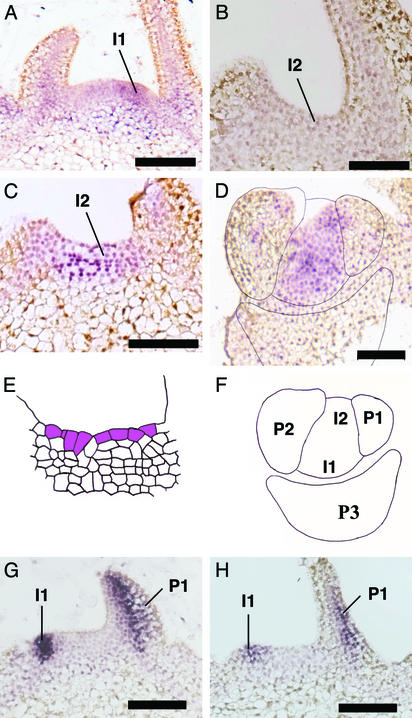
Figure 4.
Microinduction of phragmoplastin gene expression leads to an altered pattern of NTPHAN transcript accumulation. (A) Control apex hybridized with an antisense probe for NTPHAN. Transcripts accumulate at the I1 site of presumptive leaf formation. (B) Longitudinal section through the I2 position of a control apex, hybridized as in A. No signal is visible. (C) Longitudinal section through the I2 position of a Tet∷SDL12-1 apex in which microinduction was performed 16 h previously at the I2 position. Hybridization was as in A. Signal is visible. (D) Cross section through the apex of a Tet∷SDL12-1 plant treated and hybridized as in C. Signal is visible at both the I1 and I2 positions indicated in F. (E) Cellular outline of the section shown in C. Cells in the outer layer sharing anticlinal walls have been highlighted. (F) Outline of the section in D to indicate the I1 and I2 positions and primordia P3, P2, and P1. (G) Control apex hybridized with an antisense probe for NBYABBY. Signal is seen at the I1 site of presumptive leaf formation and on the abaxial side of the leaf primordium P1. (H) Longitudinal section through the apex of a Tet∷SDL12-1 plant in which microinduction was performed 16 h previously at the I2 position. The section was hybridized as in G. (Bars = 100 μm.)
The fixation of an adaxial/abaxial axis is an important step in leaf development (20). A family of YABBY transcription factors have been implicated in this process and transcripts encoding NBYABBY (from Nicotiana benthamiana) have been shown to accumulate on the abaxial side of the leaf early in development (21). We investigated the use of the NBYABBY probe as a marker for leaf adaxial/abaxial polarity in N. tabacum. The results indicated that the NBYABBY cDNA cross-hybridizes with an endogenous sequence in N. tabacum that defines the abaxial side of developing leaves, with signal first becoming visible as the primordium bulge is formed on the surface of the meristem (Fig. 4G). After local induction of phragmoplastin gene expression at the I2 position, no change in NBYABBY hybridization pattern was observed (Fig. 4H). Similarly, in situ hybridizations with a probe for transcripts encoding the small subunit of ribulose bisphosphate carboxylase (a marker for leaf differentiation) did not reveal any difference in pattern between induced and noninduced meristems (data not shown).
Induction of Phragmoplastin Gene Expression Leads to Abnormal Patterns of Cell Division in the Root Apex.
In addition to microinduction, we performed a series of experiments in which phragmoplastin gene expression was induced by the supply of Ahtet to the medium in which Tet∷SDL12 seedlings were growing. At the Ahtet concentrations used, these inductions led to no overt difference in seedling growth compared with similarly treated Tet∷GUS control seedlings (data not shown). However, analysis of the root apices of such induced Tet∷SDL12 seedlings revealed the occurrence of abnormal patterns of cell divisions compared with uninduced control seedlings. Thus, in control root apices a classic cellular architecture was observed, consisting of files of cells emanating from a distally localized organizer region (Fig. 5 A and B). This structure was disrupted in induced Tet∷SDL12-1 seedlings, leading to the occurrence of files in which cellular order was abnormal. This involved both changes in the pattern of cell division and the range of cell sizes within the root apex compared with uninduced tissue (Fig. 5 C and D). However, overall root morphology and growth was similar to that observed in control seedlings.
Figure 5.
Induction of phragmoplastin gene expression in the root leads to an abnormal pattern of cell division. (A) Longitudinal section through the root apex of a control plant. (B) Cellular outline of the section in A. Ordered cell files are observed throughout the apex. (C) Longitudinal section through the root apex of a Tet∷SDL12-1 seedling treated with Ahtet (0.02 mg/ml) for 7 days. (D) Cellular outline of section in C. Abnormal cell division patterns are observed in various regions of the apex. (Bars = 100 μm.)
Discussion
Phragmoplastin as a Tool to Manipulate Cell Division Orientation.
Several gene products have now been identified that influence cell division orientation (10). As well as providing insights into the mechanism of cytokinesis, these proteins provide potential tools for the manipulation of cell division. The data reported here indicate that the dynamin-like protein phragmoplastin can be used as such a tool. Previous data had already indicated that overexpression of phragmoplastin disrupted cytokinesis and led to abnormal orientation of new cell walls (12). However, constitutive expression of the protein was found to be deleterious for plant growth (13). We extend these studies by showing that inducible expression of phragmoplastin can be used to modify cell division pattern without plant lethality. The changes in cell division pattern that we observed in the root are comparable with (but less severe than) those described after constitutive expression of phragmoplastin (13) and are consistent with the proposed function of the protein in vesicle fusion during phragmoplast formation (12). Presumably, overexpression of phragmoplastin disrupts the process of cell-plate formation, leading to altered cell wall orientation (12). Whether regulation of phragmoplastin gene expression represents an endogenous control point for cytokinesis is unknown, but the data reported here indicate that, in conjunction with the microinduction approach, it can be used as a tool to manipulate cell division pattern in intact tissue.
Cell Division Pattern in the Meristem and Leaf Initiation.
The idea that the observed alteration in cell division pattern at the site of presumptive leaf initiation plays a causal role in this process remains embedded in reviews and textbooks. However, a body of evidence now exists that indicates that this view is no longer tenable (5, 22, 23). Our own previous work has contributed to this discussion by showing that local increase in cell division frequency does not lead to altered morphogenesis at the meristem (8). In addition, data from the manipulation of expansin gene expression provide causal data supporting an alternative mechanism in which control of cell wall extensibility plays a key role in the process of leaf initiation (7). However, these previous experiments did not directly approach the role of altered orientation of cell division in leaf initiation and the hypothesis that local alteration of this cellular parameter within the context of the otherwise highly ordered tunica layer of the meristem might provide the driving force or trigger for leaf initiation. The results reported here indicate that this is not the case and show that the occurrence of nonanticlinal cell divisions within the meristem is not sufficient to induce leaf formation. However, our data do indicate that an altered cell division pattern can influence gene expression within the meristem. As discussed below, such a linkage between division pattern and gene expression might provide a mechanism by which morphogenesis and differentiation are coordinated.
Cell Division Pattern in the Meristem and Gene Expression.
A series of powerful and elegant studies has begun to unravel the complex procedure by which an apical meristem is maintained throughout the lifetime of a plant despite repeated loss of mass by the regular generation of leaf primordia. Thus, interactions between domains within the meristem defined by gene products such as WUS, CLV3, and CLV1 are necessary for the establishment and maintenance of the meristem structure (24). These gene products (or downstream elements) are proposed to impinge on elements of tissue growth and cell division to maintain an appropriate meristem size and cell number to allow for continued meristem function. Superimposed on this molecular engine for meristem maintenance is a mechanism by which particular regions of the meristem become determined for leaf formation. How this occurs is still unclear, but elements of hormone action, cell wall extensibility, and transcription factor activity have all been implicated (7, 25, 26). Understanding how these different levels of meristem maintenance and function interact and are coordinated is a major target for research in plant development.
The data presented here provide the insight that altered parameters of cell division frequency and orientation, as well as cell wall extensibility, are capable of feeding back into cells to alter the expression of transcription factors implicated in basic aspects of leaf development. Thus, two independent tools used to manipulate different aspects of the cell division machinery, as well as modulation of expansin gene expression, led to a similar response in terms of decreased accumulation of transcripts encoding the KNOX homeobox transcription factor NTH15. Concomitant with a decrease in NTH15 mRNA accumulation was an accumulation of NTPHAN transcripts. These data are both consistent with the previously proposed function of STM-like gene products in repressing PHANTASTICA-like gene expression (19) and indicate that the observed decrease of NTH15 transcript level after our manipulations did not simply reflect a general decrease in transcriptional activity in the induced tissue. However, despite the decrease in NTH15 transcript levels (and associated increase in NTPHAN mRNA level) after modulation of phragmoplastin and cyclinA gene expression, no further downstream elements of leaf formation were observed. Thus, no ectopic expression of marker genes for leaf adaxial/abaxial polarity or leaf differentiation was detected, nor was any morphogenesis observed. These observations indicate that altered expression of STM-like and PHANTASTICA-like genes is not sufficient for leaf formation and are consistent with the idea that elements of leaf differentiation are somehow coupled with the actual formation of an organ, i.e., with morphogenesis.
How altered cell division frequency/orientation influences transcription factor gene expression is open to speculation, but it provides a potential mechanism by which morphogenesis might feed back into the differentiation of the cells incorporated within a structure undergoing change in shape, as discussed below.
A Mechanism Linking Morphogenesis and Differentiation.
Leaf initiation involves the formation of an outgrowth on the surface of the meristem. Because growth and division in the meristem are tightly coupled (9), such outgrowths on the meristem must lead to an altered pattern of cell division compared with the situation before outgrowth formation. If a mechanism were to exist by which gene expression within a group of cells was sensitive to any change in cell division within that group, then a group of cells could essentially inform itself that morphogenesis had occurred. Our data provide evidence that such a mechanism exists; i.e., we observed that altered cell division pattern in a group of cells in the meristem led to altered expression of a transcription factor implicated in leaf development. Bearing in mind recent data implicating the supracellular movement of RNA within the meristem (27), an influence of altered cell division pattern on plasmodesmatal function represents an enticing potential mechanism by which cell division pattern could influence gene expression.
Acknowledgments
We thank T. Foster (Horticulture and Food Research Institute, Mount Albert, New Zealand), Christiane Gatz (University of Goetingen, Goetingen, Germany), A. Jones (University of North Carolina), W.-H. Shen (Centre National de la Recherche Scientifique, Strasbourg, France), and D. Verma (Ohio State University) for providing vectors, cDNAs, antibodies, and seeds used in this work; N. Amrhein (Eidgenössische Technische Hochschule, Zurich) for providing laboratory space and facilities; and members of group Fleming for useful discussions. A.F. is supported by a START Fellowship from the Swiss National Science Foundation, which provided Grants 31-49337 and 31-63570.
Abbreviation
- Ahtet
anhydrotetracycline
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Steeves T A, Sussex I M. Patterns in Plant Development. Cambridge, U.K.: Cambridge Univ. Press; 1989. [Google Scholar]
- 2.Gifford E M, Corson G E. Bot Rev. 1971;37:143–229. [Google Scholar]
- 3.Lyndon R F. Plant Development: The Cellular Basis. London: Unwin Hyman; 1990. [Google Scholar]
- 4.Laufs P, Grandjean O, Jonak C, Kiêu K, Traas J. Plant Cell. 1998;10:1375–1389. doi: 10.1105/tpc.10.8.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith L G, Hake S, Sylvester A W. Development (Cambridge, UK) 1996;122:481–489. doi: 10.1242/dev.122.2.481. [DOI] [PubMed] [Google Scholar]
- 6.Meyerowitz E M. Curr Opin Genet Dev. 1996;6:475–479. doi: 10.1016/s0959-437x(96)80070-0. [DOI] [PubMed] [Google Scholar]
- 7.Pien S, Wyrzykowska J, McQueen-Mason S, Smart C, Fleming A. Proc Natl Acad Sci USA. 2001;98:11812–11817. doi: 10.1073/pnas.191380498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyrzykowska J, Pien S, Shen W-H, Fleming A J. Development (Cambridge, UK) 2002;129:957–964. doi: 10.1242/dev.129.4.957. [DOI] [PubMed] [Google Scholar]
- 9.Fleming A J. Planta. 2002;216:17–22. doi: 10.1007/s00425-002-0864-8. [DOI] [PubMed] [Google Scholar]
- 10.Verma D P S. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:751–784. doi: 10.1146/annurev.arplant.52.1.751. [DOI] [PubMed] [Google Scholar]
- 11.Gu X, Verma D P S. EMBO J. 1996;15:695–704. [PMC free article] [PubMed] [Google Scholar]
- 12.Gu X, Verma D P S. Plant Cell. 1997;9:157–169. doi: 10.1105/tpc.9.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisler-Lee C J, Hong Z L, Verma D P S. Plant Sci. 2002;163:33–42. [Google Scholar]
- 14.Gatz C, Frohbergand C, Wendenburg R. Plant J. 1992;2:397–404. doi: 10.1111/j.1365-313x.1992.00397.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 16.van den Brûle S, Müller A, Fleming A J, Smart C C. Plant J. 2002;30:649–662. doi: 10.1046/j.1365-313x.2002.01321.x. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto T, Kamiya N, Ueguchi-Tanaka M, Iwahori S, Matsuoka M. Genes Dev. 2001;15:581–590. doi: 10.1101/gad.867901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long J A, Moan E I, Medford J I, Barton M K. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 19.Byrne M, Timmermans M, Kidner C, Martienssen R. Curr Opin Plant Biol. 2001;4:38–43. doi: 10.1016/s1369-5266(00)00133-3. [DOI] [PubMed] [Google Scholar]
- 20.Golz J F, Hudson A. Plant Cell. 2002;14:S277–S288. doi: 10.1105/tpc.000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster T M, Lough T J, Emerson S J, Lee R H, Bowman J L, Forster R L S, Lucas W J. Plant Cell. 2002;14:1497–1508. doi: 10.1105/tpc.001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemerley A, De Almeida Engler J A, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. EMBO J. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan D R. Int J Plant Sci. 2001;162:465–474. [Google Scholar]
- 24.Fletcher J C, Meyerowitz E M. Curr Opin Plant Biol. 2000;3:23–30. doi: 10.1016/s1369-5266(99)00033-3. [DOI] [PubMed] [Google Scholar]
- 25.Jackson D, Veit B, Hake S. Development (Cambridge, UK) 1994;120:405–413. [Google Scholar]
- 26.Reinhardt D, Mandel T, Kuhlemeier C. Plant Cell. 2000;12:507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J Y, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Proc Natl Acad Sci USA. 2002;99:4103–4108. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]