Abstract
This study characterized the contribution of Agrobacterium tumefaciens VirB6, a polytopic inner membrane protein, to the formation of outer membrane VirB7 lipoprotein and VirB9 protein multimers required for type IV secretion. VirB7 assembles as a disulfide cross-linked homodimer that associates with the T pilus and a VirB7-VirB9 heterodimer that stabilizes other VirB proteins during biogenesis of the secretion machine. Two presumptive VirB protein complexes, composed of VirB6, VirB7, and VirB9 and of VirB7, VirB9, and VirB10, were isolated by immunoprecipitation or glutathione S-transferase pulldown assays from detergent-solubilized membrane extracts of wild-type A348 and a strain producing only VirB6 through VirB10 among the VirB proteins. To examine the biological importance of VirB6 complex formation for type IV secretion, we monitored the effects of nonstoichiometric VirB6 production and the synthesis of VirB6 derivatives with 4-residue insertions (VirB6.i4) on VirB7 and VirB9 multimerization, T-pilus assembly, and substrate transfer. A virB6 gene deletion mutant accumulated VirB7 dimers at diminished steady-state levels, whereas complementation with a plasmid bearing wild-type virB6 partially restored accumulation of the dimers. VirB6 overproduction was correlated with formation of higher-order VirB9 complexes or aggregates and also blocked substrate transfer without a detectable disruption of T-pilus production; these phenotypes were displayed by cells grown at 28°C, a temperature that favors VirB protein turnover, but not by cells grown at 20°C. Strains producing several VirB6.i4 mutant proteins assembled novel VirB7 and VirB9 complexes detectable by nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and two strains producing the D60.i4 and L191.i4 mutant proteins translocated IncQ plasmid and VirE2 effector protein substrates in the absence of a detectable T pilus. Our findings support a model that VirB6 mediates formation of VirB7 and VirB9 complexes required for biogenesis of the T pilus and the secretion channel.
Agrobacterium tumefaciens uses a type IV secretion system to deliver oncogenic transfer DNA (T-DNA) and effector proteins to plant cells during infection (13, 44). This type IV secretion system requires the activities of the integral inner membrane protein VirD4 and the 11 VirB proteins thought to assemble as a supramolecular structure that spans the cell envelope. Recent studies have indicated that VirD4 belongs to a family of translocases that mediate the delivery of conjugal DNA intermediates across the inner membrane (32). Very intriguingly, VirD4 recently was shown to localize at the cell poles of A. tumefaciens (28). A transenvelope VirB protein structure acts coordinately with VirD4 to direct substrate translocation (39), possibly specifically across the outer membrane (36). Independent of VirD4, the VirB proteins also elaborate a conjugative pilus termed the T pilus (30, 31). This extracellular filament is important for substrate transfer, most probably because it stabilizes contacts between donor and target cells. Interestingly, however, mutations of the VirB11 ATPase can abolish detectable T-pilus formation without compromising substrate transfer (39), showing that a wild-type pilus is not an obligatory component of this type IV secretion system.
VirB6 is a highly hydrophobic subunit of this secretion system that is predicted to span the inner membrane multiple times (46). Early genetic studies established the importance of VirB6 for both T-pilus biogenesis and substrate transfer (7, 21). However, its role in type IV secretion has not been defined, and two alternative models have been proposed. The first is that VirB6 is a structural component of a transenvelope VirB secretion channel. This model is supported in part by VirB6's multitransmembrane topology, which is reminiscent of inner membrane channel or transporter proteins (10, 17, 23). Additionally, VirB6 production is correlated with stabilization of putative structural subunits of the secretion channel, including VirB4, VirB9, and VirB10 (7, 24). Finally, a recent study showed that detergent-solubilized VirB6 fractionates as a high-molecular-weight species in blue native gels and by size exclusion chromatography (27). Thus, there is suggestive evidence for VirB6 oligomerization, although specific protein-protein contacts have not been reported.
The second model depicts VirB6 as an assembly factor for the T pilus (24). This model was developed from the finding that VirB6 exerts strong stabilizing effects on T-pilus-associated proteins, including VirB2 pilin, the pilus-associated proteins VirB5 and a disulfide cross-linked VirB7 homodimer species, and VirB3 which plays an unspecified role in pilus biogenesis (24). Furthermore, the Binns laboratory reported the intriguing discovery that a subset of VirB proteins can stimulate uptake of an IncQ plasmid when produced by recipient cells in matings with agrobacterial donors (8). This subset of proteins is postulated to form a stable complex, possibly a channel, at the recipient envelope that stimulates conjugal DNA transfer by 2 to 3 orders of magnitude. Though it is not known how this presumptive complex is structurally or functionally related to a type IV secretion channel, recent studies with recipient cells expressing subsets of the virB genes have demonstrated a requirement only for the VirB7 through VirB10 proteins for this stimulation effect (Z. Liu and A. N. Binns, personal communication).
In the present study, we explored the contribution of VirB6 to type IV secretion by characterizing its role in the establishment of protein-protein interactions among the VirB7, VirB8, VirB9, and VirB10 proteins. We supply evidence for interactions between VirB6, VirB7, and VirB9 independently of other VirB proteins. Furthermore, we identify correlative effects of altered VirB6 production—either by modulation of native protein steady-state levels or synthesis of VirB6 insertion mutants—on VirB7 and VirB9 multimer formation and substrate transfer or T-pilus biogenesis. We propose that VirB6 mediates assembly of the T pilus and a functional secretion machine through its effects on VirB7 and VirB9 multimerization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Table 1 lists the bacterial strains and plasmids used in this study. The conditions and media for growth of A. tumefaciens and Escherichia coli and for induction of A. tumefaciens vir genes in induction media (IM) containing 200 μM acetosyringone (AS) have been described previously (19). Plasmids were maintained in E. coli and A. tumefaciens by addition of carbenicillin (100 μg/ml), kanamycin (100 μg/ml), tetracycline (5 μg/ml), gentamicin (50 μg/ml), or spectinomycin (500 μg/ml) to the growth medium.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | λ− φ80d/lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA relA1 | Gibco-BRL |
| CJ236 | dut ung thi relA; pCJ105 (Camr) | Bio-Rad |
| BL21(DE3) | F−ompT hsdsB(rB− mB−) dcm gal λ (DE3) | Novagen |
| S17-1 | Tra genes from RP4 integrated into chromosome for plasmid mobilization | 39 |
| A. tumefaciens | ||
| A136 | Strain C58 cured of pTiC58 | 48 |
| A348 | A136 containing octopine-type Ti plasmid pTiA6NC | 22 |
| A348Spcr | A348 with Spcr by spontaneous mutagenesis | 39 |
| PC1000 | A348 derivative with deletion of the virB operon from Ti plasmid pTiA6NC | 19 |
| PC1006-PC1010 | A348 with deletions of virB6 to virB10, respectively, from Ti plasmid pTiA6NC | 7 |
| Mx358 | A348 carrying a Tn3HoHo1 insertion in virE2 | 43 |
| LBA4404 | T-DNA deletion strain | 35 |
| Plasmidsa | ||
| pBSIISK+ | Crbr; cloning vector | Stratagene |
| pBSIIKS+ | Crbr; cloning vector | Stratagene |
| pBSIISK+NdeI | Crbr; pBSIISK+ containing an NdeI site at the translational start site of lacZ | 7 |
| pBSIIKS+NdeI | Crbr; pBSIIKS+ containing an NdeI site at the translational start site of lacZ | 7 |
| pBSIIKS+NcoI | Crbr; pBSIIKS+ containing an NcoI site at the translational start site of lacZ | 7 |
| pGEX2T | Crbr; vector for construction of GST gene fusions | Pharmacia |
| pPC914KS+ | Crbr; pBSIIKS+ derivative expressing PvirB-virB1; vector for expression from PvirB by substitution of other genes for virB1 | 19 |
| pSW172 | Tetr; broad-host-range IncP plasmid containing Plac and polylinker sequence from pIC19 | 11 |
| pXZ151 | Kanr; pSW172 derivative broad-host-range IncP plasmid with the Kanr gene from pUC4K inserted at the unique StuI site | 51 |
| pML122ΔKm | Genr; mobilizable RSF1010 (IncQ) derivative | 21 |
| pXZ61 | Crbr; pBSIISK+NdeI expressing Plac-virB6 | This study |
| pSJ964 | Crbr; pPC914KS+ expressing PvirB-virB6 | This study |
| pSJ6300 | Crbr; pGEX2T carrying ∼300-bp BamHI-EcoRI fragment from pSJ6192; expresses Plac-GST-′virB6 | This study |
| pZZ11 | Crbr; pPC914KS expressing PvirB-GSTb | This study |
| pVK8 | Crbr; pZZ11 expressing PvirB-GST-virB8 by introduction of an ∼1-kb NdeI-XhoI fragment from pPC9104 | This study |
| pES8 | Crbr; pZZ11 expressing PvirB-GST-virB10 by introduction of an ∼1-kb NdeI-XhoI fragment from pPC9104 | This study |
| pAS8 | Crbr Kanr; pES8 ligated to the IncP plasmid pXZ151 | This study |
| pPC988 | Camr; pBCKS+ expressing Plac-virB8 | 7 |
| pPC9108 | Camr; pBCKS+ expressing Plac-virB10 | 7 |
| pSJ610 | Crbr; pSJ964 carrying virB6-virB11 by insertion of an ∼4-kb ClaI-EcoRV fragment from pPC801 into similarly digested pSJ964 | This study |
| pSJ6xxx series | Crbr; pXZ61 carrying in-frame tandem NcoI-BamHI or NdeI-BamHI restriction sites in virB6 at positions following the residue indicated; virB6.i4 alleles are expressed from Plac | This study |
| pSJ5xxx series | Crbr; pSJ964 with virB6.i4 alleles from pSJ6xxx plasmid series substituted for wild-type virB6; alleles are expressed from PvirB | This study |
Except where indicated, ColE1 plasmids expressing virB constructs were ligated to IncP plasmid pXZ151 for introduction into A. tumefaciens; the cointegrate plasmid in A. tumefaciens is given the ColE1 plasmid name plus a “B” (for ligation to a broad-host-range replicon).
See the text for details.
virB gene constructions.
Plasmid pXZ61 (Plac-virB6) was constructed by introducing an ∼1.1-kb SacI-BglII fragment carrying virB6 from pBB20 into SacI- and BamHI-digested pBSIISK+NdeI. Plasmid pJS964 (PvirB-virB6) was constructed by introducing an ∼1.1-kb NdeI-XhoI fragment containing virB6 from pXZ61 into similarly digested pPC914KS+. Plasmid pSJ610 expressing PvirB-virB6-virB10 was constructed by substituting an ∼4-kb ClaI-EcoRV fragment from pPC801 for virB6 in pSJ964. The construction of plasmid pPC801 carrying the intact virB operon on a ColE1 replicon will be described elsewhere. Plasmid pSJ6300 expressing Plac-GST-′virB6 was constructed by introducing the 3′ end of virB6 as a 300-bp BamHI-EcoRI fragment from pSJ6192 (see below) into similarly digested pGEX2T. IPTG (isopropyl-β-d-thiogalactopyranoside)-induced E. coli transformed with pSJ6300 served as the source of GST-′VirB6 for antibody production in rabbits (see below). Plasmid pZZ11 expressing PvirB-GST was constructed by PCR amplification of GST from pGEX2T to introduce NcoI and NdeI/XhoI sites at the 5′ and 3′ ends of GST, respectively. The resulting PCR fragment was digested with NcoI-XhoI and introduced into similarly digested pBSIIKS+NcoI. Plasmids pES10 and pVK8 expressing PvirB-GST-virB10 and PvirB-GST-virB8, respectively, were constructed by introducing the corresponding NdeI-XhoI fragments from pPC9108 and pPC988 into similarly digested pZZ11.
Plasmid pXZ61 served as a template for introduction of tandem NcoI-BamHI (CCCATGGGATCC) or NdeI-BamHI (CATATGGGATCC) restriction sites at ∼30-codon intervals along the length of virB6 by oligonucleotide-directed mutagenesis. These insertions resulted in the introduction of in-frame insertions of PMGS and HMGS residues (i4 insertions), respectively, immediately following the residue indicated. The VirB6.i4 mutant proteins, E29.i4 and D60.i4, carry PMGS insertions, and the remaining mutants carry HMGS insertions. Oligonucleotides used for the mutagenesis carried the tandem restriction sites with at least 15 bases of 5′ and 3′ flanking sequences complementary to the regions of virB6 flanking the insertion site. Mutant alleles were identified by restriction enzyme digestion, and the insertion mutations were confirmed by sequencing of the entire virB6 gene. The resulting set of 10 plasmids was designated pSJ6xxx, where xxx denotes the residue immediately preceding the 4-residue insertion. The 10 virB6.i4 alleles were introduced behind the PvirB promoter by substitution of ∼1.1-kb NarI-KpnI fragments from the pSJ6xxx plasmids for the corresponding NarI-KpnI fragment of pSJ964 to yield the pSJ5xxx plasmids, where xxx denotes the i4 insertion site. These ColE1 plasmids were introduced into A. tumefaciens by ligation to the broad-host-range plasmid pXZ151. Cointegrate plasmids constructed in this way were assigned the ColE1 plasmid name plus a “B” to designate ligation to a broad-host-range replicon.
Cellular fractionation and T-pilus isolation.
T pili were isolated as previously described (30, 40). Briefly, A. tumefaciens strains were grown to an optical density at 600 nm (OD600) of 0.5 in MG/L media (47) at 28°C. Cells were pelleted, diluted fivefold in IM, and incubated for 6 h at 22°C. Two hundred microliters of an AS-induced culture were spread on IM agar plates, and the plates were incubated for 3 days at 18°C. Cells were then gently scraped off the plates in 50 mM KPO4 buffer (pH 5.5) (buffer A) and pelleted by centrifugation at 14,000 × g for 15 min at room temperature. The supernatant was removed, and the cell pellet was resuspended in buffer A. This suspension was passed through a 25-gauge needle 10 times to collect flagella, pili, and surface proteins. The sheared bacterial cells were pelleted by centrifugation at 14,000 × g for 30 min at 4°C. The remaining supernatant was filtered through a 0.22-μm-pore-size cellulose acetate membrane to remove unpelleted cells.
T pili were harvested by centrifugation of filtered exocellular material at 100,000 × g for 1 h at 4°C. Pelleted material was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunostaining. The material was also solubilized in buffer A and loaded onto a 5-ml 20 to 70% linear sucrose density gradient. T pili were then fractionated by ultracentrifugation in an SW55 Beckman rotor at 80,000 × g for 20 h at 4°C. Fractions of 0.5 ml were collected from the bottoms of the centrifugation tube and analyzed for the presence of Vir proteins by immunoblotting and of other proteins by silver staining (40). T-pilus integrity was assessed by comparison of fractionation behaviors of VirB2 pilin and the pilus-associated proteins VirB5 and VirB7 from the exocellular fractions of strains of interest and by electron microscopy as previously described (40).
Protein analysis, immunoblotting, and cell fractionation.
Proteins were resolved by SDS-PAGE or a Tricine-SDS-PAGE system as previously described (7). Vir proteins were visualized by SDS-PAGE, protein transfer to nitrocellulose membranes, and immunoblot development with goat anti-rabbit antibodies conjugated to alkaline phosphatase and histochemical substrates. For enhanced sensitivity, blots were developed with anti-rabbit antibodies conjugated to horseradish peroxidase, and antibody-antigen interactions were visualized by chemiluminescence with an ECL kit (Amersham Pharmacia, Piscataway, N.J.). For comparisons of VirB protein abundance, cells were grown to an OD600 of 0.6, serially diluted, and plated for determinations of CFU. Proteins were loaded on SDS-polyacrylamide gels on a per cell (CFU) equivalent basis. Molecular mass markers were obtained from GIBCO-BRL (Grand Island, N.Y.). Antibody specificities were previously documented for the VirB1, VirB2, VirB4, VirB5, VirB7 through VirB11, and VirE2 proteins (39, 47, 50).
Anti-VirB6 antiserum was raised by overproduction and purification of a glutathione S-transferase (GST)-′VirB6 fusion protein from E. coli BL21(DE3, pSJ6300). Briefly, a 100-ml culture was grown to an OD600 of 0.3, IPTG (1 mM final concentration) was added to induce gst-′virB6 expression, and cells were incubated with shaking for 4 h at 37°C. GST-′VirB6 present in the soluble fraction of cell lysates was purified by passage through a glutathione Sepharose column and elution according to the manufacturer's instructions (Pharmacia). Purified GST-′VirB6 was sent to Cocalico Biologicals, Inc., (Reamstown, Pa.) for injection into New Zealand White rabbits.
Membrane solubilization and protein-protein interaction assays.
A. tumefaciens cultures (500 ml) were induced for vir gene expression in IM for 18 h at 22°C. Cells were harvested and lysed by French press treatment, and total membranes were recovered as previously described (38). Total membranes were suspended in 50 mM HEPES (pH 8.0) at a final protein concentration of 1 mg/ml. Membranes were solubilized by addition of one of the following detergents: 2% (final concentration) lauryldimethylamine oxide (LDAO), 2% deoxycholate (DOC), 2% dodecylmaltoside (DM), or 1× RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 1% DOC, 0.1% SDS, 50 mM HEPES [pH 8.0]). Treated membranes were incubated for 4 h at 4°C with gentle rocking. The solubilized material was centrifuged at 100,000 × g in a Beckman table top ultracentrifuge for 1 h at 4°C. The insoluble material was discarded, and the supernatant containing solubilized total membrane protein served as the starting material for immunoprecipitation experiments.
For immunoprecipitation, 1 ml of solubilized membrane protein at 1 mg/ml was divided into three microcentrifuge tubes, and the volumes were adjusted to 1 ml with 1× RIPA buffer or 2% LDAO in 50 mM HEPES (pH 8.0). Bovine serum albumin (5 μg/ml) was added to each sample. Anti-VirB antiserum or preimmune serum (5 μl) was added to the first two samples, and all three samples were incubated with gentle rocking for 4 h at 4°C. Protein A-Sepharose beads (3 μg) were then added to all three samples, followed by a second incubation for 4 h at 4°C. The protein A Sepharose beads were collected by centrifugation at 14,000 × g for 15 min, washed three times with 1 ml of 0.1% DOC or 0.1% LDAO in 25 mM HEPES (pH 8.0), and resuspended in 50 μl of 2× protein sample buffer. Samples were incubated at 37°C for 30 min, and 5 to 10 μl of the immunoprecipitate was analyzed by SDS-PAGE (37).
For GST pulldown assays, GST-Sepharose beads (200-μl bed volume) were added to 1 ml of solubilized membrane proteins at 1 mg/ml prepared as described above. The mixture was incubated with shaking at 4°C overnight and then centrifuged at 2,500 × g. The isolated beads were washed three times with physiologically buffered saline (pH 8.0), and bound proteins were eluted with 25 mM reduced glutathione-50 mM Tris-HCl (pH 9.0) (37).
Conjugation assays.
The RSF1010 derivative pML122ΔKm was introduced into various A. tumefaciens donor strains by diparental mating with E. coli strain S17-1(pML122ΔKm) (41). A. tumefaciens strains carrying pML122ΔKm were mated with an Spcr-derivative of A348 as previously described (21, 39). Briefly, mid-log phase (OD600 = 0.5) cells were harvested and incubated in IM for 6 h at 22°C to induce expression of the vir genes. Five microliters of preinduced donor and recipient cells were mixed on a nitrocellulose filter on an IM agar plate, and the plate was incubated for 4 days at 18°C. Mating mixtures were recovered from filters and plated onto media selective for transconjugants or serially diluted for determination of transconjugant and donor cell numbers. Frequencies of transfer were estimated as transconjugants recovered per donor. Experiments were repeated in triplicate, and results are reported for a representative experiment.
Virulence assays.
A. tumefaciens strains were tested for virulence by inoculating wound sites of Kalanchoe daigremontiana leaves. Controls for the tumorigenesis assay included the coinoculation of the same leaf with wild-type A348 and isogenic, avirulent mutant strains (7, 26). Each experiment was repeated at least three times for each strain on separate leaves.
RESULTS
Isolation of VirB protein subcomplexes by immunoprecipitation.
To characterize the role of VirB6 in assembly or function of the virB-encoded type IV secretion system, we first assayed for interactions with other VirB proteins in detergent-solubilized extracts of wild-type strain A348. In initial studies, we found that treatments of membrane fractions with 2% DM, 2% LDAO, or 1× RIPA solubilized ∼20 to 50% of each VirB protein (Fig. 1) (see Materials and Methods). An independent study reported similar findings with 2% DM (27). RIPA- and LDAO-solubilized material from total membrane fractions yielded identical results in the immunoprecipitation and GST pulldown experiments described below.
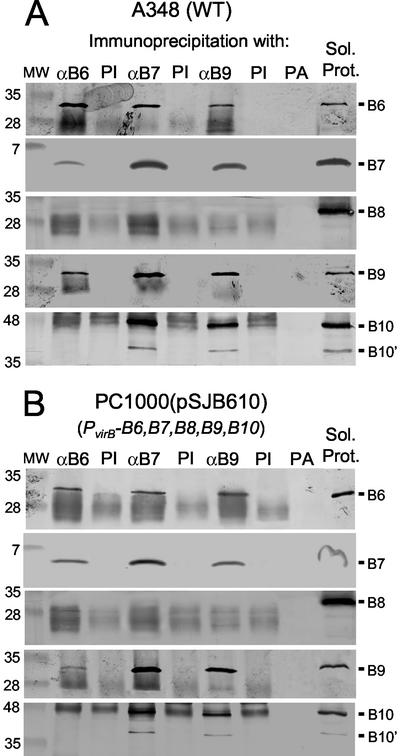
FIG. 1.
Immunoprecipitation studies with anti-VirB6, anti-VirB7, and anti-VirB9 antisera. (A) Isolation of VirB protein complexes from detergent-solubilized membrane extracts of wild-type A348. (B) VirB complexes isolated from PC1000(pSJB610). Lanes: αB6, αB7, and αB9, anti-VirB antisera; PI, preimmune serum; PA, protein A Sepharose (these were all used for precipitation); Sol. Prot., solubilized starting material for the precipitations; MW, molecular weight markers, with sizes in kilodaltons shown at left. Blots were probed with antiserum to the VirB proteins listed at the right. The cross-reactive material in the blot developed with anti-VirB10 antiserum is heavy-chain IgG, but native VirB10 (48 kDa) and VirB10′ (40 kDa) derived from translation from an internal Met were clearly distinguished from this background in the immunoblots. The IgG light chain also was immunoreactive and formed a nonspecific background in blots developed with the anti-VirB6, -VirB8, and -VirB9 antisera.
Figure 1A shows that the anti-VirB6 antiserum, but not preimmune sera or protein A Sepharose alone, precipitated native VirB6 from RIPA-solubilized extracts of wild-type A348. Of considerable interest, the anti-VirB6 antiserum also precipitated the VirB7 lipoprotein and the VirB9 protein but no detectable amounts of VirB8, VirB10 (Fig. 1A), or other VirB proteins (data not shown). Figure 1A further shows that the anti-VirB7 and anti-VirB9 antisera, but not the preimmune sera, precipitated VirB6 together with VirB7 and VirB9. Interestingly, these antisera also precipitated VirB10 but no detectable VirB8 (Fig. 1A) or other VirB proteins (data not shown). Although the polyclonal antisera used for blot development cross-reacted with the immunoglobulin G (IgG) heavy chain in the immunoprecipitates, the presence of the comigrating VirB10 (48-kDa) protein in the precipitates was easily distinguished from the cross-reactive material in the original blots. Additionally, a 40-kDa VirB10 species resulting from translation from an internal Met residue coprecipitated with the full-length protein and was easily visualized by immunostaining (Fig. 1A).
Next, we assayed for VirB6, VirB7, and VirB9 complex formation in PC1000(pSJ610), a strain producing only VirB6 through VirB10 among the VirB proteins. PC1000(pJS610) accumulated abundant levels of these VirB proteins (Fig. 1B), and as anticipated, the anti-VirB6 antiserum coprecipitated VirB6, VirB7, and VirB9 but no detectable VirB8 or VirB10 (Fig. 1B). In reciprocal experiments, both the anti-VirB7 and anti-VirB9 antisera precipitated VirB6, VirB7, and VirB9. As observed for wild-type A348, the anti-VirB7 and anti-VirB9 antisera also precipitated VirB10 but no detectable VirB8 (Fig. 1B).
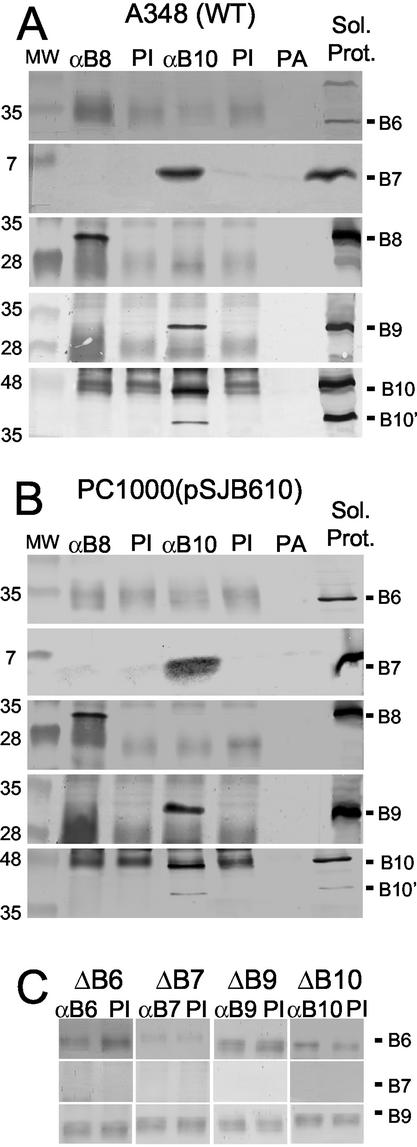
To further explore the nature of VirB8 and VirB10 protein-protein contacts, we used antisera to these proteins in immunoprecipitation studies. As shown in Fig. 2A and B, the anti-VirB10 antisera coprecipitated abundant levels of VirB7, VirB9, and VirB10 but undetectable levels of VirB6 or VirB8 from extracts of wild-type A348 and PC1000(pSJ610). By contrast, the anti-VirB8 antisera precipitated abundant levels of VirB8 but no detectable amounts of VirB6, VirB7, VirB9, or VirB10. As before, preimmune sera failed to precipitate any of the VirB proteins at detectable levels.
FIG. 2.
Immunoprecipitation studies with anti-VirB8 and anti-VirB10 antisera. (A and B) Studies of complex formation in extracts of A348 and PC1000(pSJB610), respectively. The lanes are as described in the legend to Fig. 1. Blots were probed with antiserum to the VirB proteins listed at the right. (C) Detergent-solubilized membrane extracts from each of the ΔvirB mutant strains listed at the top served as starting material for immunoprecipitation with the cognate anti-VirB antiserum to assay for nonspecific precipitation of VirB proteins. The faint immunoreactive material in all lanes is the IgG light chain.
Additional control experiments with each of the relevant virB gene deletion strains established the specificity of these precipitation studies. For example, the anti-VirB6 antiserum did not precipitate VirB6 or any other VirB protein from extracts of the ΔvirB6 mutant, PC1006. Similarly, antisera to VirB7, VirB8, VirB9, and VirB10 failed to precipitate any of these VirB proteins from the corresponding strains PC1007 (ΔvirB7), PC1008 (ΔvirB8) (data not shown), PC1009 (ΔvirB9), and PC1010 (ΔvirB10), respectively (Fig. 2C).
The results of these immunoprecipitation studies indicate that VirB6 interacts with VirB7 and VirB9 and furthermore that VirB7 interacts with VirB9 and VirB10 in A. tumefaciens. However, VirB6 does not stably associate with the presumptive VirB7-VirB9-VirB10 complex. Finally, the VirB subcomplexes composed in part of VirB6, or alternatively of VirB10, form independently of other VirB proteins, and they also lack detectable levels of VirB8.
A functional GST-VirB10 fusion protein interacts with VirB7, VirB9, and VirB10.
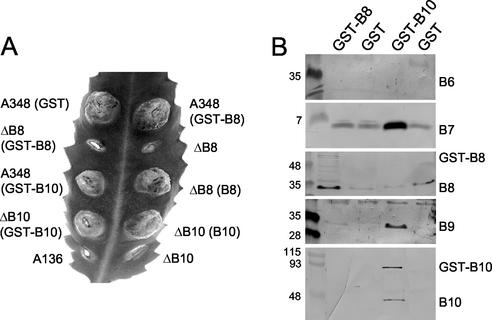
The absence of detectable associations of VirB6 and VirB8 with a putative VirB7-VirB9-VirB10 subcomplex was intriguing in view of earlier proposals that both proteins assemble as structural subunits of this type IV secretion machine (see Discussion). To complement the above-mentioned studies, we tested for interactions between the VirB6 through VirB10 proteins by fusing the GST coding sequence to the 5′ ends of virB8 and virB10 and expressing the chimeric genes from the PvirB promoter in A. tumefaciens cells. In preliminary studies, we found that both chimeric genes encode GST fusion proteins, although GST-VirB8 accumulated at very low levels that were suggestive of protein instability (Fig. 3B). GST-VirB10 was fully functional, as shown by wild-type virulence of PC1010(pAS10), a ΔvirB10 mutant trans-expressing PvirB-GST-virB10 (Fig. 3A). GST-VirB10 also was phenotypically silent in a GST-virB10/virB10 merodiploid strain, as shown by wild-type virulence of A348(pAS10). By contrast, GST-VirB8 was nonfunctional as shown by avirulence of PC1008(pVKB8), a ΔvirB8 mutant trans-expressing PvirB-GST-virB8. GST-VirB8 also was phenotypically silent in a GST-virB8/virB8 merodiploid strain. As a control, A348(pZZB11) expressing the GST protein alone exhibited wild-type virulence (Fig. 3A).
FIG. 3.
GST pulldown studies with GST-VirB10 and GST-VirB8 fusion proteins. (A) Virulence of A348 derivatives on wounded K. daigremontiana leaves. Strains: A348(GST), A348(pZZB11); ΔB8(GST-B8), PC1008(pVKB8); A348(GST-B10), A348(pAS10); ΔB10(GST-B10), PC1010(pAS10); A136, Ti plasmidless strain; A348(GST-B8), A348(pVKB8); ΔB8, PC1008; ΔB8(B8), PC1008(pPCB984); ΔB10(B10), PC1010(pPCB9104); ΔB10, PC1010. (B) GST pulldown assays with A348 derivatives producing the GST fusion proteins listed at the top of each lane. Strains: GST-B8, A348(pVKB8); GST, A348(pZZB11); GST-B10, A348(pAS10). Material eluted from GST-Sepharose was assayed by immunostaining for the presence of the VirB proteins listed at the right.
We used GST pulldown assays to screen for interactions between VirB10 and other VirB proteins. LDAO-solubilized membrane extracts from A348(pAS10) producing GST-VirB10 and A348(pZZB11) producing only GST were passed over glutathione columns, and proteins bound to the column and eluted with glutathione were analyzed by SDS-PAGE and immunostaining. The eluates from A348(pAS10) possessed abundant levels of GST-VirB10, VirB7, VirB9, and VirB10 and an undetectable amount of VirB6 (Fig. 3B). We also detected some VirB8, but eluates from the control strain, A348(pZZB11), possessed a similar amount of VirB8, suggestive of nonspecific binding to the glutathione resin. VirB7 also bound nonspecifically to some extent, yet an abundant amount coeluted with GST-VirB10. We also assayed A348(pVKB8) producing GST-VirB8 for complex formation. These eluates possessed GST-VirB8 and VirB8, the latter possibly as a result of nonspecific binding to the column, but undetectable amounts of VirB6, VirB7, or VirB10 (Fig. 3B). These findings supply further evidence for assembly of a VirB7-VirB9-VirB10 complex that is devoid of detectable amounts of VirB6 or VirB8. The results of our complementation studies leave open the possibility that the GST moiety interferes with interactions between GST-VirB8 and other VirB proteins, although this cannot be the case for GST-VirB10. We also acknowledge that the detergent solubilization conditions employed in these studies might have disrupted weak affinity interactions between VirB6 and VirB8 and the presumptive complex(es) composed of VirB7, VirB9, and VirB10.
Nonstoichiometric VirB6 production disrupts assembly of VirB7 and VirB9 complexes and interferes with substrate transfer.
We evaluated the in vivo biological importance of VirB6 interactions with VirB7 and VirB9 initially by assaying for correlative effects of nonstoichiometric native VirB6 production on VirB7 and VirB9 complex formation, T-pilus production, and substrate transfer. Very interestingly, these studies showed, first, that VirB6 synthesis is important for accumulation of a disulfide cross-linked VirB7 homodimer in cells producing other VirB proteins and, second, that VirB6 overproduction induces assembly of higher-order VirB9 complexes and also exerts a temperature-dependent block on substrate transfer without detectably disrupting biogenesis of the T pilus.
Strain PC1006 (ΔvirB6) lacks detectable VirB6, and complementation with plasmid pXZB61, a multicopy IncP replicon expressing virB6 from a moderately weak Plac promoter in A. tumefaciens (11), resulted in the accumulation of VirB6 at a steady-state level approximating that detected in wild-type A348 (Fig. 4A). However, complementation with pSJB964, a multicopy IncP replicon expressing virB6 from the strong PvirB promoter (11), yielded a significantly higher level of VirB6 than that in A348. Cellular proteins were applied to the gels on a per cell equivalent basis, and as expected, VirE2 accumulated at comparable levels in each of these strains (Fig. 4A). Also, as reported previously, the ΔvirB6 mutation was correlated with a reduction in levels of several VirB proteins (e.g., VirB7, VirB9, and VirB10) and this destabilizing effect was exacerbated by growth at 28°C, a temperature favoring VirB protein turnover (Fig. 4A) (4, 7, 24). Interestingly, however, PC1006(pXZB61) producing VirB6 at wild-type levels—but not the VirB6-overproducing strain—showed enhanced steady-state levels of VirB7, VirB9, and VirB10 at the elevated growth temperature (Fig. 4A). Note also that VirB8 levels were unaffected either by growth temperature or by nonstoichiometric synthesis of VirB6 (Fig. 4A) (see Discussion).
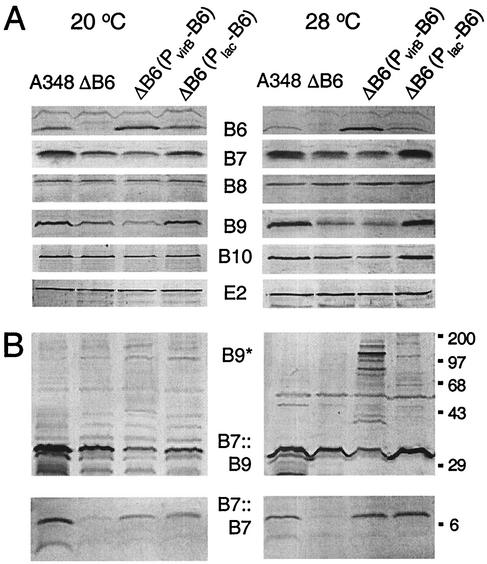
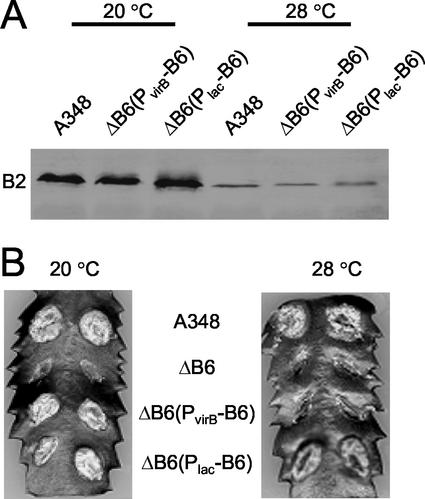
FIG. 4.
Accumulation of the VirB6 through VirB10 proteins as a function of altered VirB6 production and growth temperature. (A) Immunoblot analysis of total protein samples electrophoresed under reducing conditions; blots were developed with antisera specific for the proteins listed in the center. (B) Immunoblot analysis of protein samples electrophoresed under nonreducing conditions; top panels were developed with anti-VirB9 antiserum to show the VirB7::VirB9 heterodimer (B7::B9) and higher-order VirB9 complexes (B9*), and bottom panels were developed with anti-VirB7 antiserum to show the VirB7 homodimer (B7::B7). Strains: A348, wild-type; ΔB6, PC1006; ΔB6(PvirB-B6), PC1006(pSJB964); ΔB6(PlacZ-B6), PC1006(pXZB61). Total cellular proteins from equivalent numbers of cells were subjected to SDS-PAGE and immunoblot analysis. Molecular size markers (in kilodaltons) are shown at right.
PC1006 accumulated the VirB7 homodimer at an undetectable level when assayed at <12 h after induction of the vir genes (Fig. 4B) and at a low but detectable level upon >14 h of vir gene induction (data not shown). A similar virB6 deletion mutation of the nopaline strain C58 also was correlated with an apparent absence of VirB7 homodimer (24). trans-expression of virB6 from the Plac or the PvirB promoter in strain PC1006 restored the abundance of the cross-linked homodimer to an appreciably higher level within 12 h of vir gene induction, establishing that VirB6 protein synthesis contributes to assembly or stability of the VirB7 homodimer (Fig. 4B).
In addition to a cross-linked VirB7 homodimer, A348 cells assemble a cross-linked VirB7-VirB9 heterodimer that is critical for biogenesis of the secretion machine (1, 5, 6, 20, 42). Interestingly, PC1006 also accumulated a diminished steady-state level of the VirB7-VirB9 heterodimer compared with wild-type A348 (Fig. 4B). Complementation with Plac-virB6 partially restored the abundance of the heterodimer, whereas complementation with PvirB-virB6 resulted in a further reduction in the heterodimer level at both induction temperatures. Correspondingly, the VirB6-overproducing strain accumulated a number of high-molecular-weight VirB9 complexes or aggregates; these species were devoid of VirB7 or other VirB proteins (Fig. 4B and data not shown). Both the reduction in heterodimer abundance and the accumulation of higher-order VirB9 species accompanying VirB6 overproduction were very pronounced upon vir induction at 28°C and only barely evident at 20°C.
To assess whether the appearance of novel VirB9 species accompanying VirB6 overproduction affected biogenesis or activity of this secretion system, we assayed for assembly of the T pilus and substrate translocation. Others have shown that wild-type A. tumefaciens grown at 20°C produces abundant amounts of T pilus, whereas growth at 28°C or higher temperatures interferes with pilus production (4, 30). Strain PC1006 is defective for pilus production at any temperature (21), but interestingly, both PC1006(pXZB61) (PvirB-virB6) and PC1006(pSJB964) (PvirB-virB6) accumulated the T pilus at levels comparable to that of wild-type cells at both 20 and 28°C (Fig. 5A). In the experiments shown in Fig. 5A, the presence of the T pilus was monitored by assaying for exocellular VirB2 pilin. Further studies established that the T pili isolated from the two mutant strains and T pili from A348 displayed identical fractionation profiles in sucrose gradients and that the T pili from all three strains were morphologically indistinguishable by electron microscopy (data not shown). Therefore, VirB6 overproduction did not detectably disrupt production of the T pilus at either induction temperature.
FIG. 5.
Effects of altered VirB6 synthesis on T-pilus production and virulence at 20 and 28°C. (A) Exocellular VirB2 pilin content as a measure of T-pilus production of strains grown at 20 and 28°C. A348, wild-type; ΔB6(PvirB-B6), PC1006(pSJB964); ΔB6(PlacZ-B6), PC1006(pXZB61); ΔB6 (PC1006) elaborates no T pilus (21, 30, 39). (B) Virulence of corresponding strains on K. daigremontiana leaves maintained at the temperatures shown.
PC1006(pXZB61) (Plac-virB6) displayed wild-type virulence at both 20 and 28°C, whereas in striking contrast, the VirB6-overproducing strain displayed wild-type virulence at 20°C but was completely avirulent at 28°C (Fig. 5B). In the assays shown in Fig. 5B, both PC1006 derivatives and wild-type A348 were inoculated at ∼108 CFU on wounded plant tissues, and further studies established that PC1006(pSJB964) still failed to incite tumor formation even with a very heavy inoculum of 1011 CFU. Though dilution studies have shown that A348 displays attenuated virulence when inoculated at low CFU on plants maintained at 28°C compared to 20°C (4), we found that this strain elicits similar tumorigenic responses when inoculated at ≥106 CFU on plants maintained at 20 and 28°C (Fig. 5A and data not shown). Thus, VirB6 overproduction induces both the formation of higher-order VirB9 species and blocks type IV secretion under experimental conditions in which A348 is fully transfer competent. VirB6 overproduction from PvirB did not perturb cell growth or viability, as shown by growth rate determinations and CFU counts throughout growth cycles of cultures incubated at 20 or 28°C (data not shown). The deleterious effect of VirB6 overproduction on substrate transfer at 28°C therefore cannot be attributed to nonspecific effects on membrane integrity.
VirB6.i4 mutant proteins disrupt VirB7 and VirB9 complex formation, T-pilus production, and substrate transfer.
We constructed a set of 4-residue (HMGS or PMGS) insertions at ∼25-residue intervals in hydrophilic regions of VirB6 for further structure-function studies. Initial studies showed that all of the mutant proteins accumulated at levels comparable to that of native VirB6 produced by A348 when synthesized from the PvirB promoter (Fig. 6A). All but one of the mutations rendered VirB6 nonfunctional with respect to pilus biogenesis and T-DNA transfer (see below), and none of the mutant proteins exerted dominant effects over the native protein in virB6/virB6.i4 merodiploid strains.
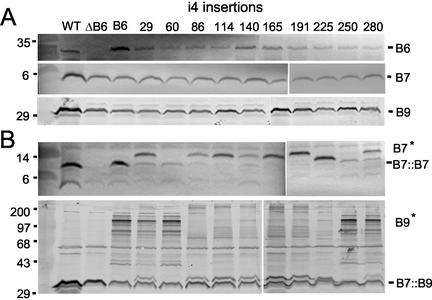
FIG. 6.
Effects of VirB6.i4 synthesis on VirB7 and VirB9 complex formation. (A) Protein samples from strains grown at 28°C were electrophoresed under reducing conditions. Blots were developed with antisera specific for the VirB proteins listed at right, and the top immunoblot shows steady-state levels of the VirB6.i4 derivatives. (B) Protein samples electrophoresed under nonreducing conditions. The upper blot was developed with anti-VirB7 antiserum to show VirB7 homodimer and VirB7* species. The lower blot was developed with anti-VirB9 antiserum to show the VirB7::VirB9 heterodimer and higher-order VirB9* species. Strains: WT, A348; ΔB6, PC1006; B6, PC1006(pSJB964); i4 insertions, PC1006(pSJB5xxx) strains producing VirB6.i4 derivatives with insertions after the residue indicated. Molecular size markers (in kilodaltons) are listed at left.
PC1006 derivatives producing the VirB6 mutant proteins possessed two phenotypes of interest. First, several strains accumulated novel VirB7 and VirB9 species detectable by electrophoresis of protein samples under nonreducing conditions (Fig. 6B). For example, whereas strains producing two mutant proteins, D60.i4 and I250.i4, accumulated the VirB7 homodimer, other strains accumulated a novel complex, termed VirB7*, that migrated at a position in gels corresponding roughly to an increase of ∼7-kDa over that of the ∼9-kDa homodimer. Additionally, strains synthesizing four mutant proteins, E29.i4, D60.i4, I250.i4, and A280.i4, accumulated higher-order VirB9 complexes resembling those detected in the VirB6 overproducing strain. The remaining strains accumulated higher-order VirB9 complexes or aggregates that failed to enter the nonreducing gels (Fig. 6B). Of further interest, the higher-order VirB9 complexes were abundant in extracts of strains grown at 28°C and less abundant in extracts of strains grown at 20°C (Fig. 6B and data not shown) (see below). The VirB7* and higher-order VirB9 complexes or aggregates were composed only of VirB7 and VirB9, respectively, among the VirB proteins, but further studies are needed to assay for complex formation with other cellular proteins. It is also possible that the VirB7* species correspond to structural variants of the VirB7 homodimer that display anomalous migration in the nonreducing gels. Of final interest, most of the i4 insertions located within the central region of VirB6 induced a similar spectrum of higher-order VirB7* and VirB9* complexes in nonreducing gels, whereas those located at the termini induced a distinct spectrum (Fig. 6B). The central region of VirB6 is predominantly extracytoplasmic (S. Jakubowski, unpublished data), and thus our ongoing studies are exploring the importance of this region for VirB6-VirB7-VirB9 complex formation.
Second, though most i4 mutations abolished VirB6 function, three i4 mutations were permissive either for substrate transfer or biogenesis of the T pilus (Table 2). PC1006(pSJB5280) producing the A280.i4 derivative was reported above to accumulate higher-order VirB9 complexes at 28°C. This strain showed additional properties reminiscent of phenotypes associated with native-protein overproduction, e.g., no observable effect on T-pilus production, but interference of T-DNA transfer at 28°C. VirB6.280.i4 is not visibly overproduced (Fig. 6A), though it is possible that this mutant protein is aggregated and fails to enter the protein gels. Conversely, strains producing the Q140.i4 or L191i4 mutant protein failed to elaborate T pili but were capable of exporting substrates. Although T-DNA transfer was undetectable, both strains efficiently transferred two other substrates to recipient cells (Table 2). Previous work has shown that this type IV secretion system translocates the VirE2 effector protein independently of T-DNA (35, 44). Accordingly, the Q140.i4- and L191.i4-producing strains transferred VirE2 to plant cells, as shown by a mixed infection assay (35) (Table 2). These mixed infection assays yielded tumors similar in size and time course of appearance to those of control mixed infections with a virE2 mutant (Mx358) and a T-DNA deletion strain (LBA4404), suggestive of efficient VirE2 export. Additionally, this type IV secretion system mobilizes non-selftransmissible IncQ plasmids, and both the Q140.i4- and L191.i4-producing strains delivered the IncQ plasmid, pML122, to agrobacterial recipients (Table 2). In both cases, the IncQ plasmid transfer frequencies were only ∼1 order of magnitude lower than that of a wild-type A348 donor and well above the frequency of reversion to Genr (<108) by recipient cells. These findings indicate that the Q140.i4 and L191.i4 mutations block biogenesis of the wild-type T pilus but support translocation of certain substrates. As discussed below, similar types of “uncoupling” mutations have been identified previously for the VirB11 ATPase (39).
TABLE 2.
Effects of i4 insertions on substrate transfer and pilus production
| Straina | Virulenceb | Transfer of:
|
Exocellular VirB2f | ||
|---|---|---|---|---|---|
| T-DNAc | VirE2d | pML122ΔKme | |||
| A348 | +++ (++) | NA | NA | 1.5 × 10−4 | +++ (+) |
| PC1006 (ΔB6) | − | − | − | <1 × 10−8 | − |
| Plac-virB6 | +++ (++) | NA | NA | 6.8 × 10−5 | +++ (+) |
| PvirB-virB6 | +++ (−) | NA | NA | 1.7 × 10−4 | +++ (+) |
| E29 | − | − | − | <1 × 10−8 | − |
| D60 | − | − | − | <1 × 10−8 | − |
| Q85 | − | − | − | <1 × 10−8 | − |
| T114 | − | − | − | <1 × 10−8 | − |
| Q140 | − | − | ++ | 1.2 × 10−5 | − |
| L165 | − | − | − | <1 × 10−8 | − |
| L191 | − | − | ++ | 9.8 × 10−6 | − |
| F225 | − | − | − | <1 × 10−8 | − |
| I250 | − | − | − | <1 × 10−8 | − |
| A280 | +++ (−) | NA | NA | 5.8 × 10−5 | +++ (+) |
PC1006 strains expressed native virB6 from the Plac or PvirB promoter or virB6.i4 alleles from PvirB.
Monitored by tumor formation at inoculated wound sites of Kalanchoe leaves. Pluses denote virulence of strains maintained on plants at 20°C, and pluses in parentheses denote virulence of strains maintained on plants at 28°C. See the text for experimental details.
Mixed infections with the T-DNA deletion strain LBA4404 (35). NA, not applicable (strains exhibiting wild-type virulence cannot be tested for export of individual substrates by mixed infection assays).
Mixed infections with the virE2 mutant mx358.
Conjugation assays with i4 mutant strains as donors and A348 Spcr as recipient. Values are transconjugants per donor cell.
T-pilus production as monitored by abundance of exocellular VirB2 pilin. Pluses denote relative abundance of exocellular pilin isolated from strains induced for vir gene expression at 20°C, and pluses in parentheses denote the relative abundance of exocellular pilin from strains induced at 28°C.
DISCUSSION
Biogenesis of the A. tumefaciens T-DNA type IV secretion system is a dynamic process requiring the sequential export, localization, and multimerization of the VirB structural subunits. Prior work has established that the VirB7 lipoprotein plays a central role in this assembly pathway, partly through the formation of a disulfide bridge with VirB9 (1, 5, 20, 42). Dimerization serves to stabilize both proteins (20, 42) and also several inner membrane proteins including the bitopic protein VirB10 (6). Dimerization also induces assembly of VirB10 oligomers as revealed by chemical cross-linking (6). In addition to these biochemical properties, yeast dihybrid screens have supplied evidence for pairwise contacts between VirB7 and VirB9 and between VirB8, VirB9, and VirB10 (16, 45). Thus, it has been proposed that VirB7 through VirB10 form a transenvelope core structure upon which this type IV system is built (13, 16, 27). In the present study, we sought to gain further biochemical evidence for interactions between these proteins in A. tumefaciens, and we evaluated the contribution of the polytopic inner membrane protein VirB6 to VirB complex formation. Collectively, our findings show that VirB7, VirB9, and VirB10 form a stable VirB subassembly in vivo that, interestingly, is devoid of detectable levels of VirB6 or VirB8. Furthermore, our results support a proposal originally set forth by Hapfelmeier et al. (24) that VirB6 mediates the formation of VirB subcomplexes required for biogenesis of this secretion system.
VirB6, a mediator of T-pilus biogenesis and assembly or activity of the type IV secretion channel.
VirB6 is a highly hydrophobic inner membrane protein (46), which likely explains the inability of a comprehensive yeast dihybrid screen to identify VirB6 contacts with other VirB proteins (45). The VirB proteins can, however, be solubilized with nonionic detergents (15, 27; this study), permitting the use of immunoprecipitation and protein affinity tags for protein-protein interaction studies. Here, we report the first biochemical evidence for heterotypic interactions between VirB6 and other VirB proteins, specifically, VirB7 and VirB9, in detergent extracts of wild-type A348 as well as a strain producing only VirB6 through VirB10 among the VirB proteins. Though this latter strain produces VirB8 or VirB10, we were unable to detect interactions between VirB6 and these proteins. These results have been replicated using extracts from LDAO, DOC, DM, and RIPA solubilization as starting material for the protein interaction studies (S. Jakubowski, data not shown). These findings indicate that VirB6 interacts with VirB7 and VirB9 independently of other VirB proteins, though we presently cannot rule out the possibility that VirB6 contacts with VirB7 and VirB9 are mediated by mutual interactions with another cellular factor.
We further gained evidence for causal relationships between altered VirB6 synthesis, misassembly of VirB7 and VirB9 multimers, and disruption of substrate transfer or pilus biogenesis. For example, characterization of a ΔvirB6 mutant and corresponding complementation studies demonstrated the importance of VirB6 protein synthesis for formation or stability of the VirB7 homodimer (Fig. 4). This species is a component of the T pilus, and thus our findings support a proposal by Hapfelmeier et al. (24) that VirB6 participates in biogenesis of the T pilus. However, two additional lines of study strongly suggest that VirB6 also participates in assembly or function of the secretion channel at least in part through its effects on VirB9 multimerization. First, we determined that VirB6 overproduction was correlated with a reduction in steady-state abundance of the cross-linked VirB7-VirB9 heterodimer and the accumulation of higher-order VirB9 complexes in cells grown at 28°C, a temperature that also rendered this strain avirulent. Conversely, the higher-order VirB9 complexes were less abundant at 20°C, a temperature at which this strain exhibited wild-type virulence. Also importantly, despite these effects on virulence, VirB6 overproduction did not detectably disrupt biogenesis of the T pilus. Possible mechanisms for how VirB6 might influence channel activity through its effects on VirB9 multimerization are discussed below.
Second, i4 insertion mutations of VirB6 also induced formation of novel, higher-order VirB7 and VirB9 complexes and, correspondingly, disrupted substrate transfer, pilus biogenesis, or both processes. Of special interest, a mutant strain producing the A280.i4 derivative was not detectably overproduced but displayed phenotypes similar to the VirB6-overproducing strain, e.g., higher-order VirB9 complex formation and a temperature-dependent block on substrate transfer without a detectable effect on piliation. Conversely, two other mutations, Q140.i4 and L191.i4, induced the formation of VirB9 complexes or aggregates that failed to enter the protein gels and blocked biogenesis of the wild-type T pilus. Yet, the mutant proteins supported translocation of two substrates, an IncQ plasmid and the VirE2 effector protein. Though the ability to detect substrate export might depend on the relative sensitivities of the corresponding transfer assays, it is clear the mutant proteins support the efficient translocation of at least certain substrates in the absence of pilus production, for which an assay based on detection of exocellular VirB2 pilin is a highly sensitive indicator (30, 31, 39, 40). This capacity to uncouple pilus biogenesis from substrate transfer by mutagenesis was first demonstrated with the VirB11 ATPase (39). Presently, we cannot distinguish whether these types of mutations arrest pilus biogenesis altogether or simply block pilus elongation, reminiscent of mutations identified in TraU (34) and TraH, TraF, and TraW (2) of the F plasmid. Nevertheless, the isolation of such mutations constitutes strong genetic evidence that VirB6 and VirB11 each supply functions that can alternatively drive assembly of a wild-type T pilus or a functional secretion channel.
It should be noted that the type IV “machine” is often depicted as a single supramolecular structure for which there is some experimental evidence (36), yet such a structure has not yet been visualized. The uncoupling mutations of VirB6 and VirB11 might exert their effects on this structure, for example, by supporting T-pilus assembly while sterically blocking substrate passage or vice versa. However, other findings suggest that the VirB and VirD4 proteins might assemble as distinct surface structures or organelles. For example, recent cell biological studies have demonstrated a polar localization for the putative VirD4 translocase (28), whereas VirB8, VirB9, and VirB10 (29) distribute as distinct foci around the cell envelope. In addition, it is notable that type IV secretion systems of other bacterial species can function independently of detectable pilus production. For example, in Helicobacter pylori, the Cag type IV system assembles as a channel for secretion of CagA to mammalian cells without contributions of homologs for the VirB2 or VirB5 pilin proteins (14). The Bordetella pertussis Ptl system also secretes multisubunit PT toxin in the absence of VirB5, and efforts to detect a Ptl pilus have thus far been unsuccessful (9). Recent studies also have identified competence systems composed of homologs of putative type IV structural subunits but exclusive of pilus proteins (3, 25). Indeed, recent studies by Liu and Binns suggest that a VirB7-VirB9-VirB10 complex stimulates DNA uptake independently of the T pilus or other VirB proteins when synthesized in recipient cells in matings with agrobacterial donors and thus might also assemble as a functional conduit for DNA (Z. Liu and A. N. Binns, personal communication). If the VirB/VirD4 proteins in fact can assemble as distinct organelles, then the VirB6 and VirB11 uncoupling mutants might interact productively to drive assembly of one such structure while interfering with the formation or action of the second.
Chen et al. (12) recently presented biochemical findings that were interpreted as evidence for an alternative export route for type IV secretion substrates, distinct from that mediated by the VirB/D4 proteins. Such a route is not consistent with a large amount of genetic data (7, 44), yet it is formally possible that the uncoupling mutations promote utilization of an alternative secretion pathway. If so, however, it is difficult to envision why only very few mutations in the VirB proteins thus far have been shown to display such uncoupling phenotypes.
We envision three possible mechanisms by which VirB6 might exert its effects on pilus biogenesis and channel assembly or activity through its effects on VirB7 and VirB9 multimerization. Originally, we considered that VirB6 serves as a scaffold upon which newly secreted VirB7 and VirB9 monomers dock to optimize the alignment of reactive Cys residues for assembly of stabilizing intermolecular disulfide cross-links. This mechanism is supported by the findings that VirB6 production is important for accumulation of the VirB7 homodimer and VirB6 overproduction induces formation of higher-order VirB9 complexes or aggregates that are devoid of VirB7 (Fig. 4). However, our group (42) and others (Z. Liu and A. N. Binns, personal communication) have shown that VirB7 and VirB9 can still dimerize, albeit at low levels, when produced in a strain with deletions of other VirB proteins. The low abundance of the VirB7 dimers independently of VirB6 could be due to the absence of a VirB6 scaffold function but suggests a second possible mechanism whereby VirB6 interacts with the disulfide cross-linked forms of these proteins to direct later stages of machine assembly. In this regard, it is notable that the VirB7 homodimer sorts across the outer membrane where it associates with the T pilus and also is recovered at high levels in the culture supernatant (40), whereas the VirB7-VirB9 heterodimer sorts to the outer membrane (19, 20). Interestingly, the A. tumefaciens genome is devoid of homologs for all of the Lol proteins that recently have been shown to direct lipoprotein sorting in E. coli (49), with the possible exception of a LolD homolog (GenBank accession no. gi25384591). A. tumefaciens therefore almost certainly uses a Lol-independent pathway for sorting of its lipoproteins, and VirB6 might be recruited for sorting of the VirB7 multimers.
Third, though we favor a morphogenetic role for VirB6, especially for the T pilus, the available data do not exclude the possibility of a regulatory role for channel activity through modulation of the conformational and/or oligomeric status of VirB7 and VirB9. If so, however, VirB6 most probably exerts its regulatory action as a peripherally associated component of the secretion channel, as depicted in recent models (27, 45). By use of two independent biochemical approaches, we were unable to demonstrate a stable association of VirB6 with VirB10, which is a strong candidate for functioning as a structural subunit of the secretion machine (6, 7, 8, 16, 20). Moreover, we have determined that all of the VirB6.i4 mutant proteins characterized in this study as well as a series of VirB6 fusions to reporter proteins to be described elsewhere are phenotypically silent when produced in otherwise wild-type A348 cells. These recessive phenotypes contrast strongly with the strong negative dominance, as well as destabilizing effects on other VirB proteins accompanying expression of virB7-phoA and virB10-phoA chimeras in otherwise wild-type A348 cells (19; S. Jakubowski, unpublished data). Additionally, although VirB6 forms higher-order complexes detectable by gel filtration chromatography, its fractionation profile is clearly distinct from the profiles of VirB7, VirB9, and VirB10 (27). Finally, recent studies by Liu and Binns suggest that VirB6 is not required for VirB-mediated stimulation of DNA uptake in agrobacterial recipient cells during conjugation, consistent with the notion that VirB6 configures the VirB proteins as a dedicated export machine (Z. Liu and A. N. Binns, personal communication). It is of interest that several properties of VirB6—its polytopic membrane topology, probable peripheral association with the type IV machine, and proposed activity as a mediator of organellar biogenesis—are reminiscent of features ascribed to the YidC/Axl/Oxb family of proteins, whereby the prokaryotic homolog, YidC, mediates assembly of the SecYEG translocon (33).
VirB8, a presumptive nucleation factor.
Recently reported yeast (16) and E. coli (18) dihybrid data supplied evidence for VirB8 interactions with VirB9 and VirB10. Additionally, Kumar et al. (29) have shown that the assembly of VirB9 and VirB10 as foci on the cell surface is dependent on VirB8, prompting a proposal that VirB8 functions as a nucleating center for this type IV secretion machine. However, we were unable to demonstrate interactions between VirB8 and other VirB proteins in detergent extracts of A. tumefaciens membranes. We also did not detect strong stabilizing effects of the putative structural subunits, e.g., VirB7, VirB9, and VirB10, on VirB8 or vice versa (Fig. 4 and data not shown) (7, 20), and previously our group supplied evidence that VirB8 can function substoichiometrically to mediate substrate transfer (20). The recent gel filtration studies of Krall et al. (27) also showed that VirB8 displays a distinct and much broader fractionation profile than VirB7, VirB9, or VirB10, suggestive of formation of heterogeneous complexes. Collectively, these observations suggest that VirB8 might mediate assembly of this secretion system via transient interactions with different VirB proteins or protein subassemblies. In this light, it is intriguing to note that a recent dihybrid screen supplied evidence for an interaction between VirB8 and VirB1, a putative transglycosylase (46). It is interesting to speculate that VirB8 functions as a morphogenetic factor by successively recruiting VirB1 for localized lysis of the murein layer and then the VirB structural subunits for machine assembly.
Summary.
In summary, we propose that VirB6 coordinates its activities with VirB8 to direct formation and positioning of VirB7 and VirB9 complexes required for biogenesis of a type IV secretion channel and T pilus. VirB6 might further participate in the dynamics of substrate secretion through a weak-affinity association with the secretory apparatus. Finally, VirB6's contributions to channel assembly or function and the biogenesis of a wild-type T pilus can be separated by mutation.
Acknowledgments
We thank laboratory members for helpful discussions and Evgeni Sagulenko, Vita Sagulenko, and Zhenming Zhao for excellent technical help.
Work in our laboratory is supported by NIH grant GM48746.
REFERENCES
- 1.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. USA 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, K. G., W. A. Klimke, J. Manchak, and L. A. Frost. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C., N. Domke, M. Beinhofer, and S. Hapfelmeier. 2001. Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 183:6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron, C., Y. R. Thorstenson, and P. C. Zambryski. 1997. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol. 179:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beaupré, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohne, J., A. Yim, and A. N. Binns. 1998. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc. Natl. Acad. Sci. USA 95:7057-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 10.Calamia, J., and C. Manoil. 1990. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc. Natl. Acad. Sci. USA 87:4937-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C.-Y., and S. C. Winans. 1991. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J. Bacteriol. 173:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., C. M. Li, and E. W. Nester. 2000. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc. Natl. Acad. Sci. USA 97:7545-7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally-related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 15.Dang, T. A., X.-R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol. Microbiol. 32:1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, A., and Y.-H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dassa, E., and S. Muir. 1993. Membrane topology of MalG, an inner membrane protein from the maltose transport system of Escherichia coli. Mol. Microbiol. 7:29-38. [DOI] [PubMed] [Google Scholar]
- 18.Ding, Z., Z. Zhao, S. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, D., T. A. Dang, G. M. Spudich, X.-R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez, D., G. M. Spudich, X.-R. Zhou, and P. J. Christie. 1996. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J. Bacteriol. 178:3168-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullner, K. J. 1998. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J. Bacteriol. 180:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 23.Gouffi, K., C. L. Santini, and L. F. Wu. 2002. Topology determination and functional analysis of the Escherichia coli TatC protein. FEBS Lett. 525:65-70. [DOI] [PubMed] [Google Scholar]
- 24.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 26.Jin, S. G., T. Komari, M. P. Gordon, and E. W. Nester. 1987. Genes responsible for the supervirulence phenotype of Agrobacterium tumefaciens A281. J. Bacteriol. 169:4417-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, R. B., Y. H. Xie, and A. Das. 2000. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 36:608-617. [DOI] [PubMed] [Google Scholar]
- 30.Lai, E. M., O. Chesnokova, L. M. Banta, and C. I. Kado. 2000. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J. Bacteriol. 182:3705-3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai, E. M., and C. I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180:2711-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Luirink, J., T. Samuelsson, and J. W. de Gier. 2001. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501:1-5. [DOI] [PubMed] [Google Scholar]
- 34.Moore, D., K. Maneewannakul, S. Maneewannakul, J. H. Wu, K. Ippen-Ihler, and D. E. Bradley. 1990. Characterization of the F-plasmid conjugative transfer gene traU. J. Bacteriol. 172:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otten, L., H. De Greve, J. Leemans, R. Hain, P. Hooykaas, and J. Schell. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195:159-163. [Google Scholar]
- 36.Pantoja, M., L. Chen, Y. Chen, and E. W. Nester. 2002. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 45:1325-1335. [DOI] [PubMed] [Google Scholar]
- 37.Rashkova, S. 1998. Ph.D. thesis. The University of Texas-Houston Medical School, Houston.
- 38.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Mutational analysis of the Agrobacterium tumefaciens VirB11 ATPase: identification of functional domains and evidence for multimerization. J. Bacteriol. 179:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:37-45. [Google Scholar]
- 42.Spudich, G. M., D. Fernandez, X.-R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. USA 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stachel, S. E., and E. W. Nester. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti. EMBO J. 5:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. de Vlaam, T. J. Regensburg-Tuink, and P. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979-982. [DOI] [PubMed] [Google Scholar]
- 45.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Inaugural article: peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward, J. E., D. E. Akiyoshi, D. Reiger, A. Datta, M. P. Gordon, and E. W. Nester. 1988. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J. Biol. Chem. 263:5804-5814. [PubMed] [Google Scholar]
- 47.Ward, J. E., Jr., E. M. Dale, P. J. Christie, E. W. Nester, and A. N. Binns. 1990. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J. Bacteriol. 172:5187-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson, B., T. C. Currier, M. P. Gordon, M. D. Chilton, and E. W. Nester. 1975. Plasmid required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 123:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yakushi, T., K. Masuda, S. Narita, S. Matsuyama, and H. Tokuda. 2000. A new ABC transporter mediating the detachment of lipid-modified proteins from membranes. Nat. Cell Biol. 2:212-218. [DOI] [PubMed] [Google Scholar]
- 50.Zhao, Z., E. Sagulenko, Z. Ding, and P. J. Christie. 2001. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J. Bacteriol. 183:3855-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, X.-R., and P. J. Christie. 1999. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 181:4342-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]