Development is not restricted to so-called higher organisms. Developmental processes in bacteria include differentiation of a single cell, such as the swarmer-to-stalk transition by Caulobacter crescentus and spore formation by Bacillus subtilis. Other microbes, such as Myxococcus xanthus, produce specialized cells within a population to form sporulating fruiting bodies (19). As we learn more about microbial biofilm formation, it is becoming clear that this is yet another example of a bacterial developmental process (4, 12). Like other developmental systems, building a biofilm requires a series of discrete and well-regulated steps. While the exact molecular mechanisms may differ from organism to organism, the stages of biofilm development appear to be conserved among a wide range of microbes. These stages include attachment of cells to a substrate, the growth and aggregation of cells into microcolonies, and the maturation and maintenance of architecture (3-5) (Fig. 1). In this issue of the Journal of Bacteriology, Finelli and colleagues at the Hospital for Sick Children in Toronto demonstrate the use of a new tool for further dissecting the development of bacterial biofilms (8).
FIG. 1.
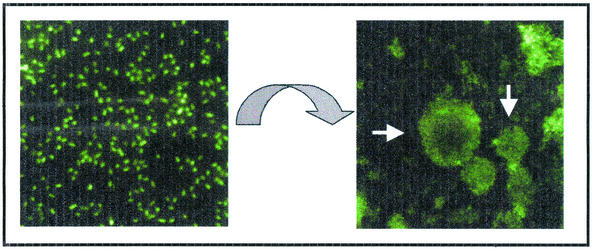
How do bacteria control the transition from free-swimming planktonic cells to life in a multibacterial community? On the left is shown fluorescently labeled planktonic P. aeruginosa at ×1,400 magnification. Within a few days, these individual microbes can form the community pictured on the right. This top-down fluorescent image shows a mature 5-day-old biofilm grown in a flowing system at ×230 magnification. The macrocolonies shown here (white arrows) vary from ∼35 to more than 50 μm in height.
Bacterial developmental processes contribute to the success of a microbe in its environment. In the case of M. xanthus, development of the fruiting body includes a division of labor between sporulating cells, which give rise to the seeds of future generations, and the stalk cells, whose final act is to lift these spores above the substratum and aid in their dispersal. Biofilms also enable specialization of cells within a population, as was shown by Branda et al., who demonstrated spore-specific gene expression within localized regions of B. subtilis biofilms (1). Despite recent progress, it is apparent that we still have a great deal to learn about the mechanisms of bacterial biofilm construction and the interplay among phenotypically distinct subpopulations within these communities.
Finelli and colleagues took advantage of a well-known technique called IVET (for “in vivo expression technology”) (10) and adapted the system to study genes expressed in a mature biofilm. Three new genes required for biofilm formation by Pseudomonas aeruginosa were identified by their application of this system, called IBET (for “in-biofilm expression technology”). A key element of the IBET system is a strict nutritional requirement to allow the selection of biofilm-expressed promoters that rescue this auxotrophy. These researchers looked for promoters whose expression rescued the adenine deficiency of a P. aeruginosa ΔpurKE mutant that grows in a biofilm but not on minimal agar medium lacking adenine. Their approach was validated by mutational analysis when they found that three of the five biofilm-expressed genes were required for biofilm formation but played no role in planktonic growth. This study represents the first time that such an approach has been used to study development in a microbial system.
Like other developmental pathways, P. aeruginosa biofilm development is controlled by a number of different regulators, including LasR, RhlR, GacA, RpoS, Crc, and PvrR (6, 23; reviewed in reference 4). A new regulator can now be added to this list, encoded by the open reading frame designated PA3782 (based on the P. aeruginosa genome project, www.Pseudomonas.com), which appears to code for a transcriptional regulator of the AraC-XylS family. These transcription factors have been well studied as regulators of carbon metabolism and are also known to control virulence factor expression by numerous pathogens (7). While there is at least one example of a AraC-XylS family regulator participating in microbial developmental—the PA3782 homolog AdpA is required for the formation of aerial hyphae in Streptomyces griseus (11)—a role for this large family of transcription factors in microbial development may be underappreciated. Of the ∼800 potential family members identified as of 2002 (7), many of which have no known function (24), it would come as no surprise if at least some participated in the regulation of biofilm formation and/or other developmental pathways.
One important problem facing biofilm researchers is trying to understand the changes in metabolism bacteria undergo as they adapt to life in these communities. The second gene required for biofilm development identified by Finelli and colleagues, PA3701, is predicted to encode an alcohol dehydrogenase or oxidoreductase. It is likely that this enzyme is necessary for the physiological adaptation of P. aeruginosa to the biofilm mode of life. This protein can be added to a long list of other enzymes involved in amino acid biosynthesis, carbon metabolism, and respiration that were previously identified by genomic and proteomic approaches as being expressed within the context of a biofilm (18, 26). While understanding the role that such enzymes play in biofilms is often difficult, and more often ignored, this information may be key to getting a firm grip on the changes microbes undergo as they adapt to life in a community.
The third biofilm gene identified (PA0240, designated opdF) codes for a predicted porin based on its similarity to the OprD family of porins. Although the function of OpdF in biofilm formation remains to be elucidated, potential roles for this membrane protein include transport of signaling molecules, nutrients, or metabolic products. Alternatively, this protein may be needed to adapt to conditions particular to a biofilm, such as high osmolarity (15), or may serve as adhesin, as has been predicted for other surface structures, such flagella, pili, the predicted cup-encoded fimbriae, and the AggA outer membrane protein (OMP) (2, 16, 17, 20, 25). Whatever its function, OpdF is not the first porin or OMP required for biofilm development by pseudomonads. Yoon and colleagues demonstrated that the OMP OprF is necessary for the anaerobic growth of P. aeruginosa biofilms (27). Furthermore, proteomic and microarray studies suggested that other OMPs are up-regulated in biofilm versus planktonic cells, although there is no direct evidence that these surface factors are needed for biofilm development (18, 26).
Pseudomonads, and in particular P. aeruginosa, are among the best-studied biofilm formers, due in large part to the long history of study of these organisms, their genetic tractability, and the availability of the genome sequence of the laboratory strain PAO1 (22). Many tactics have been employed to get a handle on the genes required for biofilm development by P. aeruginosa. Genetic approaches have been successful at identifying the factors involved in the early steps in biofilm development, such as attachment to a surface and the formation of microcolonies (14, 25). While proteomic and genomic studies have examined gene and protein expression at all stages of biofilm formation (18, 26), the functional roles of these factors are still being characterized. Finelli and colleagues have taken expression studies a step further by demonstrating that three of the five genes identified in the original IBET screen as being expressed in a biofilm are also necessary for biofilm development. The next goal should be to determine the specific stages in the development of a biofilm wherein these genes are required. That is, what functions do these cells lack that confer the observed defects in biofilm formation—are these mutant bacteria unable to establish early surface contacts, are these contacts unstable, or have cell-to-cell interactions been disrupted, etc.? The phenotype of each of these mutants is somewhat different, indicating that they may participate in different aspects of community formation.
The genes identified in this screen are by necessity expressed throughout biofilm development because the bacteria are required to produce endogenous adenine throughout the 5-day period of biofilm growth and maturation. Genes, and their products, that play only a transient role in biofilm formation would probably not have been uncovered. Expression of the IBET-identified genes throughout development suggests that they may be important at multiple steps in the biofilm pathway. These observations and the experiences of other developmental microbiologists illustrate the need to be able to turn genes on and off throughout biofilm formation to study their function at discrete steps throughout this process. The technology to perform such studies awaits future development.
One fascinating note is the lack of overlap between the genes identified in this study and genes found by using other genetic, genomic, and/or proteomic approaches. I suspect that this observation means two things. First, we are just scratching the surface of the functions required for making and maintaining a biofilm. Second, bacteria have more than one way to make a biofilm. It seems likely that particular environmental stimuli (growth media or the substratum, etc.) trigger different developmental pathways, all culminating with the same end point, a mature biofilm. Earlier studies suggested that there are multiple pathways for biofilm development in Pseudomonas fluorescens and that these pathways may be, in part, regulated by the availability of particular carbon sources or other nutrients (13). We should keep this concept of multiple biofilm pathways in mind as we consider more complex communities, some of which may be comprised of multiple microbial species. For example, recent work by Hogan and Kolter demonstrated biofilm formation by P. aeruginosa on the fungus Candida albicans (9), and Singh and colleagues provided evidence for biofilms of P. aeruginosa in the cystic fibrosis lung (21). Are these biofilms the same as or different from a community formed on an abiotic surface? It is quite possible that there is more than one way to make a biofilm, and I think it would be unwise to discount that such complexity is outside the repertoire of the microorganism! When the information from multiple biofilm studies utilizing a variety of approaches is integrated, we'll gain a more complete view of biofilm development by P. aeruginosa.
Acknowledgments
Many thanks go to D. A. Hogan for her input on this commentary.
Work in the O'Toole lab is supported by the NSF (9984521); the Cystic Fibrosis Foundation (O'TOOLE01GO); Microbia, Inc.; and the Pew Charitable Trusts. G.A.O. is a Pew Scholar in the Biomedical Sciences.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Branda, S. S., J. E. Gonzalez-Pastor, S. Ben-Yehuda, R. Losick, and R. Kolter. 2001. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:11621-11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buell, C. R., and A. J. Anderson. 1992. Genetic analysis of the aggA locus involved in agglutination and adherence of Pseudomonas putida, a beneficial fluorescent pseudomonad. Mol. Plant Microbol Interact. 5:154-162. [DOI] [PubMed] [Google Scholar]
- 3.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davey, M. E., and G. O. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 6.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 7.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finelli, A., C. V. Gallant, K. Jarvi, and L. L. Burrows. 2003. Use of in-biofilm expression technology to identify genes involved in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:2700-2710 [DOI] [PMC free article] [PubMed]
- 9.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 10.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 11.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 12.O'Toole, G. A., H. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 13.O'Toole, G. A., and R. Kolter. 1998. The initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 14.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms, p. 91-109. In R. J. Doyle (ed.), Methods in enzymology, vol. 310. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 15.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Eschericia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prince, A. 1992. Adhesins and receptors of Pseudomonas aeruginosa associated with infection of the respiratory tract. Microb. Pathog. 13:251-260. [DOI] [PubMed] [Google Scholar]
- 17.Saiman, L., K. Ishimoto, S. Lory, and A. Prince. 1990. The effects of piliation and exoproduct expression on the adherence of Pseudomonas aeruginosa to respiratory epithelial monolayers. J. Infect. Dis. 161:541-548. [DOI] [PubMed] [Google Scholar]
- 18.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimkets, L. J., and Y. V. Brun. 1999. Prokaryotic development: Strategies to enhance survival, p. 1-7. In L. J. Shimkets and Y. V. Brun (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 20.Simpson, D. A., R. Ramphal, and S. Lory. 1995. Characterization of Pseudomonas aeruginosa fliO, a gene involved in flagellar biosynthesis and adherence. Infect. Immun. 63:2950-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 22.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 23.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 27.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms. Relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]