Abstract
An enzymatic pathway for synthesis of 5-phospho-d-ribosyl α-1-diphosphate (PRPP) without the participation of PRPP synthase was analyzed in Escherichia coli. This pathway was revealed by selection for suppression of the NAD requirement of strains with a deletion of the prs gene, the gene encoding PRPP synthase (B. Hove-Jensen, J. Bacteriol. 178:714-722, 1996). The new pathway requires three enzymes: phosphopentomutase, ribose 1-phosphokinase, and ribose 1,5-bisphosphokinase. The latter activity is encoded by phnN; the product of this gene is required for phosphonate degradation, but its enzymatic activity has not been determined previously. The reaction sequence is ribose 5-phosphate → ribose 1-phosphate → ribose 1,5-bisphosphate → PRPP. Alternatively, the synthesis of ribose 1-phosphate in the first step, catalyzed by phosphopentomutase, can proceed via phosphorolysis of a nucleoside, as follows: guanosine + Pi → guanine + ribose 1-phosphate. The ribose 1,5-bisphosphokinase-catalyzed phosphorylation of ribose 1,5-bisphosphate is a novel reaction and represents the first assignment of a specific chemical reaction to a polypeptide required for cleavage of a carbon-phosphorus (C—P) bond by a C-P lyase. The phnN gene was manipulated in vitro to encode a variant of ribose 1,5-bisphosphokinase with a tail consisting of six histidine residues at the carboxy-terminal end. PhnN was purified almost to homogeneity and characterized. The enzyme accepted ATP but not GTP as a phosphoryl donor, and it used ribose 1,5-bisphosphate but not ribose, ribose 1-phosphate, or ribose 5-phosphate as a phosphoryl acceptor. The identity of the reaction product as PRPP was confirmed by coupling the ribose 1,5-bisphosphokinase activity to the activity of xanthine phosphoribosyltransferase in the presence of xanthine, which resulted in the formation of 5′-XMP, and by cochromatography of the reaction product with authentic PRPP.
NAD biosynthesis in Escherichia coli usually proceeds by consumption of 5-phospho-d-ribosyl α-1-diphosphate (PRPP). NAD is synthesized from aspartate and dihydroxyacetone phosphate. A de novo pathway and a number of salvage pathways for the reutilization of nicotinamide mononucleotide and nicotinamide exist, as shown in Fig. 1 (32). Two of the enzymatic reactions, the reactions catalyzed by quinolinate and nicotinate phosphoribosyltransferases, require PRPP. PRPP-less mutants with a deletion of the prs gene, encoding PRPP synthase, consequently require NAD or nicotinamide mononucleotide. Δprs strains also require guanosine, uridine, histidine, and tryptophan, which are likewise synthesized with PRPP as an intermediate (14, 15). Nevertheless, mutants that suppress the NAD requirement are easily obtained by selecting for growth of Δprs cells on medium lacking NAD. These mutants still require guanosine, uridine, histidine, and tryptophan. All such NAD-suppressed mutants were previously shown to have lesions in the pst-phoU operon (17), which leads to high-level constitutive expression of genes belonging to the phosphate (Pho) regulon (39, 40). Furthermore, pst, phoU, or phoR also suppressed the NAD requirement of Δprs strains. Suppression by pst, phoU, or phoR mutations was eliminated by a phoB mutation, while suppression by a phoR mutation was also eliminated by a creC mutation. These data show that synthesis of a Pho regulon gene product is apparently responsible for suppression of the NAD requirement (17).
FIG. 1.
NAD metabolism of E. coli. The de novo and salvage pathways are shown. Abbreviations: Asp, aspartate; DHAP, dihydroxyacetone phosphate; Gln, glutamine; Glu, glutamate; NA, nicotinate; NAm, nicotinamide; NAMN, nicotinate mononucleotide; NAAD; nicotinate adenine dinucleotide; NMN, nicotinamide mononucleotide; QA, quinolinate; Rib-5P, ribose 5-phosphate. Relevant enzyme-catalyzed reactions are indicated by gene designations (nadA, quinolinate synthase; nadC, quinolinate phosphoribosyltransferase; pncB, nicotinate phosphoribosyltransferase).
We show here that suppression of the NAD requirement requires at least three polypeptides, one of which is specified by the phnN gene. The phnN gene lies within a 14-cistron operon, phnCDEFGHIJKLMNOP, which encodes proteins for uptake, breakdown, and regulation of phosphonate degradation by carbon-phosphorus (C-P) lyase (7, 29, 39). In this paper we describe the first detection of an in vitro activity associated with a component of a C-P lyase.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The E. coli K-12 strains used are listed in Table 1. Cells were grown in AB minimal medium (8) or in NZY medium (containing [per liter] 10 g of NZ-amin [Struers, Copenhagen, Denmark], 5 g of yeast extract [Oxoid], and 5 g of NaCl; pH adjusted to 7.5) (18). Carbon sources were used at a concentration of 0.2% (glucose, galactose, melibiose) or 0.1% (thymidine). Thiamine (1 mg/liter) was routinely added to minimal media. The following other compounds were added when necessary: guanosine (30 mg/liter), uridine (20 mg/liter), and amino acids and δ-aminolevulinate (40 mg/liter each). NAD was added to both NZY and AB media at a concentration of 25 mg/liter. Ampicillin was used at a concentration of 25 or 100 mg/liter, whereas kanamycin and tetracycline were used at concentrations of 30 and 10 mg/liter, respectively. Cell growth was monitored by determining the optical density at 436 nm (OD436) with an Eppendorf PCP6121 spectrophotometer. An OD436 of 1 (path length, 1 cm) corresponds to approximately 3 × 1011 cells per liter.
TABLE 1.
Bacterial strains
| Strain | Genotype | Source, reference, or construction |
|---|---|---|
| BW12057 | araD Δ(lac)U169 rpsL aroB crp creC thi psiD31::lacZ (MudI-1734) | Laboratory strain |
| BW14001 | Δ(lac)χ74 ΔphoA532 Δ(mel-proP-phnCDEFGHIJKLMNOP)2::Tn5seq1/132(tet) | 28 |
| BW14894 | Δ(lac)χ74 Δ(phnC?DEFGHIJKLMNOP)33-30 | 43 |
| BW15089 | Δ(lac)χ74 robA creC Δ(gal-attλ-bio)76 zbh-283::Tn10 | Laboratory strain |
| BW17471 | Δ(lac)χ74 ΔphoA532 phnM28::TnphoA′-1 phn-10::uidA2-aadA | Laboratory strain |
| BW17562 | Δ(lac)χ74 ΔphoA532 phnF23::TnphoA′-9 phn-10::uidA2-aadA | Laboratory strain |
| BW18524 | Δ(lac)χ74 ΔphoA532 phnCpΔ(phnCDE)59 | Laboratory strain |
| BW20274 | Δ(lac)χ74 ΔphoA532 phnN45::TnphoA′-3 phn-10::uidA2-aadA | Laboratory strain |
| BW24388 | Δ(lac)χ74 Δ(phnC?DEFGHIJKLMNOP)33-30 attλ::pCAH56 | Laboratory strain |
| BW24389 | Δ(lac)χ74 Δ(phnC?DEFGHIJKLMNOP)33-30 attλ::pAH126 (Ptac-phnN) | Laboratory strain |
| CAG5051 | thi relA spoT supQ nadA::Tn10 | C. Grossb |
| CAG18430 | zjj-202::Tn10 | C. Grossb |
| HO340 | —a | 31 |
| HO402 | metB zcg-2402::Tn10 | 13 |
| HO650 | supF relA spoT rpsL lamB metB deoD udp gsk-3 pncA hemA | 15 |
| HO677 | zcg::Tn10 supF relA spoT rpsL lamB metB deoD udp gsk-3 pncA hemA | P1(HO402) × HO650, Tetr |
| HO770 | deoD gsk-3 udpa | 33 |
| HO773 | deoD gsk-3 udp Δ(prs-4)::Kanra | 33 |
| HO861 | deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | 17 |
| HO878 | deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001 nadA::Tn10a | 17 |
| HO959 | zjj-202::Tn10 Δ(deoCABD) Δlac thi upp udp ton | P1(CAG18430) × SØ928, Tetr |
| HO965 | zjj-202::Tn10 Δ(deoCABD) gsk-3 udp Δ(prs-4)::Kanra | P1(HO959) × HO773, Tetr |
| HO967 | zjj-202::Tn10 Δ(deoCABD) gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(HO959) × HO861, Tetr |
| HO1077 | supF relA spoT rpsL lamB metB guaA deoD deoB thyA zjj-202::Tn10 | P1(CAG18430) × SØ489, Tetr |
| HO1088 | deoD gsk-3 udp Δ(prs-4)::Kanr/F zzf::Tn10 lacIq | 23 |
| HO1123 | zjj-202::Tn10 thi galE Δ(attλ-bio) deoC56 deoB::λcI857 | P1(CAG18430) × SØ1055, Tetr |
| HO1150 | zjj-202::Tn10 Δ(gal-deoR) thi bio upp deoC+A+B+ Δ(deoD-thr) | P1(CAG18430) × SØ1068, Tetr |
| HO1156 | zjj-202::Tn10 Δ(deoD-thr) gsk-3 udp Δ(prs-4)::Kanra | P1(HO1150) × HO861, Tetr |
| HO1171 | zjj-202::Tn10 thi galE Δ(attλ-bio) deoA103 deoC lysA argA cytR | P1(CAG18430) × SØ1108, Tetr |
| HO1183 | zjj-202::Tn10 deoA103 deoD gsk-3 udpa | P1(HO1171) × HO770, Tetr |
| HO1187 | zjj-202::Tn10 deoA103 deoD gsk-3 udp Δ(prs-4)::Kanra | P1(HO1183) × HO861, Tetr |
| HO1357 | zjj-202::Tn10 deoCa | P1(HO1123) × HO340, Tetr |
| HO1358 | zjj-202::Tn10 deoC deoD gsk-3 udpa | P1(HO1357) × HO770, Tetr |
| HO1359 | zjj-202::Tn10 deoC deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(HO1358) × HO861, Tetr |
| HO1403 | hemA zcg::Tn10 deoD gsk-3 udp (pst-phoU)1001a | P1(HO677) × HO861, Tetr |
| HO1404 | psiD31::lacZ (MudI-1734) hemA zcg::Tn10 deoD gsk-3 udp (pst-phoU)1001a | P1(BW12057) × HO1403, Kanr |
| HO1408 | psiD31::lacZ (MudI-1734) deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(HO773) × HO1404, Hem+ |
| HO1494 | Δ(mel-proP-phnCDEFGHIJKLMNOP)2::Tn5seq1/132(tet) deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW14001) × HO861, Tetr |
| HO1496 | phnCpΔ(phnCDE)59 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW18524) × HO1494, Mel+ |
| HO1559 | phnM28::TnphoA′-1 phn-10::uidA2-aadA deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW17471) × HO1494, Mel+ |
| HO1565 | phnF23::TnphoA′-9 phn-10::uidA2-aadA deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW17562) × HO1494, Mel+ |
| HO1569 | phnN45::TnphoA′-3 phn-10::uidA2-aadA deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW20274) × HO1494, Mel+ |
| HO1652 | Δ(gal-attλ-bio)76 zbh-283::Tn10 deoD gsk-3 udp Δ(prs-4)::Kanra | P1(BW15089) × HO773, Tetr |
| HO1654 | Δ(phnC?DEFGHIJKLMNOP)33-30 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW14894) × HO1494, Mel+ |
| HO1655 | Δ(gal-attλ-bio)76 zbh-283::Tn10 Δ(phnC?DEFGHIJKLMNOP)33-30 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW15089) × HO1654, Tetr |
| HO1877 | attλ::pCAH56 Δ(phnC?DEFGHIJKLMNOP)33-30 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW24388) × HO1655, Gal+ |
| HO1878 | attλ::pAH126 (Ptac-phnN) Δ(phnC?DEFGHIJKLMNOP)33-30 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(BW24389) × HO1655, Gal+ |
| HO1879 | attλ::pCAH56 deoD gsk-3 udp Δ(prs-4)::Kanra | P1(BW24388) × HO1652, Gal+ |
| HO1902 | attλ::pAH126 (Ptac-phnN) deoD gsk-3 udp Δ(prs-4)::Kanr | P1(BW24389) × HO1652, Gal+ |
| HO1908 | nadA::Tn10 attλ::pAH126 (Ptac-phnN) Δ(phnC?DEFGHIJKLMNOP)33-30 deoD gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(CAG5051) × HO1878, Tetr |
| HO1909 | nadA::Tn10 attλ::pAH126 (Ptac-phnN) deoD gsk-3 udp Δ(prs-4)::Kanra | P1(CAG5051) × HO1902, Tetr |
| HO1910 | zjj-202::Tn10 Δ(deoCABD) attλ::pAH126 (Ptac-phnN) Δ(phnC?DEFGHIJKLMNOP)3-30 gsk-3 udp Δ(prs-4)::Kanr (pst-phoU)1001a | P1(HO959) × HO1878, Tetr |
| HO1911 | zjj-202::Tn10 Δ(deoCABD) attλ::pAH126 (Ptac-phnN) gsk-3 udp Δ(prs-4)::Kanra | P1(HO959) × HO1902, Tetr |
| HO1935 | Δ(deoBD)::KanrrecD1903::mini-Tet Δ(lac)χ74 ΔphoA phnCp(phnCDE)59 | Homologous recombination with linearized DNA of pHO370, Kanr, via TP1904 |
| HO1938 | Δ(deoBD)::KanrhemA zcg::Tn10 gsk-3 udp (pst-phoU)1001a | P1(HO1935) × HO1403, Kanr |
| HO1949 | Δ(prs-4)::Kanr Δ(deoBD)::Kanrgsk-3 udp (pst-phoU)1001a | P1 (HO773) × HO1938, Hem+ |
| MC1061 | araD139 Δ(ara-leu) Δ(lac)χ74 galE15 galK16 rpsL | 6 |
| SØ489 | supF relA spoT rpsL lamB metB guaA deoD deoB thyA | P. Nygaard, University of Copenhagen |
| SØ928 | Δ(deoCABD) Δlac thi upp udp ton | K. Hammerc |
| SØ1055 | thi galE Δ(attλ-bio) deoC56 deoB::λcI857 | K. Hammer, Danish Technical University |
| SØ1068 | Δ(gal-deoR) thi bio upp deoC+A+B+ Δ(deoD-thr) | K. Hammer, Danish Technical University |
| SØ1108 | thi galE Δ(attλ-bio) deoA103 deoC lysA argA cytR | K. Hammer, Danish Technical University |
| TP1904 | recD1903::mini-Tet Δ(lac)χ74 ΔphoA phnCp(phnCDE)59 | T. Poulsen, University of Copenhagen |
The strain also contains araC(Am) araD Δ(lac)U169 trp(Am) mal(Am) rpsL relA thi supF.
See reference 36.
See reference 38.
Genetic techniques.
Previously described procedures were used for transduction (30) and transformation (26). Gene conversion by homologous recombination was performed by using a recD strain (35). Integration of pAH126 at attλ was performed as described elsewhere (10).
DNA technology and plasmids.
Plasmid DNA was isolated by the procedure of Birnboim and Doly (4) or by commercial procedures (Qiagen). Restriction and ligation of DNA were performed as described by the vendors of restriction endonucleases (Amersham, Promega, and Roche) or T4 DNA ligase (Amersham). The plasmids containing deo cistrons were constructed as follows. Plasmid pBRdeo, provided by J. Neuhard (University of Copenhagen), consists of the entire deo operon (deoC+ deoA+ deoB+ deoD+) as a 5,660-bp EcoRI-PvuII-digested DNA fragment ligated to similarly digested DNA of pBR322 (5). To delete deoD, DNA of pBRdeo was digested by restriction endonucleases PvuII and HpaI, and this was followed by ligation, resulting in pHO365 (deoC+ deoA+ deoB+). To delete deoB and deoD, DNA of pBRdeo was digested by restriction endonucleases PvuII and HindIII, and this was followed by S1 nuclease treatment and ligation, resulting in pHO366 (deoC+ deoA+). To delete deoA, deoB, and deoD, DNA of pBRdeo was digested by restriction endonucleases PvuII and NcoI, and this was followed by S1 nuclease treatment and ligation, resulting in pHO367 (deoC+). The deo operon is expressed from the deoP2 promoter in these plasmids (37). To construct a plasmid containing only deoB, DNA of pBRdeo was digested by restriction endonuclease BclI. This treatment liberated a 1,551-bp DNA fragment containing deoB, as well as four other DNA fragments. This DNA was ligated to BamHI-digested DNA of pBR322. Recombinant plasmids containing deoB+ were obtained by transformation of the ligated DNA to strain HO1077 (deoB) and selection for deoB+ on medium with thymidine as the carbon source. The resulting plasmid, which presumably expressed deoB from the tet promoter, was pHO368 (deoB+). To replace deoB and deoD with a kanamycin resistance-encoding DNA fragment, DNA of pBRdeo was digested by restriction endonuclease HindIII, and this was followed by S1 nuclease treatment and digestion with HpaI. This DNA was ligated to DNA of pUC4-K (44) previously digested with HindII, which liberated the kanamycin resistance-encoding DNA fragment, as well as two other DNA fragments. The desired plasmid was obtained by transformation of the ligated DNA to strain HO1183 (deoA) and selection for deoA+ on medium containing kanamycin and thymidine as the carbon source. This procedure resulted in pHO370 (deoC+ deoA+ ΔdeoBD::Kanr). This plasmid contained approximately 1,000 bp downstream of the HpaI site to allow for homologous recombination.
Plasmids containing phnN were constructed as follows. Wild-type phnN was PCR amplified by using the oligodeoxyribonucleotides 5′-GAAGATCTCAT-ATGATGGGAAAAACTGATTTGG and 5′-GCGAGCTCT-ACAAGCAGGCATGGTGTTTC as the primers, DNA of pBW120 (41) as the template, the four deoxyribonucleotides, and Vent DNA polymerase (New England Biolabs) (the nucleotides preceding hyphens indicate noncomplementary extensions). The PCR product was cloned into pSK50-ΔuidA2 (9) by using NdeI and SacI, resulting in pCP-phnN. The insert was verified by automated DNA sequencing of both strands. The phnN gene was then subcloned into the conditional-replication and modular plasmid pCAH56 (10) by using NdeI and NheI, resulting in pAH126. This plasmid contained phnN under control of Ptac and could be readily integrated into the chromosome. In addition, phnN was amplified with the oligodeoxyribonucleotides 5′-GAGAATTCATTAAAGAGGAGAAATTAACT-ATGATGGGAAAACTGATTTGGTTAATGG (phnNupUHE) and 5′-TGGTTGGGATCCCGAGCCATGGTTATTACAAG-CAGGCATGGTGTTTCTCC as the primers, DNA of pAH126 as the template, the four deoxyribonucleotides, and DNA polymerase of Pyrococcus furiosum (Gibco-BRL). The resulting DNA was digested by restriction endonucleases EcoRI and NcoI, ligated to similarly digested DNA of pUHE23-2 (H. Bujard, personal communication), and transformed to strain MC1061, resulting in pHO500. A variant of phnN, specifying a polypeptide with a six-histidine tail at the carboxy-terminal end, was constructed by PCR with the oligodeoxyribonucleotides phnNupUHE and 5′-TGGTTGGGATCCCGAGCCATGGTTATTAATGGTGATGGTGATGGTGCAAG-CAGGCATGGTGTTTCTCC as the primers and DNA of pAH126 as the template, and this was followed by EcoRI and NcoI digestion, ligation, and transformation as described above for pHO500. The resulting plasmid was designated pTR553. The correct inserts of the plasmids were confirmed by automated nucleotide sequencing. Plasmid pGD248, a derivative of pSU18 (3), harbors a lac promoter-expressed xapAB operon, which encodes xanthosine phosphorylase and xanthosine permease (G. Dandanell, personal communication).
Expression of recombinant phnN and purification of histidine-tailed ribose 1,5-bisphosphokinase.
Strain HO1088 (Δprs)/pTR553 (phnN) was grown in NZY broth supplemented with ampicillin and tetracycline. At an OD436 of approximately 1, isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added at a concentration of 0.5 mM to induce expression of the phnN gene. After 2 h of incubation cells were harvested by centrifugation, washed with unsupplemented AB minimal medium, and broken by ultrasonic treatment. Debris was removed by centrifugation, and the supernatant fluid was applied to a Co2+-containing Talon metal affinity resin. After the column was washed, ribose 1,5-bisphosphokinase was eluted with imidazole as described by the resin supplier (Clontech).
Enzyme assays.
The activity of ribose 1,5-bisphosphokinase was assayed as follows. A 100-μl reaction mixture contained 50 mM Tris-HCl (pH 7.5), 0.6 mM ribose 1,5-bisphosphate, 1 mM (120-GBq/mol) [γ-32P]ATP, and 2 mM magnesium chloride. The reaction was initiated by the addition of ribose 1,5-bisphosphokinase (0.8 nmol/min). Samples (10 μl) were removed after 1, 3, and 10 min of incubation at 30°C and mixed with 5 μl of 0.33 M formic acid. This 15 μl was applied to polyethyleneimine-coated cellulose thin-layer chromatographic plates (Baker-flex; J. T. Baker). After drying, the chromatogram was developed in 0.85 M KH2PO4 which had been previously adjusted to pH 3.4 with 0.85 M H3PO4 (20). Radioactivity was quantitated with an Instant Imager (Packard). Occasionally, 1 mM (120-GBq/mol) [α-32P]ATP or 1 mM (120-GBq/mol) [α-32P]GTP was used instead of [γ-32P]ATP. Production of PRPP with purified E. coli PRPP synthase was performed as previously described (42). When unlabeled ribose 1,5-bisphosphokinase reaction product or PRPP synthase reaction product was synthesized, the same procedures were employed, except that the labeled nucleotide was omitted, and heating at 90°C for 1 min terminated the reaction. Consumption of labeled ribose 1,5-bisphosphokinase reaction product or PRPP synthase reaction product by xanthine phosphoribosyltransferase (XPRTase) was performed as follows. A 50-μl reaction mixture contained 40 mM Tris-HCl (pH 7.5), 0.1 mM xanthine, 32P-labeled ribose 1,5-bisphosphokinase reaction product or 32P-labeled PRPP (approximately 0.1 kBq), 10 mM magnesium chloride, and XPRTase (30 nmol/min). A 10-μl sample was removed after 10 min of incubation at 37°C, mixed with formic acid, and examined by thin-layer chromatography as described above. The formation of 5′-XMP with unlabeled ribose 1,5-bisphosphokinase reaction product or PRPP synthase reaction product was assayed by a similar procedure. A 50-μl reaction mixture contained 40 mM Tris-HCl (pH 7.5), 0.1 mM (185-GBq/mol) xanthine, unlabeled ribose 1,5-bisphosphokinase reaction product or PRPP synthase reaction product (the same volume as described above), 10 mM magnesium chloride, and XPRTase (30 nmol/min). A 10-μl sample was removed after 10 min of incubation at 37°C and applied to a polyethyleneimine thin-layer plate. After drying, the chromatogram was developed in methanol (2 cm), followed by 1 M acetic acid (2 cm) and 0.9 M acetic acid-0.3 M lithium chloride (16 cm) (16, 34). Protein content was determined as previously described (23).
Materials.
[α-32P]ATP (15 TBq/mmol) and [α-32P]GTP (110 TBq/mmol) were obtained from Amersham-Pharmacia Biotech. [γ-32P]ATP was prepared as previously described (20). [8-14C]xanthine was obtained from New England Nuclear. Unlabeled nucleotides were obtained from Roche. Xanthine, adenine, ribose, ribose 1-phosphate, ribose 5-phosphate, and PRPP were obtained from Sigma. Ribose 1,5-bisphosphate was supplied by H. Klenow (University of Copenhagen) (22). Purified recombinant Bacillus subtilis XPRTase was supplied by K. F. Jensen (University of Copenhagen).
RESULTS
Genes necessary for the NAD suppression phenotype.
It has been shown previously that the NAD suppression phenotype requires a wild-type allele of the nadC gene, which encodes quinolinate phosphoribosyltransferase. Similarly, utilization of nicotinate by cells with the NAD suppression phenotype requires a wild-type allele of the pncB gene, which encodes nicotinate phosphoribosyltransferase (17).
Involvement of deoB.
In general, conversion of purine or pyrimidine bases to the corresponding ribonucleoside 5′-monophosphates may occur by either of two pathways: (i) by phosphoribosylation or (ii) by ribosylation followed by phosphorylation. The former reaction, catalyzed by base-specific phosphoribosyltransferases, requires PRPP. The latter reaction occurs, with guanine as an example, as follows: guanine + ribose 1-phosphate → guanosine + Pi. The latter reaction is catalyzed by purine nucleoside phosphorylase. In a second reaction, guanosine is phosphorylated to GMP by guanosine kinase. Ribose 1-phosphate may be generated from ribose 5-phosphate in an isomerization reaction catalyzed by phosphoribomutase, which is encoded by the deoB gene. To test if the NAD suppression phenotype requires phosphoribomutase, several mutant strains and plasmids were constructed (Table 2). First, removal of the entire deo operon resulted in generation of an NAD requirement (compare strains HO861 and HO967). Although the extent of the deo deletion is unknown, introduction of a Δ(deoD-thr) allele (strain HO1156), a deoC allele (HO1359), or a deoA allele (HO1187) had no effect on the NAD phenotype, as these strains, like HO861, were NAD prototrophs. In contrast, the deoB strain HO1949 was an NAD auxotroph, indicating that deoB is required for the NAD suppression phenotype. Second, plasmids bearing various deo cistrons were introduced into strain HO967 [Δdeo(CABD)]. These results showed that either deoB or deoA restored the NAD suppression phenotype to strain HO967 (compare strains HO967, HO967/pHO365, HO967/pHO366, and HO967/pHO368). The phosphoribomutase (deoB) may provide ribose 1-phosphate by isomerization of ribose 5-phosphate. Thymidine phosphorylase (deoA) may also provide ribose 1-phosphate, although by a different mechanism, namely, by phosphorolysis of uridine: uridine + Pi → uracil + ribose 1-phosphate. It has been shown that thymidine phosphorylase has specificity for uridine (11). Finally, we showed that xanthosine phosphorylase (xapA) was able to restore the NAD suppression phenotype (compare strains HO967/pSU18 and HO967/pGD248). Xanthosine phosphorylase has specificity for guanosine, as well as for xanthosine, and it catalyzes the following reaction: guanosine + Pi → guanine + ribose 1-phosphate. Both uridine and guanosine were present in the growth medium, and the strains are wild type for deoA and xapA in the chromosome. However, apparently due to the low affinity of thymidine phosphorylase towards uridine, the deoA gene must be present in a high-copy-number plasmid to have an effect on the NAD suppression phenotype. While xanthosine phosphorylase is normally synthesized only in the presence of xanthosine (12), xapA gene expression is independent of xanthosine in pGD248, in which it is behind a lac promoter.
TABLE 2.
Effect of deo alleles on NAD suppression phenotypea
| Strain | Genotype and/or plasmid | Pyridine source
|
|
|---|---|---|---|
| None | NAD | ||
| HO773 | Δprs deoD | − | + |
| HO861 | Δprs pst-phoU deoD | + | + |
| HO967 | Δprs pst-phoU Δ(deoCABD) | − | + |
| HO1156 | Δprs pst-phoU Δ(deoD-thr) | + | + |
| HO1359 | Δprs pst-phoU deoD deoC56 | + | + |
| HO1187 | Δprs pst-phoU deoD deoA103 | + | + |
| HO1949 | Δprs pst-phoU Δ(deoBD) | − | + |
| HO967 | pHO365 (deoC+A+B+bla+) | + | + |
| HO967 | pHO366 (deoC+A+bla+) | + | + |
| HO967 | pHO367 (deoC+bla+) | − | + |
| HO967 | pHO368 (deoB+bla+) | + | + |
| HO967 | pBR322 (bla+) | − | + |
| HO967 | pGD248 (xapA+B+cat+) | + | + |
| HO967 | pSU18 (cat+) | − | + |
| HO773 | pHO368 (deoB+bla+) | − | + |
| HO773 | pGD248 (xapA+B+cat+) | − | + |
Cells were grown at 32°C in glucose minimal medium supplemented with guanosine, uridine, histidine, tryptophan, and NAD as indicated. Plasmid-containing strains were grown with ampicillin or chloramphenicol present, +, growth; −, no growth.
Involvement of phnN.
The NAD suppression phenotype results from Pho regulon-controlling mutations (17). We therefore sought to identify which component of the Pho regulon is responsible for the suppression phenotype. First, several mutant phosphate starvation-inducible (psi) alleles were introduced into strain HO861, including psiC, psiD, psiE, and psiF alleles. One of these, psiD31, restored the NAD requirement (Table 3). The others had no effect. The psiD31 allele (also called phnD31) is a polar insertion at the phn locus (27), encoding enzymes involved in degradation of phosphonates (7, 41). As expected, deletion of the phn operon also restored the NAD requirement (strain HO1655). We therefore tested null alleles of every phn cistron, most of which are nonpolar (28), in the NAD suppression strain. Introduction of phnM28 or phnN45 eliminated suppression; i.e., both the Δprs pst-phoU phnM and Δprs pst-phoU phnN strains were NAD auxotrophs (Table 3). Among the entire collection of phn mutations analyzed, the phnM mutation was the only polar mutation. We therefore inferred that the effect of phnM was due to polarity on phnN. Furthermore, a mutation eliminating the phosphonate transport system (ΔphnCDE) had no effect on the NAD suppression phenotype (strain HO1496). This was also the case for a mutation in phnF (HO1565) (Table 3), mutations in phnGHIJKL, and mutations in phnO and phnP (data not shown).
TABLE 3.
Effect of phn mutations on NAD suppression phenotypea
| Strain | Genotype | Pyridine source
|
||
|---|---|---|---|---|
| None | Quinolinate or nicotinate | NAD | ||
| HO861 | Δprs pst-phoU | + | + | + |
| HO1408 | Δprs pst-phoU psiD31 | − | NTb | + |
| HO1655 | Δprs pst-phoU Δphn | − | − | + |
| HO1559 | Δprs pst-phoU phnM28 | − | NT | + |
| HO1569 | Δprs pst-phoU phnN45c | − | NT | + |
| HO1496 | Δprs pst-phoU Δ(phnC-E) | + | NT | + |
| HO1565 | Δprs pst-phoU phnF23c | + | NT | + |
| HO1877 | Δprs pst-phoU Δphn attλ::pCAH56 | − | − | + |
| HO1878 | Δprs pst-phoU Δphn attλ::pAH126 (Ptac-phnN) | + | + | + |
| HO1908 | Δprs pst-phoU Δphn attλ::pAH126 (Ptac-phnN) nadA | − | + | + |
| HO878 | Δprs pst-phoU nadA | − | + | + |
| HO1910 | Δprs pst-phoU Δphn attλ::pAH126 (Ptac-phnN) Δdeo | − | − | + |
| HO967 | Δprs pst-phoU Δdeo | − | − | + |
| HO773 | Δprs | − | − | + |
| HO1879 | Δprs attλ::pCAH56 | − | − | + |
| HO1902 | Δprs attλ::pAH126 (Ptac-phnN) | + | + | + |
| HO1909 | Δprs attλ::pAH126 (Ptac-phnN) nadA | − | + | + |
| HO1911 | Δprs attλ::pAH126 (Ptac-phnN) Δdeo | − | − | + |
| HO965 | Δprs Δdeo | − | − | + |
Cells were grown at 32°C in glucose minimal medium supplemented with guanosine, uridine, histidine, tryptophan, and the pyridine source indicated. Growth was read after 48 h of incubation; +, growth; −, no growth.
NT, not tested.
The phnF and phnN alleles are nonpolar.
To show that phnN was sufficient for establishing the NAD suppression phenotype, phnN was cloned into a plasmid (pAH126), and a single copy of this plasmid was subsequently integrated into the chromosome of the NAD auxotrophic strain HO1655 (Δprs pst-phoU Δphn). Introduction of phnN alone (strain HO1878) resulted in NAD prototrophy, whereas the vector had no effect (HO1877). Similarly, insertion of phnN into the NAD auxotrophic strain HO773 (Δprs [pst-phoU+]) resulted in NAD prototrophy. As expected, introduction of a mutant nad allele resulted in a pyridine requirement that could be satisfied by quinolinate or nicotinate in addition to NAD, whereas introduction of a Δdeo(CABD) allele resulted in a pyridine requirement that could be satisfied only by NAD (Table 3). Finally, a plasmid-harbored phnN allele, either a wild-type allele (pHO500) or a mutant allele specifying a histidine-tailed version of PhnN (pTR553), also suppressed the NAD requirement.
We conclude that the NAD suppression phenotype is caused by derepression of expression of the phn operon, which results in synthesis of the phnN gene product. This gene product, together with quinolinate phosphoribosyltransferase and phosphoribomutase, is necessary for the NAD suppression phenotype. The latter two enzymes were also present in the unsuppressed parental strain.
Reaction catalyzed by the phnN gene product.
The ribose moiety of ribose 1-phosphate formed by phosphoribomutase eventually ends up as the ribose moiety of nicotinate mononucleotide (NAMN) in reactions which are catalyzed by the phnN gene product and by quinolinate phosphoribosyltransferase. One or more additional enzymes might be involved as well. We therefore considered the possibility that phnN encodes an enzyme with ribose 1-phosphokinase activity (ribose 1-phosphate + ATP → ribose 1,5-bisphosphate + ADP) or with ribose 1,5-bisphosphokinase activity (ribose 1,5-bisphosphate + ATP → PRPP + ADP). To do this, the phnN gene was altered to specify a version with a histidine tail, and this variant protein was purified as described in Materials and Methods. The activity of the enzyme was assayed by incubation with the ribose compounds of interest in the presence of [γ-32P]ATP, followed by separation of the reaction mixture by thin-layer chromatography and finally by identification of novel radioactive compounds. Indeed, incubation of PhnN with [γ-32P]ATP and ribose 1,5-bisphosphate resulted in the formation of a radioactive compound, which comigrated with PRPP in one-dimensional thin-layer chromatography (Fig. 2A). This compound also comigrated with PRPP in two-dimensional thin-layer chromatography (data not shown). In contrast, no novel radioactive compound could be detected by incubation of PhnN with [γ-32P]ATP and ribose, ribose 1-phosphate, or ribose 5-phosphate (Fig. 2A).
FIG. 2.
Identification of PRPP as the product of the PhnN-catalyzed reaction. Incubation and one-dimensional chromatography were performed as described in Materials and Methods. Abbreviations: Appl., application line; F, solvent front. (A) Incubation of PhnN with various ribose compounds and [γ-32P]ATP. All of the incubation mixtures contained [γ-32P]ATP. Lanes 1 and 6 to 9 contained PhnN, whereas PhnN was omitted from lanes 2 to 5. Other additions were as follows: lane 1, no ribose compound; lanes 2 and 6, ribose; lanes 3 and 7, ribose 1-phosphate; lanes 4 and 8, ribose 5-phosphate; and lanes 5 and 9, ribose 1,5-bisphosphate. The positions of ATP, PRPP, ADP, and Pi are indicated on the right. In the reaction mixture shown in lane 9 a small amount of ADP was formed in addition to PRPP, presumably due to the presence of traces of [β-32P]ATP in the [γ-32P]ATP preparation. (B) Consumption of 32P-labeled PhnN reaction product (lanes 1 to 5) or PRPP synthase-generated [32P]PRPP (lanes 6 to 10) by XPRTase. Production of labeled compounds and incubation with XPRTase were performed as described in Materials and Methods. The reaction mixture analyzed in each lane contained reaction cocktail with the following supplement(s): lanes 1 and 6, no supplement; lanes 2 and 7, xanthine; lanes 3 and 8, XPRTase; lanes 4 and 9, xanthine and XPRTase; lanes 5 and 10, xanthine, XPRTase, and unlabeled PRPP (5 mM). The positions of ATP, PRPP, and Pi are indicated on the right. Lanes 1 to 5 contained appreciable amounts of ATP, whereas no ATP was present in lanes 6 to 10. We were unable to identify conditions that allowed complete consumption of ATP by PhnN, whereas ATP was readily consumed by PRPP synthase. (C) Conversion of unlabeled PhnN reaction product or PRPP to 5′-[14C]XMP in the presence of XPRTase and [14C]xanthine. The assay conditions are described in Materials and Methods. The reaction mixture analyzed in each lane contained reaction cocktail consisting of [14C]xanthine with the following addition(s): lane 1, no addition; lane 2, XPRTase; lane 3, PhnN reaction product; lane 4, PRPP (synthesized by PRPP synthase); lane 5, XPRTase and PhnN reaction product; lane 6, XPRTase and PRPP (synthesized by PRPP synthase); lane 7, XPRTase and commercial PRPP (5 mM); lane 8, XPRTase and a sample of PhnN assay mixture to which no PhnN enzyme was added (the mixture contained ribose 1,5-bisphosphate and ATP); and lane 9, XPRTase and a sample of PRPP synthase assay to which no PRPP synthase was added (the mixture contained ribose 5-phosphate and ATP). The positions of xanthine and 5′-XMP are indicated on the right. Spots were visualized under a UV mineral lamp at 254 nm.
To confirm that the compound formed was PRPP, we used a purified preparation of B. subtilis XPRTase, which catalyzes the reaction xanthine + PRPP → 5′-XMP + PPi. Thus, if the compound synthesized by PhnN is PRPP, it should be consumed by XPRTase in the presence of xanthine. This is exactly what was observed (Fig. 2B, lanes 1 to 4). Furthermore, addition of unlabeled PRPP prevented consumption of the radiolabeled PhnN-synthesized compound by XPRTase (Fig. 2B, lane 5). Similar results were obtained by incubation of authentic PRPP synthesized by using highly purified E. coli PRPP synthase (Fig. 2B, lanes 5 to 10). In these experiments PRPP was presumably labeled in the β position of the diphosphoryl group. Upon reaction with XPRTase, this diphosphoryl group was released as PPi. Eventually, the formation of small amounts of PPi could be seen (Fig. 2B), although most of the PPi was presumably converted to Pi by a pyrophosphatase present in the XPRTase preparation.
To reinforce the finding that the compound synthesized by PhnN was a substrate for XPRTase, the compound was synthesized in unlabeled form and again used as a substrate, together with [8-14C]xanthine and XPRTase; in this way 5′-[14C]XMP was generated (Fig. 2C, lane 5). Similarly, 5′-[14C]XMP was generated by incubation of [8-14C]xanthine and XPRTase with PRPP synthesized by E. coli PRPP synthase or with commercial PRPP (Fig. 2C, lanes 6 and 7). The results of some additional control reactions are shown in Fig. 2C. Formation of 5′-XMP required PRPP and XPRTase (Fig. 2C, lanes 1 to 4). Including either ribose 1,5-bisphosphate and ATP or ribose 5-phosphate and ATP (i.e., the substrates of ribose 1,5-bisphosphokinase and PRPP synthase, respectively) in the XPRTase assay mixture resulted in abolishing 5′-XMP production at levels above background levels (Fig. 2C, compare lanes 8 and 9 to lane 1). Thus, the XPRTase preparation contained no PRPP-forming contaminant. In a similar way we showed that the compound synthesized by PhnN could be converted to [14C]UMP in the presence of highly purified uracil phosphoribosyltransferase and [14C]uracil (data not shown). When ribose 1,5-bisphosphate and PhnN were incubated with [α-32P]ATP, the labeled nucleotide product was ADP rather than AMP. This showed that the enzyme transferred a phosphoryl group rather than a diphosphoryl group. The PhnN protein was unable to use GTP as a phosphoryl donor, as measured with [α-32P]GTP, in which case no labeled compound besides GTP appeared on the thin-layer chromatogram (data not shown). We conclude from these experiments that the product of the PhnN-catalyzed reaction is PRPP and consequently, that PhnN is an enzyme with ribose 1,5-bisphosphokinase activity.
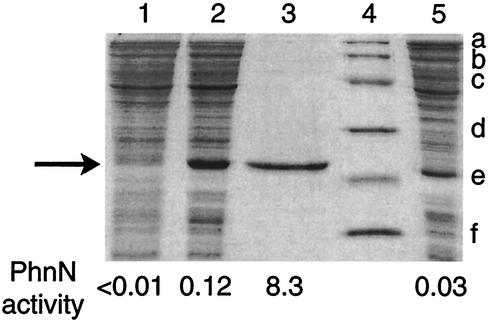
To further show that phosphorylation of ribose 1,5-bisphosphate was catalyzed by the phnN gene product, we compared the band intensity of the PhnN polypeptide during purification with the ribose 1,5-bisphosphokinase activity of the same fractions (Fig. 3). We were unable to determine the ribose 1,5-bisphosphokinase activity in a crude extract of strain HO1088/pTR553 grown in the absence of IPTG (i.e., the activity was less than 0.01 μmol/min/mg of protein), and similarly, no PhnN band was visible in the gel. In contrast, appreciable specific activity was observed in a crude extract of cells grown in the presence of IPTG, and there was a further increase in the specific activity of the purified enzyme. The relative intensity in the gel of the PhnN band increased from the crude extract to the purified enzyme. Thus, there was clear agreement between enzyme activity and band intensity of the PhnN polypeptide following gel electrophoresis in dodecyl-containing polyacrylamide. We therefore concluded that the band identified in Fig. 3 represents PhnN and that this polypeptide is responsible for ribose 1,5-bisphosphate phosphorylation. The relative molecular weight of this polypeptide was 23,500, which is in agreement with the value (21,408) calculated from the deduced nucleotide sequence of the allele specifying the histidine-tailed polypeptide. Lane 5 in Fig. 3 contained extract of a strain expressing wild-type phnN. As expected, the PhnN polypeptide was slightly smaller than the histidine-tailed polypeptide. The relative intensity of the PhnN band was somewhat less than that of the histidine-tailed PhnN (Fig. 3, lane 2). However, the ribose 1,5-bisphosphokinase activity of the former was significantly lower than that of the latter. We therefore concluded that the wild-type PhnN and the histidine-tailed PhnN have similar specific activities.
FIG. 3.
Purification and activity of histidine-tailed ribose 1,5-bisphosphokinase. Cells of strain HO1088 (Δprs)/pTR553 (specifying histidine-tailed PhnN) were grown in NZY medium, phnN gene expression was induced by IPTG, and ribose 1,5-bisphosphokinase was purified and assayed as described in Materials and Methods. In addition, a culture sample was removed before addition of IPTG. Furthermore, strain HO1088/pHO500 (specifying wild-type PhnN) was grown in parallel with strain HO1088/pTR553. Extracts and purified enzyme were subjected to gel electrophoresis in 17.5% polyacrylamide containing sodium dodecyl sulfate. Lane 1, crude extract (10 μg of protein) of HO1088/pTR553 cells before IPTG addition; lane 2, crude extract (10 μg) of HO1088/pTR553 cells after IPTG addition; lane 3, purified PhnN protein (1.3 μg); lane 4, molecular weight markers (positions indicated on the right, as follows: a, phosphorylase b [molecular weight, 97,400]; b, serum albumin [66,200]; c, ovalbumin [45,000]; d, carbonic anhydrase [31,000]; e, trypsin inhibitor [21,500]; f, lysozyme [14,400]); lane 5, crude extract (10 μg) of HO1088/pHO500 cells after IPTG addition. The arrow on the left indicates the position of the PhnN band. The ribose 1,5-bisphosphokinase activity of each fraction (in micromoles per minute per milligram of protein) is indicated at the bottom.
Formation in vivo of PRPP in NAD suppression mutants.
It was shown previously that the PRPP content of an E. coli Δprs pst-phoU strain was less than 5% of that of a wild-type strain (i.e., PRPP was undetectable) (17). Strain HO1088 (Δprs)/pTR553 (phnN) is NAD prototrophic, presumably due to increased expression of phnN. However, in the presence of adenine (100 mg/liter) this strain became NAD auxotrophic. Adenine is a potent scavenger of PRPP in cells that contain a wild-type allele of the apt gene, which encodes adenine phosphoribosyltransferase (2). In addition, the strain formed colonies in the absence of tryptophan, although at a much lower rate than it formed colonies in the absence of NAD. These data suggest that PRPP indeed is synthesized in strain HO1088/pTR553. We were unable to cultivate strain HO1088/pTR553 in a low-phosphate medium, and thus, we were unable to determine the sizes of the intracellular pools of PRPP or nucleotides.
DISCUSSION
Pathway for NAMN synthesis in NAD suppression mutants.
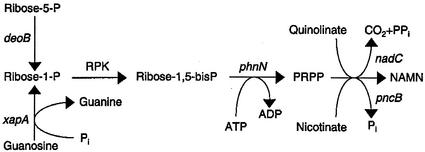
Our results show that PRPP can be synthesized from ribose 5-phosphate by the following pathway: ribose 5-phosphate → ribose 1-phosphate → ribose 1,5-bisphosphate → PRPP in reactions catalyzed by the sequential action of phosphoribomutase (deoB), ribose 1-phosphokinase, and ribose 1,5-bisphosphokinase (phnN) (Fig. 4). The activity of the enzyme catalyzing the second step, ribose 1-phosphokinase, remains to be identified, along with the corresponding gene. However, our data leave no doubt that ribose 1-phosphate is indeed involved in the pathway. NAD suppression strictly requires the production of ribose 1-phosphate, an intermediate of cellular metabolism only under certain conditions, as follows: either (i) when ribose 5-phosphate and phosphoribomutase are present (ribose 5-phosphate → ribose 1-phosphate) or (ii) when a ribonucleoside can be phosphorolyzed (e.g., guanosine in the presence of xanthosine phosphorylase [guanosine + Pi → guanine + ribose 1-phosphate]). Both of these reactions are included in Fig. 4.
FIG. 4.
Proposed pathway for conversion of ribose 5-phosphate to the 5′-phosphoribosyl moiety of NAMN. Abbreviations: P, phosphate; RPK, ribose 1-phosphokinase. Other enzyme-catalyzed reactions are indicated by gene designations (deoB, phosphoribomutase; xapA, xanthosine phosphorylase; phnN, ribose 1,5-bisphosphokinase activity; nadC, quinolinate phosphoribosyltransferase; pncB, nicotinate phosphoribosyltransferase.)
The physiological role of ribose 1,5-bisphosphate is poorly understood. Its only known function is as a coenzyme for phosphopentomutase of E. coli and other organisms (22, 24). The pathway for ribose 1,5-bisphosphate synthesis has not been described previously, except that macrophages synthesize the compound by dephosphorylation of PRPP (21).
Role of the phnN gene product in phosphonate degradation by the C-P lyase pathway.
We have shown that the PhnN protein is capable of catalyzing the phosphorylation of ribose 1,5-bisphosphate to PRPP; i.e., it is a ribose 1,5-bisphosphokinase. This is not only the first assignment of a chemical reaction to a specific polypeptide specified by the phn operon, but it is the first report of characterization of an enzyme with this activity. The specific activity of the enzyme with ribose 1,5-bisphosphate and ATP as substrates was rather low (i.e., undetectable) in an extract of uninduced cells. This low activity presumably provides an explanation for the retention in the NAD suppression mutants of requirements for purine and pyrimidine compounds. In wild-type E. coli cells approximately 80% of the PRPP formed is spent on purine and pyrimidine nucleotide synthesis, approximately 10% is spent on synthesis of the amino acid histidine, approximately 10% is spent on synthesis of the amino acid tryptophan, and approximately 1% is spent on NAD synthesis (19). Thus, only a small amount of PRPP may be required to satisfy the need for NAD and tryptophan biosynthesis. In addition, the elevated ribose 1,5-bisphosphokinase activity may not increase the PRPP pool to an extent that satisfies the amount required for nucleotide and histidine synthesis, as the level of the enzyme's substrate, ribose 1,5-bisphosphate, may not be elevated.
The biochemistry of phosphonate degradation by the E. coli C-P lyase pathway is poorly understood. Methyl phosphonate is converted to methane and some other unknown phosphorus-containing compound(s). Furthermore, methane formation appears to occur at the outside of the cell as the reaction may occur in cells which are deficient in the phosphonate ABC transporter specified by phnCDE (43; W. W. Metcalf and B. L. Wanner, unpublished data). It has also been shown that wild-type E. coli K-12 cells, which are unable to degrade phosphonates due to a transport defect (25), accumulate [32P]α-1-(ethylphosphono)ribose, when they are supplied with [32P]ethyl phosphonate; i.e., the ethyl phosphonate is attached to ribose at the C-1 hydroxyl as a phosphate ester. Most of this ribose derivative is found in the culture fluid. Phosphonate degradation-proficient cells fail to accumulate the compound, presumably because the compound is further degraded (1). A relationship between this phosphono-ribose compound and the activity of the phnN gene product, if any, remains to be established. However, it is possible that the α-1-(ethylphosphono)ribose is a dephosphorylation product of 5-phospho-α-1-(ethylphosphono)ribose. As noted above, α-1-(ethylphosphono)ribose was detected primarily in the culture fluid, possibly resulting from dephosphosphorylation of 5-phospho-α-1-(ethylphosphono)ribose by a nonspecific periplasmic phosphatase. Hypothetical reactions involving ribose 1,5-bisphosphokinase are shown in Fig. 5. We suggest that 5-phospho-α-1-(methylphosphono)ribose is an intermediate in methylphosphonate utilization. 5-Phospho-α-1-(methylphosphono)ribose may be demethylated by the C-P lyase, yielding methane and ribose 1,5-bisphosphate. The latter compound then may be phosphorylated to PRPP by PhnN, as was shown to occur in vitro in the present work. Alternatively, PhnN may phosphorylate 5-phos-pho-α-1-(methylphosphono)ribose to methylphosphono-PRPP, which in turn may be demethylated by the C-P lyase, yielding methane and PRPP.
FIG. 5.
Hypothetical reactions involved in the degradation of methylphosphonate by the C-P lyase pathway. An intermediate in the pathway is 5-phospho-α-1-(methylphosphono)ribose. This compound may be either demethylated to ribose 1,5-bisphosphate or phosphorylated to methylphosphono-PRPP. Ribose 1,5-bisphosphate or methylphosphono-PRPP may be phosphorylated or demethylated, which results in the formation of PRPP.
Acknowledgments
We are grateful to Hans Klenow for providing ribose 1,5-bisphosphate, to K. Frank Jensen for supplying purified XPRTase, to Carol Gross, Karin Hammer, and Tim Poulsen for supplying bacterial strains, and to Jan Neuhard and Gert Dandanell for providing DNA of pBRdeo and pGD248, respectively. We thank Tonny D. Hansen for expert technical assistance.
Financial support was provided by the Danish Natural Science Research Council to B.H.-J. and by the National Institutes of Health to B.L.W.
REFERENCES
- 1.Avilla, L. Z., K. M. Draths, and J. W. Frost. 1991. Metabolites associated with organophosphonate C-P bond cleavage: chemical synthesis and microbial degradation of [32P]-ethylphosphonic acid. Bioorg. Med. Chem. Lett. 1:51-54. [Google Scholar]
- 2.Bagnara, A. S., and L. R. Finch. 1974. The effects of bases and nucleosides on the intracellular contents of nucleotides and 5-phosphoribosyl 1-pyrophosphate in Escherichia coli. Eur. J. Biochem. 41:421-430. [DOI] [PubMed] [Google Scholar]
- 3.Bartolomé, B., Y. Jubete, E. Martinez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heynecker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 6.Casadaban, M. J., A. Martinez-Arias, S. K. Shapiro, and J. Chou. 1983. β-Galactosidase gene fusion for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 100:293-308. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C.-M., Q.-Z. Ye, Z. Zhu, B. L. Wanner, and C. T. Walsh. 1990. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J. Biol. Chem. 265:4461-4471. [PubMed] [Google Scholar]
- 8.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 9.Haldimann, A., M. K. Prahalad, S. L. Fisher, S.-K. Kim, C. T. Walsh, and B. L. Wanner. 1996. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc. Natl. Acad. Sci. USA 93:14361-14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies in bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer-Jespersen, K. 1983. Nucleoside catabolism, p. 203-258. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, Inc., London, United Kingdom.
- 12.Hammer-Jespersen, K., R. S. Buxton, and T. D. Hansen. 1980. A second purine nucleoside phosphorylase in Escherichia coli K-12. II. Properties of xanthosine phosphorylase and its induction by xanthosine. Mol. Gen. Genet. 179:341-348. [DOI] [PubMed] [Google Scholar]
- 13.Hove-Jensen, B. 1983. Chromosomal location of the gene encoding phosphoribosylpyrophosphate synthetase in Escherichia coli. J. Bacteriol. 154:177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hove-Jensen, B. 1988. Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine and tryptophan in Escherichia coli. J. Bacteriol. 170:1148-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hove-Jensen, B. 1989. Phosphoribosylpyrophosphate (PRPP)-less mutants of Escherichia coli. Mol. Microbiol. 3:1487-1492. [DOI] [PubMed] [Google Scholar]
- 16.Hove-Jensen, B. 1992. Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine 1-phosphate uridyltransferase in Bacillus subtilis. J. Bacteriol. 174:6852-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hove-Jensen, B. 1996. Phosphoribosyl diphosphate synthetase-independent NAD de novo synthesis in Escherichia coli: a new phenotype of phosphate regulon mutants. J. Bacteriol. 178:714-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hove-Jensen, B., and M. Maigaard. 1993. Escherichia coli rpiA gene encoding ribose phosphate isomerase A. J. Bacteriol. 175:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen, K. F. 1983. Metabolism of 5-phosphoribosyl 1-pyrophosphate (PRPP) in Escherichia coli and Salmonella typhimurium, p. 1-25. In A. Munch-Petersen (ed.), Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Academic Press, Inc., London, United Kingdom.
- 20.Jensen, K. F., U. Houlberg, and P. Nygaard. 1979. Thin-layer chromatographic methods to isolate 32P-labelled 5-phosphoribosyl-α-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal. Biochem. 98:254-263. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi, T., T. L. Veech, and K. Uyeda. 2001. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphoshate and ribose 1,5-bisphosphate. J. Biol. Chem. 276:28554-28561. [DOI] [PubMed] [Google Scholar]
- 22.Klenow, H. 1953. Some properties of the phosphoribomutase reaction. Arch. Biochem. Biophys. 46:186-200. [DOI] [PubMed] [Google Scholar]
- 23.Krath, B. N., and B. Hove-Jensen. 2001. Class II recombinant phosphoribosyl diphosphate synthase from spinach. Phosphate independence and diphosphoryl donor specificity. J. Biol. Chem. 276:17851-17856. [DOI] [PubMed] [Google Scholar]
- 24.Leer, J. C., and K. Hammer-Jespersen. 1975. Multiple forms of phosphodeoxyribomutase from Escherichia coli. Physical and chemical characterization. Biochemistry 14:599-607. [DOI] [PubMed] [Google Scholar]
- 25.Makino, K., S.-K. Kim, H. Shinagawa, M. Amemura, and A. Nakata. 1991. Molecular analysis of the cryptic and functional phn operons for phosphonate use in Escherichia coli K-12. J. Bacteriol. 173:2665-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel, M., and A. Higa. 1970. Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53:159-162. [DOI] [PubMed] [Google Scholar]
- 27.Metcalf, W. W., P. M. Steed, and B. L. Wanner. 1990. Identification of phosphate-starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mud1) transcriptional fusions. J. Bacteriol. 172:3191-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., and B. L. Wanner. 1991. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J. Bacteriol. 173:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf, W. W., and B. L. Wanner. 1993. Evidence for a fourteen-gene, phnC to phnP locus for phosphonate metabolism in Escherichia coli. Gene 129:27-32. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics. Cold Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Nilsson, D., and B. Hove-Jensen. 1987. Phosphoribosylpyrophosphate synthetase of Bacillus subtilis. Cloning, characterization and chromosomal mapping of the prs gene. Gene 53:247-255. [DOI] [PubMed] [Google Scholar]
- 32.Penfound, T., and J. W. Foster. 1996. Biosynthesis and recycling of NAD, p. 721-730. In F. C. Neidhart, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 33.Post, D. A., R. L. Switzer, and B. Hove-Jensen. 1996. The defective phosphoribosyl diphosphate synthase in a temperature-sensitive prs-2 mutant of Escherichia coli is compensated by increased enzyme synthesis. Microbiology 142:359-365. [DOI] [PubMed] [Google Scholar]
- 34.Randerath, K., and E. Randerath. 1965. Ion-exchange thin-layer chromatography. XIV. Separation of nucleotide sugars and nucleoside monophosphates on PEI-cellulose. Anal. Biochem. 13:575-579. [DOI] [PubMed] [Google Scholar]
- 35.Shevell, D. E., A. M. Abou-Zamzam, B. Demple, and G. C. Walker. 1988. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J. Bacteriol. 170:3294-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping in Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valentin-Hansen, P., H. Aiba, and D. Schümperli. 1982. The structure of tandem regulatory regions in the deo operon of Escherichia coli K12. EMBO J. 1:317-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentin-Hansen, P., B. A. Svenningsen, A. Munch-Petersen, and K. Hammer-Jespersen. 1978. Regulation of the deo operon in Escherichia coli. The double negative control of the deo operon by the cytR and deoR repressors in a DNA directed in vitro system. Mol. Gen. Genet. 159:191-202. [DOI] [PubMed] [Google Scholar]
- 39.Wanner, B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357-1381. In F. C. Neidhart, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 40.Wanner, B. L. 1997. Phosphate signaling and the control of gene expression in Escherichia coli, p. 104-128. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman and Hall, New York, N.Y.
- 41.Wanner, B. L., and J. A. Boline. 1990. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J. Bacteriol. 172:1186-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willemoës, M., B. Hove-Jensen, and S. Larsen. 2000. Steady state kinetic model for the binding of substrates and allosteric effectors to Escherichia coli phosphoribosyl-diphosphate synthase. J. Biol. Chem. 275:35408-35412. [DOI] [PubMed] [Google Scholar]
- 43.Yakovleva, G. M., S.-K. Kim, and B. L. Wanner. 1998. Phosphate-independent expression of the carbon-phosphorus activity of Escherichia coli. Appl. Microbiol. Biotechnol. 49:573-578. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]