The recent flood of genomic (13), transcriptomic (29), and other high-throughput data (21, 33, 45) makes the need to interpret this information in a systemic fashion increasingly pressing. The construction of in silico models represents a way to interpret these data and place them in the context of cellular physiology. A variety of in silico modeling approaches in biology have been developed, including detailed kinetic models, cybernetic models, stochastic models, metabolic control analysis, biochemical systems theory, and constraint-based methods. Many of these methodologies have been reviewed elsewhere (5, 20, 25, 40, 46, 52, 62).
Modern modeling approaches in biology need to be easily scalable and able to integrate available “-omics” data (38) that may contain tens of thousands of measurements. A constraint-based modeling approach (5, 14, 62) meets these criteria and at present is the only methodology by which genome-scale models have been constructed. The few parameters used in a constraint-based framework enable models to be built quickly and to encompass a larger portion of biochemical reaction networks than the portion currently encompassed by other modeling methodologies. To date, constraint-based models account for the largest metabolic models in terms of numbers of genes and reactions and have proven to be predictive of some types of data, including phenomic data (15, 26, 63), qualitative transcriptomic data (9), and gene knockout data (16, 54).
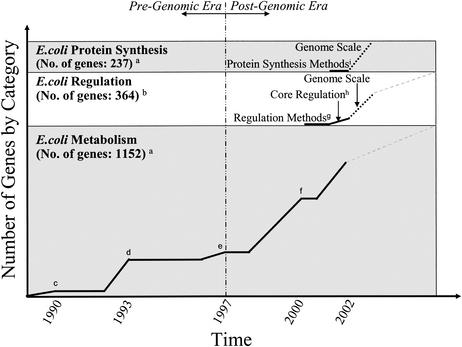
Escherichia coli is a well-studied organism, and much is known about its metabolism, regulation, and physiology. Constraint-based models of E. coli have been under development for the past 13 years. The continual growth in the size and scope of constraint-based E. coli models, as shown in Fig. 1, illustrates the iterative nature of in silico model building and how such models expand in scope and completeness over time. While many modeling approaches have been used to study E. coli (2, 12, 57, 65), in this minireview we focus on the development of successive constraint-based models that have been formulated to describe the metabolic network of E. coli and summarize the models' abilities to predict or explain phenotypic behavior. The principles that have been developed and the experiences that have been gained from modeling E. coli can be directly applied to modeling other organisms; this process has begun for Haemophilus influenzae (17), Helicobacter pylori (48), Saccharomyces cerevisiae (8, 23), and Methylobacterium extorquens (58), and more models are expected to emerge soon. The scope of constraint-based in silico models should continue to grow, and these models are likely to have a variety of uses in the near future.
FIG. 1.
Development of successive constraint-based FBA models of E. coli. Constraint-based models of E. coli first focused on metabolism. By the time the complete genome was sequenced (1997), only 26% of metabolic genes were accounted for in FBA models. Over the next 5 years the number grew to include nearly 80% of the metabolic genes. Methods for incorporating transcriptional regulation have been developed and implemented in a core metabolic model of E. coli, as have methods for including protein synthesis. Expanding the regulatory and protein synthesis models to the genome scale can be accomplished by using information that is known today (indicated by dotted lines). Further functional analysis of genes should increase the size of models (dashed lines). These three components can be combined to form an integrated model (E. coli i2K) that accounts for nearly 2,000 genes. The superscript letters indicate references, as follows: a, reference 4; b, reference 55; c, reference 32; d, references 59 and 60; e, references 42 and 43; f, reference 16; g, reference 11; h, reference 9; and i, reference 1.
PRINCIPLES OF CONSTRAINT-BASED MODELING
Reviews that describe the constraint-based modeling procedure have appeared previously (5, 14, 19, 62). While kinetic models may ultimately provide a detailed understanding of integrated cellular functions, they are limited by the current availability of the information needed to construct them and by the fact that kinetic constants can vary across a population and change over time through evolution. The constraint-based modeling procedure does not strive to find a single solution but rather finds a collection of all allowable solutions to the governing equations that can be defined. Solutions that violate any of the imposed constraints are excluded from the collection, which mathematically is called a solution space. The subsequent application of additional constraints further reduces the solution space and, consequently, reduces the number of allowable solutions that a cell can utilize. The constraints that have been used in the first generation of constraint-based models include stoichiometric constraints (mass balance), thermodynamic constraints (regarding the reversibility of a reaction), and enzymatic capacity constraints (using an appropriate Vmax value).
Stoichiometric constraints can be represented by the matrix equation Sv = 0, where S is the stoichiometric matrix describing all the reactions in the network and v is a vector describing the fluxes through each of the reactions. Each column of S corresponds to an individual reaction, and the rows of S correspond to the different metabolites. The stoichiometric coefficients of a reaction are then represented as elements in column (i.e., Sij corresponds to the stoichiometric coefficient of the ith metabolite in the jth reaction). The equation Sv = 0 imposes the restriction that the total rate of production for any metabolite must equal the total rate of consumption for that metabolite. In addition to stoichiometric or mass balance constraints, thermodynamic constraints and enzyme capacity constraints place limits on the range of values for individual fluxes (vj) in the network. Enzyme capacity constraints place an upper limit on the values that a given flux can take. Application of thermodynamic constraints further restricts the range of flux values. If a reaction is irreversible, the corresponding flux must be greater than or equal to zero; however, reversible reactions can have positive or negative flux values. A more systematic representation of thermodynamic constraints has appeared (3, 44). Stoichiometric, enzyme capacity, and thermodynamic constraints represent hard inviolable physicochemical constraints that cells must abide by.
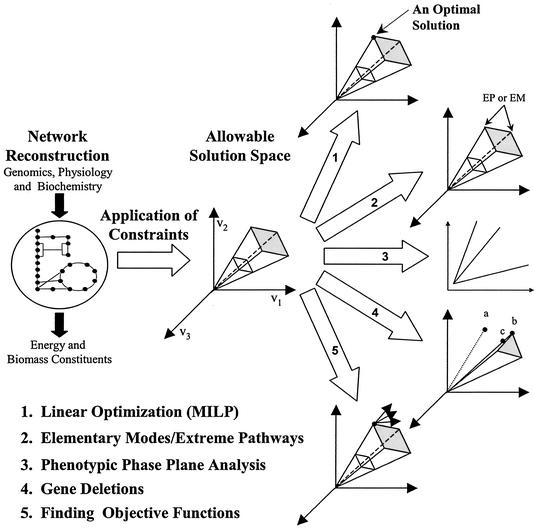
Given the governing constraints, the next step involves characterizing the allowable solution space and predicting which solution a cell is likely to use. Different techniques exist under the constraint-based modeling framework, including extreme pathway analysis (50), elementary mode analysis (52, 53), flux balance analysis (FBA) (5, 14, 19, 62), and minimization of metabolic adjustments (54), which aid in this process. Figure 2 illustrates the different constraint-based modeling techniques used to characterize the solution space defined by the network and the applied constraints.
FIG. 2.
Constraint-based modeling. Application of constraints to a reconstructed metabolic network leads to a defined solution space in which a cell's network must operate. From this solution space a number of methods have been developed that help predict or explain phenotypic behavior. Linear optimization can be used to find solutions in the space that maximize or minimize a given objective (5, 14, 19, 62), and mixed-integer linear programming (MILP) can be used to find multiple optima if they exist (30, 41). Elementary mode analysis (52, 53) and extreme pathway analysis (50) can be used to characterize vectors in the solution space; the edges of the space correspond to extreme pathways (EP) and are a subset of the elementary modes (EM). Phenotypic phase plane analysis shows for what conditions the metabolic network operates under different limitations (18). The effects of gene deletions can also be computed. In the diagram the old optimal solution (point a) does not lie in the new solution space. A new optimum can be calculated (point b), or a suboptimal solution that is closest to the old optimum can be calculated (point c) (54). In addition, work has been done by using experimental flux measurements (indicated by a point) to back-calculate objective functions (indicated by vectors) (6a).
Characterizing the solution space.
Extreme pathways and elementary modes represent sets of vectors that describe the solution space and are themselves biochemically valid flux distributions through a defined metabolic network. Calculation of elementary modes and extreme pathways has been described elsewhere (50, 53). Elementary modes are unique vectors that characterize the solution space. An elementary mode is defined as a “minimal set of enzymes that could operate at steady-state with all irreversible reactions proceeding in the appropriate direction” (52). Extreme pathways are related to elementary modes and correspond directly to the edges of the convex solution space. Both approaches have been applied to core metabolic networks of E. coli whose sizes are listed in Table 1. Positive linear combinations of these vectors can be used to generate any valid steady-state flux solution under the governing constraints. These analysis methods are useful for characterizing the solution space, and the next step is to try to determine what solution in the solution space the cell actually chooses to use.
TABLE 1.
Elementary mode analysis and extreme pathway analysis models
| Model | Year | No. of metabolic reactions | No. of metabolites |
|---|---|---|---|
| Liao et al. | 1996 | 28 | 20 |
| Schilling et al. | 2000 | 78 | 53 |
| Stelling et al. | 2002 | 110 | 89 |
Calculating optimal phenotypes.
Linear optimization has been used to find a particular solution within the allowable solution space that maximizes or minimizes a particular objective function. The objective function can be used to explore the content of a solution space, to determine capabilities (such as optimal metabolic by-product yields), or to guess at likely phenotypic functions. This approach is referred to as FBA and has been reviewed elsewhere (5, 14, 19, 62). Some commonly used objective functions include production of ATP, production of a desired by-product, or growth rate (as defined by the weighted consumption of metabolites needed to make biomass). The size of the metabolic networks used to model E. coli metabolism by FBA has expanded over time from 14 to 929 metabolic reactions, as shown in Table 2.
TABLE 2.
Flux balance models
| Model | Year(s) | No. of metabolic reactions | No. of metabolites |
|---|---|---|---|
| Majewski and Domach | 1990 | 14 | 17 |
| Varma and Palssona | 1993-1995 | 146 | 118 |
| Pramanik and Keaslingb | 1997-1998 | 300 (317) | 289 (305) |
| Edwards and Palsson | 2000 | 720 | 436 |
| Covert and Palssonc | 2002 | 113 | 77 |
| Reed and Palsson | 2003 | 929 | 626 |
Varma and Palsson's model contained a catabolic and biosynthetic network. The values shown are the values for the combined networks.
Pramanik and Keasling developed two models. The second model, the number of parameters of which is indicated in parentheses, included reactions from the first model plus an additional 17 reactions.
Regulated model of central metabolism.
Construction of constraint-based models is iterative in nature, and while correct predictions are desirable, the incorrect predictions are the ones that lead to an iterative refinement of the models (37). Over the past 13 years the constraint-based modeling approach has been used to study, explain, and predict the phenotypic behavior of E. coli. The individual models can be characterized by the type of analysis performed and by the time frame in which they were developed.
ELEMENTARY MODE AND EXTREME PATHWAY ANALYSES
A representation of E. coli's central metabolic network was constructed and analyzed by using elementary mode analysis (31). For this study modes that produce 3-deoxy-d-arabinoheptulosonate 7-phosphate, a precursor of the aromatic amino acids, and/or ATP were calculated with xylose or glucose as the carbon source. When glucose was the carbon source, all possible steady-state flux distributions (i.e., solutions which obey stoichiometric and thermodynamic constraints) could be written as the sum of 11 calculated elementary modes. Examination of the properties of the calculated elementary modes, such as 3-deoxy-d-arabinoheptulosonate 7-phosphate yields, provided insight into experimental results (31).
The extreme pathways for a larger network containing 78 reactions and 53 metabolites were calculated for two different carbon sources, glucose and succinate. The extreme pathway vectors calculated for the conditions when the biomass precursors were included in the network as a growth reaction were correlated with results from FBA when growth was used as the objective function (49). More recently, the elementary modes in a larger representation of E. coli's metabolic network (containing 110 reactions and 89 metabolites) were calculated and analyzed (56). In this study five different carbon sources were used, and again a reaction representing the drain of metabolic precursors needed for growth was added to the network. Elementary mode analysis was used to determine which of 90 genes were essential for growth; a reported 90% of the predictions agreed with experimental data when phenotypes were classified as either growth or no growth. The number of elementary modes varied depending on the carbon source and ranged from 598 when acetate was the carbon source to 27,099 for glucose. The modes were further analyzed to identify which enzymes would most likely be regulated for changing growth conditions (i.e., different carbon sources). A good correlation between regulatory predictions and measured mRNA expression data was found (56).
The metabolic networks studied by elementary mode or extreme pathway analysis are smaller than the networks studied by FBA. This limitation is due to the computational complexity associated with calculating these vectors and not from limitations on known reaction stoichiometry (28).
Direct determination of optimality properties can be accomplished by using optimization procedures that circumvent the exhaustive enumeration of extreme pathway analysis or elementary mode analysis. Linear programming has been used extensively to determine the optimality properties of reconstructed E. coli networks.
PREGENOME ERA MODELS
Initial model of core metabolism.
In 1990, Majewski and Domach formulated an FBA model that included 14 metabolic reactions (tricarboxylic acid [TCA] cycle, partial glycolysis, and respiratory chain) and four load fluxes that served as drains for the metabolic precursors needed for cell growth (oxaloacetate, α-ketoglutarate [αKG], pyruvate, and acetyl coenzyme A) (32). In their analysis production of high-energy phosphate bonds on ATP and GTP was used as the objective function.
Majewski and Domach studied the optimal behavior of the metabolic network under two different types of constraints, enzymatic capacity constraints and electron transport chain constraints. Both types of constraints placed an upper limit on the value for fluxes through different reactions in the network. By maximizing the utilization of the network for the production of high-energy phosphate bonds under either an enzymatic constraint (αKG dehydrogenase) or an electron transport chain capacity constraint, equations describing the onset of acetate overflow and the rate of acetate production could be derived. The experimentally determined secretion patterns for acetate overflow in E. coli agreed with the network operating under enzymatic capacity constraints (32). The model led to the conclusion that the electron transport chain capacity is a constraint only when the growth rate approaches the maximum achievable growth rate (32).
Model built on Neidhardt's compendium.
Varma and Palsson's reconstruction of E. coli's metabolic network contained both anabolic and catabolic reactions based on previously published information (34, 35). The model contained 53 catabolic reactions and 94 biosynthetic reactions that produce the amino acids, nucleic acids, and cell membrane and cell wall constituents found in cell biomass (59, 60). Several network properties were calculated from this model, such as optimal production of cofactors (ATP, NADPH, NADH) (60), optimal production of metabolic precursors (such as pyruvate or succinate) (60), maximal theoretical yields for amino acid and nucleic acid production (59), and evaluation of constraints (energy, redox, or stoichiometric) that restrict production of metabolites or cofactors (59, 60), as well as optimal flux distributions for biomass production (61). Model predictions agreed with experimental results when E. coli was grown on glucose minimal medium under aerobic and anaerobic conditions and if the metabolic network was optimized for the production of biomass constituents (63). Sensitivity analysis of the predicted growth rate with respect to the biomass composition was conducted, and it was found that the sensitivity of the biomass yield to changes in metabolite requirements was low (61).
Effects of growth rate-dependent biomass composition.
Pramanik and Keasling's metabolic network was an expansion of Varma and Palsson's model and consisted of 300 reactions and 289 metabolites (an additional 17 reactions and 16 metabolites were added later) (42, 43). The biomass composition of E. coli varies with the carbon source and the growth rate (36). Varma and Palsson's model had used a fixed biomass composition based on previously published data, while Pramanik and Keasling's model used derived equations relating growth rate to biomass requirements, allowing for changes in biomass composition in a growth rate-dependent manner (43).
To analyze the accuracy of the model, 13 experimentally measured flux values (64) were compared to fluxes calculated by using Pramanik and Keasling's model (43). Three experimental conditions were examined: anaerobic growth on glucose, aerobic growth on acetate, and aerobic growth on acetate plus glucose. For aerobic growth on acetate plus glucose the average difference between experimental flux measurements and model predictions was 16%. Similar results were found for aerobic growth on acetate (17%). No experimental flux values were available for comparison for anaerobic glucose, but a branched TCA cycle (with an oxidative branch producing αKG and a reductive branch producing succinyl coenzyme A) had been observed experimentally; the model's prediction agreed with this observation. Further analysis of the model also indicated that the predicted flux values were sensitive to biomass composition (42, 43). Other studies have been conducted by using slight modifications of Pramanik and Keasling's model to investigate the effects of large-scale gene deletions or additions (6) and to calculate a minimal gene set (7).
POSTGENOME ERA MODELS
Finally, a genome-scale model.
The complete genome of E. coli K-12 strain MG1655 was sequenced and annotated in 1997 (4). Genomic data allow systematic assessment of the capabilities of the organism; in addition, genes encoding metabolic enzymes that have not yet been biochemically characterized can be included in a model based on sequence similarity. For less-studied organisms the genome plays a more significant role in network reconstruction, and many of the enzymes are assigned based on sequence homology and await biochemical characterization. In the case of E. coli, the genomic information, coupled with physiological and biochemical data (10), enabled reconstruction of a genome-scale metabolic model describing the E. coli metabolic network (16).
Edwards and Palsson's model included 720 reactions and 436 metabolites involved in glycolysis, the TCA cycle, the pentose phosphate pathway, respiration, anaplerotic reactions, fermentative reactions, amino acid biosynthesis and degradation, nucleotide biosynthesis and interconversions, fatty acid biosynthesis and degradation, phospholipid biosynthesis, cofactor biosynthesis, and metabolite transport. This model was validated with mutant data (16), was used to design quantitative experiments (15), and was found to predict the outcome of adaptive evolution (26).
The results of in silico gene deletion studies were compared with growth data obtained with known mutants. The in vivo growth characteristics of a series of E. coli mutants on several different carbon sources were examined and compared to the in silico deletion results. In this analysis, 68 of 79 (86%) of the in silico predictions were consistent with the experimental observations (16). The predictions of the in silico E. coli model were highly consistent with phenotypes of known mutants and knockouts.
Phenotypic phase plane analysis was developed (18) and applied to Edwards and Palsson's E. coli model. Quantitative predictions regarding optimal usage of a carbon source and oxygen to maximize the growth rate were made and tested for a variety of substrates. Growth on M9 minimal medium with acetate, malate, or succinate as the primary carbon source agreed with the computational hypothesis (15, 26); however, glycerol supported only suboptimal growth of E. coli. Repeated exponential balanced growth batch cultures on glycerol were then incubated for 60 days (around 900 generations), and the cells reproducibly evolved towards the a priori predicted optimal growth behavior (26).
Incorporating effects of transcriptional regulation.
A shortcoming of the purely stoichiometric metabolic models is that they do not account for transcriptional regulation, so all the gene products are assumed to be available to the cell to optimize its performance in a defined environment. This assumption is based on the rationale that E. coli would have evolved its regulatory network to allow optimal growth under conditions to which the microorganism was already adapted. Some instances where this assumption might not be true are for E. coli mutants or for growth on multiple carbon sources. It has also been found that some carbon sources do no initially support optimal behavior, although limited adaptive evolution data do suggest that over time E. coli adjusts its metabolic fluxes to find the optimal solution (26; S. S. Fong and B. O. Palsson, unpublished data).
To address this issue, the effects of transcriptional regulation have recently been included in a constraint-based model of E. coli central metabolism (9). The method for modeling transcriptional regulation is based on Boolean logic, where the genes can be either on or off, and their status is evaluated based on conditional if statements. The regulatory network has been reconstructed for central metabolism in E. coli by using this method. Covert and Palsson's metabolic and regulatory network includes 149 genes (16 are regulatory genes), which take part in 113 metabolic reactions, 45 of which are regulated by 16 regulatory proteins (9).
Covert and Palsson's regulated model has been used to compare regulated model predictions of gene deletions with mutant data and predictions from an unregulated model (9). The unregulated network correctly predicted 97 of 116 cases correctly (83.6%), while the regulated network predicted 106 of 116 cases correctly (91.4%); thus, incorporating regulation improved the accuracy of in silico knockout predictions. The model was also used to calculate time courses of batch growth. The in silico predictions were in agreement with the experimental by-product (acetate, formate, and ethanol) secretion patterns, as well as the glucose uptake and biomass production patterns (9).
FUTURE DIRECTIONS
Expanding current models.
Annotation updates to the E. coli genome (55), coupled with previously published data and organism-specific databases, including EcoCyc (27), have enabled expansion of Edwards and Palsson's metabolic model. So far, an additional 242 genes have been added to Edwards and Palsson's model, and the metabolic network now contains 929 different metabolic reactions (J. L. Reed and B. O. Palsson, unpublished data). Genome-scale maps of E. coli metabolism have been constructed and are available (http://gcrg.ucsd.edu/organisms/ecoli/html). Work is now focused on filling in the gaps in the metabolic network and validating the model. This model should continue to grow as more metabolic genes are identified and characterized.
Recent work has also been done to include the regulatory constraint-based model of E. coli (9). The expansion of this regulatory model to include the effects of regulation on larger genome-scale models of metabolism is one of the next foreseeable steps in building more accurate constraint-based models of E. coli. Tools that should aid in this effort include RegulonDB, a database containing information about gene regulation in E. coli that is publicly available (47), and methods that extract gene regulatory networks from transcriptomic data (24).
Finding objective functions.
There are many issues that remain involving the selection of an objective function. Biomass compositions have often been used to compute the basis for optimal growth objective function. The effects of growth rate-dependent changes in biomass composition have already been accounted for (43). In addition to these advances, work is being done to back-calculate objective functions based on measured flux data, so that utilization of a calculated objective function yields a solution that minimizes the error between predicted and experimentally measured fluxes (6a).
Alternative solutions.
A single linear optimization identifies one solution in the solution space; however, alternate optimal solutions can exist in the allowable solution space. These equivalent solutions can be calculated by using a variety of techniques, such as mixed-integer linear programming (30, 41), extreme pathway analysis (39), and elementary mode analysis (52); which optimal solution is actually used by the cell is still not known.
The in silico finding that the same phenotype can be attained in more than one way with the same underlying network gives rise to the possibility that it may be difficult to determine the true state of a cell. Preliminary experimental data support this expectation since strains that evolve to have the same growth phenotype (26) are not identical (Fong and Palsson, unpublished). Further, evolution of phosphotransferase system knockouts in E. coli also support this expectation (22).
Gene knockouts.
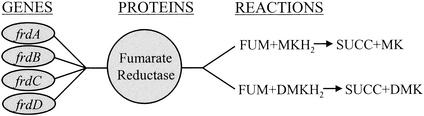
The in silico representation of biological associations among genes, proteins, and reactions is important when the effects of gene deletions are modeled. Enzyme subunits and enzyme complexes need to be taken into account when associations among genes, enzymes, and reactions are made (Fig. 3). Deleting a gene in constraint-based models results in removing the reactions associated with the protein from the network, unless other isozymes are present. Removal of reactions changes the solution space, and some wild-type solutions might be eliminated (Fig. 2). Knocking out essential genes from the model produces no solutions which allow for cellular growth under the governing constraints.
FIG. 3.
Gene-protein-reaction associations. The association between the enzyme fumarate reductase and the genes which code for its subunits is shown. All four gene products come together to make a functional enzyme. This enzyme is capable of carrying out two reactions, (i) the transfer of electrons from menaquinol (MKH2) to fumarate (FUM) and (ii) the transfer of electrons from demethylmenaquinol (DMKH2) to FUM. The products of both reactions are succinate (SUCC) and either menaquinone (MK) or demethylmenaquinone (DMK). Deletion of any of the subunits would eliminate the functional enzyme. This is simulated by removing the two reactions from the network (unless an isozyme exists).
In previous studies researchers have focused on determining if genes or reactions are essential using FBA (16, 48), elementary mode analysis (56), and extreme pathway analysis (51). Methods for predicting suboptimal solutions in gene knockout studies are being developed because an optimal solution might not be biologically achievable due to regulatory or kinetic effects. One such method, diagrammed in Fig. 2, has already been developed and finds the solution in the knockout solution space by minimizing the difference in fluxes between the wild-type optimal solution and a solution residing in the knockout solution space (54).
Application of additional constraints.
Significant progress has been made in the last 13 years towards building constraint-based models of E. coli; they are now genome-scale models. Enhancing the predictive capabilities of these models in the future should be accomplished by broadening the scope of the models (including other cellular processes), as well as exploring the use of additional constraints. The utilization of other physicochemical constraints, such as the conservation of energy, kinetic constraints, osmotic balances, or electroneutrality, should further reduce the allowable solution space, resulting in more accurate predictions. A framework for implementing energy balance constraints (3, 44) has already been developed and applied to the central metabolic network of E. coli (3).
Integrated models.
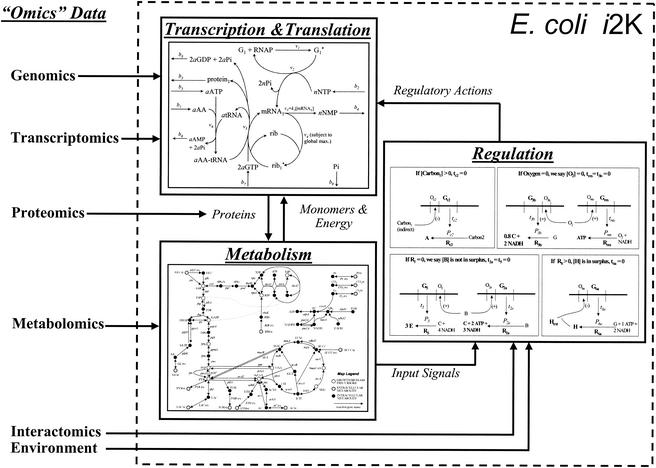
Other cellular processes can be described in a constraint-based modeling framework based on the genome sequence, such as transcription (1), translation (1), and DNA replication. These processes place direct metabolite and energy demands (i.e., through the objective function) on the metabolic network. These processes are coupled, and metabolism affects the rates of transcription, translation, and replication; these processes, in return, direct metabolism (Fig. 4). The development of constraint-based models that include metabolism, regulation, and protein synthesis should allow simultaneous reconciliation of diverse “-omics” data (such as proteomic, metabolomic, transcriptomic, and phenomic data) and back-calculation of biological parameters (such as promoter strengths).
FIG. 4.
Integrated constraint-based model of E. coli: the E. coli i2K model. Constraint-based modeling frameworks have been developed for metabolism (5, 14, 19, 50, 52, 62), regulation (9), transcription, and translation (1). The connectivity among the three modeling components is shown here. Integration of these three modeling components should produce an integrated model of E. coli that accounts for nearly 2,000 genes, referred to as the E. coli i2K model. This model can be used to reconcile diverse “-omics” data and utilize the data to more accurately predict a cellular phenotype.
CONCLUSION
A shift in biology from a component-based perspective to a systems view of the cell is occurring as a result of high-throughput data generation. This shift in thinking requires construction of in silico models in order to understand systemic behavior of complete biological processes. Genome-scale models need to be built to evaluate and study the intrinsic biological properties that emerge from the system as a whole. The challenge is to develop methodologies to construct and study such models. Constraint-based models are the first step towards achieving this goal. Within this approach, models can be easily scaled up, and “-omics” data can be integrated. E. coli has proven to be a useful biological model organism, and constraint-based models have been developed for this organism over the last 13 years. The current E. coli models already include almost 1,000 genes, and 2,000-gene models are within reach (Fig. 1). An integrated 2,000-gene model (Fig. 4), accounting for the core functions of metabolism, transcription, translation, and regulation, can serve as a foundation on which other biological processes, such as growth and motility, can be simulated. In the foreseeable future, such constraint-based in silico models are expected to be commonly used in microbiology for hypothesis building and testing, driving both in silico and in vivo experimentation.
Acknowledgments
We acknowledge the support of grants from NSF (grant BES 01-20363) and NIH (grant GM 57089).
REFERENCES
- 1.Allen, T. E., and B. O. Palsson. 2003. Sequenced-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220:1-18. [DOI] [PubMed] [Google Scholar]
- 2.Arkin, A., J. Ross, and H. H. McAdams. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics 149:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard, D. A., S. D. Liang, and H. Qian. 2002. Energy balance for analysis of complex metabolic networks. Biophys. J. 83:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett 3rd, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Bonarius, H. P. J., G. Schmid, and J. Tramper. 1997. Flux analysis of underdetermined metabolic networks: the quest for the missing constraints. Trends Biotechnol. 15:308-314. [Google Scholar]
- 6.Burgard, A. P., and C. D. Maranas. 2001. Probing the performance limits of the Escherichia coli metabolic network subject to gene additions or deletions. Biotechnol. Bioeng. 74:364-375. [DOI] [PubMed] [Google Scholar]
- 6a.Burgard, A. P., and C. D. Maranas. 2002. An optimization based framework for inferring and testing hypothesized metabolic objective functions. Biotechnol. Bioeng, in press. [DOI] [PubMed]
- 7.Burgard, A. P., S. Vaidyaraman, and C. D. Maranas. 2001. Minimal reaction sets for Escherichia coli metabolism under different growth requirements and uptake environments. Biotechnol. Prog. 17:791-797. [DOI] [PubMed] [Google Scholar]
- 8.Carlson, R., D. Fell, and F. Srienc. 2002. Metabolic pathway analysis of a recombinant yeast for rational strain development. Biotechnol. Bioeng. 79:121-134. [DOI] [PubMed] [Google Scholar]
- 9.Covert, M. W., and B. O. Palsson. 2002. Transcriptional regulation in constraints-based metabolic models of Escherichia coli. J. Biol. Chem. 277:28058-28064. [DOI] [PubMed] [Google Scholar]
- 10.Covert, M. W., C. H. Schilling, I. Famili, J. S. Edwards, I. I. Goryanin, E. Selkov, and B. O. Palsson. 2001. Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26:179-186. [DOI] [PubMed] [Google Scholar]
- 11.Covert, M. W., C. H. Schilling, and B. Palsson. 2001. Regulation of gene expression in flux balance models of metabolism. J. Theor. Biol. 213:73-88. [DOI] [PubMed] [Google Scholar]
- 12.Domach, M. M., S. K. Leung, R. E. Cahn, G. G. Cocks, and M. L. Shuler. 2000. Computer model for glucose-limited growth of a single cell of Escherichia coli B/r-A. Biotechnol. Bioeng. 67:827-840. [DOI] [PubMed] [Google Scholar]
- 13.Drell, D. 2002. The Department of Energy Microbial Cell Project: a 180° paradigm shift for biology. OMICS J. Integr. Biol. 6:3-9. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, J. S., M. Covert, and B. Palsson. 2002. Metabolic modelling of microbes: the flux-balance approach. Environ. Microbiol. 4:133-140. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, J. S., R. U. Ibarra, and B. O. Palsson. 2001. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat. Biotechnol. 19:125-130. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, J. S., and B. O. Palsson. 2000. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. USA 97:5528-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards, J. S., and B. O. Palsson. 1999. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J. Biol. Chem. 274:17410-17416. [DOI] [PubMed] [Google Scholar]
- 18.Edwards, J. S., R. Ramakrishna, and B. O. Palsson. 2002. Characterizing the metabolic phenotype: a phenotype phase plane analysis. Biotechnol. Bioeng. 77:27-36. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, J. S., R. Ramakrishna, C. H. Schilling, and B. O. Palsson. 1999. Metabolic flux balance analysis, p. 13-57. In S. Y. Lee and E. T. Papoutsakis (ed.), Metabolic engineering. Marcel Dekker, New York, N.Y.
- 20.Fell, D. 1996. Understanding the control of metabolism. Portland Press, London, United Kingdom.
- 21.Fiehn, O., J. Kopka, P. Dormann, T. Altmann, R. N. Trethewey, and L. Willmitzer. 2000. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18:1157-1161. [DOI] [PubMed] [Google Scholar]
- 22.Flores, S., G. Gosset, N. Flores, A. A. de Graaf, and F. Bolivar. 2002. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by (13)C labeling and NMR spectroscopy. Metab. Eng. 4:124-137. [DOI] [PubMed] [Google Scholar]
- 23.Forster, J., I. Famili, P. C. Fu, B. O. Palsson, and J. Nielsen. 2003.. Genome-scale reconstruction of the Saccharomyces cerevisiae metabolic network. Genome Res. 13:244-253. [DOI] [PMC free article] [PubMed]
- 24.Hartemink, A. J., D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2002. Combining location and expression data for principled discovery of genetic regulatory network models. Pac. Symp. Biocomput., p. 437-449. [PubMed]
- 25.Hasty, J., D. McMillen, F. Isaacs, and J. J. Collins. 2001. Computational studies of gene regulatory networks: in numero molecular biology. Nat. Rev. Genet. 2:268-279. [DOI] [PubMed] [Google Scholar]
- 26.Ibarra, R. U., J. S. Edwards, and B. O. Palsson. 2002. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature 420:186-189. [DOI] [PubMed] [Google Scholar]
- 27.Karp, P. D., M. Riley, M. Saier, I. T. Paulsen, J. Collado-Vides, S. M. Paley, A. Pellegrini-Toole, C. Bonavides, and S. Gama-Castro. 2002. The EcoCyc Database. Nucleic Acids Res. 30:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klamt, S., and J. Stelling. 2002. Combinatorial complexity of pathway analysis in metabolic networks. Mol. Biol. Rep. 29:233-236. [DOI] [PubMed] [Google Scholar]
- 29.Lander, E. S. 1999. Array of hope. Nat. Genet. 21:3-4. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S., C. Phalakornkule, M. M. Domach, and I. E. Grossmann. 2000. Recursive MILP model for finding all the alternate optima in LP models for metabolic networks. Comp. Chem. Eng. 24:711-716. [Google Scholar]
- 31.Liao, J. C., S. Y. Hou, and Y. P. Chao. 1996. Pathway analysis, engineering and physiological considerations for redirecting central metabolism. Biotechnol. Bioeng. 52:129-140. [DOI] [PubMed] [Google Scholar]
- 32.Majewski, R. A., and M. M. Domach. 1990. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35:732-738. [DOI] [PubMed] [Google Scholar]
- 33.Naaby-Hansen, S., M. D. Waterfield, and R. Cramer. 2001. Proteomics-post-genomic cartography to understand gene function. Trends Pharmacol. Sci. 22:376-384. [DOI] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 35.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell. Sinauer Associates, Inc., Sunderland, Mass.
- 36.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 37.Palsson, B. O. 2000. The challenges of in silico biology. Nat. Biotechnol. 18:1147-1150. [DOI] [PubMed] [Google Scholar]
- 38.Palsson, B. O. 2002. In silico biology through “omics.” Nat. Biotechnol. 20:649-650. [DOI] [PubMed] [Google Scholar]
- 39.Papin, J. A., N. D. Price, J. S. Edwards, and B. O. Palsson. 2002. The genome-scale metabolic extreme pathway structure in Haemophilus influenzae shows significant network redundancy. J. Theor. Biol. 215:67-82. [DOI] [PubMed] [Google Scholar]
- 40.Patnaik, P. R. 2001. Microbial metabolism as an evolutionary response: the cybernetic approach to modeling. Crit. Rev. Biotechnol. 21:155-175. [DOI] [PubMed] [Google Scholar]
- 41.Phalakornkule, C., S. Lee, T. Zhu, R. Koepsel, M. M. Ataai, I. E. Grossmann, and M. M. Domach. 2001. A MILP-based flux alternative generation and NMR experimental design strategy for metabolic engineering. Metab. Eng. 3:124-137. [DOI] [PubMed] [Google Scholar]
- 42.Pramanik, J., and J. D. Keasling. 1998. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol. Bioeng. 60:230-238. [DOI] [PubMed] [Google Scholar]
- 43.Pramanik, J., and J. D. Keasling. 1997. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 56:398-421. [DOI] [PubMed] [Google Scholar]
- 44.Price, N. D., I. Famili, D. A. Beard, and B. O. Palsson. 2002. Extreme pathways and Kirchhoff's second law. Biophys. J. 83:2879-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raamsdonk, L. M., B. Teusink, D. Broadhurst, N. Zhang, A. Hayes, M. C. Walsh, J. A. Berden, K. M. Brindle, D. B. Kell, J. J. Rowland, H. V. Westerhoff, K. van Dam, and S. G. Oliver. 2001. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19:45-50. [DOI] [PubMed] [Google Scholar]
- 46.Reich, J. G., and E. E. Sel'kov. 1981. Energy metabolism of the cell: a theoretical treatise. Academic Press, London, United Kingdom.
- 47.Salgado, H., A. Santos-Zavaleta, S. Gama-Castro, D. Millán-Zárate, E. Díaz-Peredo, F. Sánchez-Solano, E. Pérez-Rueda, C. Bonavides-Martínez, and J. Collado-Vides. 2001. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 29:72-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schilling, C. H., M. W. Covert, I. Famili, G. M. Church, J. S. Edwards, and B. O. Palsson. 2002. Genome-scale metabolic model of Helicobacter pylori 26695. J. Bacteriol. 184:4582-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilling, C. H., J. S. Edwards, D. Letscher, and B. O. Palsson. 2000. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol. Bioeng. 71:286-306. [PubMed] [Google Scholar]
- 50.Schilling, C. H., D. Letscher, and B. O. Palsson. 2000. Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J. Theor. Biol. 203:229-248. [DOI] [PubMed] [Google Scholar]
- 51.Schilling, C. H., and B. O. Palsson. 2000. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J. Theor. Biol. 203:249-283. [DOI] [PubMed] [Google Scholar]
- 52.Schuster, S., T. Dandekar, and D. A. Fell. 1999. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 17:53-60. [DOI] [PubMed] [Google Scholar]
- 53.Schuster, S., and C. Hilgetag. 1994. On elementary flux modes in biochemical reaction systems at steady state. J. Biol. Syst. 2:165-182. [Google Scholar]
- 54.Segre, D., D. Vitkup, and G. M. Church. 2002. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 99:15112-15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2:35.1-35.7. [Online.] [DOI] [PMC free article] [PubMed]
- 56.Stelling, J., S. Klamt, K. Bettenbrock, S. Schuster, and E. D. Gilles. 2002. Metabolic network structure determines key aspects of functionality and regulation. Nature 420:190-193. [DOI] [PubMed] [Google Scholar]
- 57.Van Dien, S. J., and J. D. Keasling. 1998. A dynamic model of the Escherichia coli phosphate-starvation response. J. Theor. Biol. 190:37-49. [DOI] [PubMed] [Google Scholar]
- 58.Van Dien, S. J., and M. E. Lidstrom. 2002. Stoichiometric model for evaluating the metabolic capabilities of the facultative methylotroph Methylobacterium extorquens AM1, with application to reconstruction of C(3) and C(4) metabolism. Biotechnol. Bioeng. 78:296-312. [DOI] [PubMed] [Google Scholar]
- 59.Varma, A., B. W. Boesch, and B. O. Palsson. 1993. Biochemical production capabilities of Escherichia coli. Biotechnol. Bioeng. 42:59-73. [DOI] [PubMed] [Google Scholar]
- 60.Varma, A., and B. O. Palsson. 1993. Metabolic capabilities of Escherichia coli. I. Synthesis of biosynthetic precursors and cofactors. J. Theor. Biol. 165:477-502. [DOI] [PubMed] [Google Scholar]
- 61.Varma, A., and B. O. Palsson. 1993. Metabolic capabilities of Escherichia coli. II. Optimal growth patterns. J. Theor. Biol. 165:503-522. [DOI] [PubMed] [Google Scholar]
- 62.Varma, A., and B. O. Palsson. 1994. Metabolic flux balancing: basic concepts, scientific and practical use. Bio/Technology 12:994-998. [Google Scholar]
- 63.Varma, A., and B. O. Palsson. 1994. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60:3724-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walsh, K. J., and D. E. Koshland. 1985. Branch point control by the phosphorylation state of isocitrate dehydrogenase. A quantitative examination of fluxes during a regulatory transition. J. Biol. Chem. 260:8430-8437. [PubMed] [Google Scholar]
- 65.Wong, P., S. Gladney, and J. D. Keasling. 1997. Mathematical model of the lac operon: inducer exclusion, catabolite repression, and diauxic growth on glucose and lactose. Biotechnol. Prog. 13:132-143. [DOI] [PubMed] [Google Scholar]