Abstract
The bundle-forming pilus (BFP) is an important virulence factor for enteropathogenic Escherichia coli (EPEC). Genes involved in its biogenesis and regulation are tightly regulated by PerA (BfpT), a member of the AraC/XylS family of transcriptional regulators. The aim of this work was to purify PerA and determine its association with bfpA and perA (bfpT) regulatory regions by electrophoretic mobility shift and DNase I footprinting assays. PerA was purified as a maltose-binding protein (MBP) fusion, which was capable of complementing bfpA expression and which was able to restore the localized adherence phenotype of an EPEC perA mutant strain. Upstream of bfpA and perA, MBP-PerA recognized with similar affinity asymmetric nucleotide sequences in which a 29-bp-long AT-rich consensus motif was identified. These DNA motifs share 66% identity and were previously shown, by deletion analysis, to be involved in the PerA-dependent expression of both genes. Interestingly, in perA, this motif spans the sequence between positions −75 and −47, approximately one helix turn upstream of the −35 promoter sequence, while in bfpA, it spans the sequence between positions −83 and −55, approximately two helix turns upstream from the promoter. An additional PerA binding site was identified at the 5′ end of the bfpA structural gene, which was not required for its activation. Experiments with LexA-PerA fusions suggested that PerA acts as a monomer to activate the transcription of both perA and bfpA, in contrast to what has been documented for other members of this family of transcriptional regulators.
Enteropathogenic Escherichia coli (EPEC) strains are a major cause of acute and persistent diarrhea in children and a leading cause of infant death in developing countries (28). EPEC infections are characterized by two distinctive phenotypes: localized adherence (LA) and formation of attaching-and-effacing lesions (A/E) (11, 53). The LA phenotype involves the initial adherence of EPEC to the intestinal brush border in a nonintimate fashion, forming discrete microcolonies on the surface of epithelial cells. This phenotype is mediated by the bundle-forming pilus (BFP), a long flexible filament that forms surface organelles (bundles) that interconnect individual bacteria to create tridimensional clusters. The genes required for the biogenesis of the BFP are encoded by the EAF plasmid (42, 45, 48), while those responsible for the A/E phenotype are chromosomally encoded within the locus for enterocyte effacement (LEE) (10). The expression of BFP is under the positive control of PerA (also known as BfpT), an AraC/XylS-like transcriptional activator encoded by the per operon, which contains the genes perA, perB, and perC (14, 49). Expression of the bfp operon is modulated by environmental and physiological cues, such as temperature, the presence of ammonium, and the growth medium (35). PerA also autoregulates its expression in a positive manner (23) and has been involved in the regulation of ler, a positive regulator of genes encoded within and outside the LEE (9, 24), as well as that of trcA, a gene encoded within the locus for improving microcolony formation (LIM) (50). perA expression has also been shown to be modulated positively by a quorum-sensing mechanism (44) and negatively by GadX, an activator of glutamate descarboxylase genes, under acidic conditions (41). The essential role of PerA in EPEC pathogenesis was established in a study with volunteers in which a perA mutant strain showed significantly reduced virulence (3).
The nucleotide sequence alignment of the bfpA and perA regulatory regions revealed significant identity in a 40-bp-long AT-rich tract segment located upstream from the putative promoters, which was previously shown by deletion analysis to be necessary for the PerA-dependent activation of both promoters (5, 23). However, the precise binding site had not been defined, mainly due to the difficulties in purifying PerA.
PerA contains two putative helix-turn-helix (HTH) motifs at the C-terminal domain and is more related to those members of the AraC family that regulate virulence factors (13). Rns, the transcriptional regulator of expression of CS1 and CS2 pili in enterotoxigenic E. coli, and ToxT, a transcriptional regulator of cholera toxin and toxin-coregulated pilus in Vibrio cholerae, are two of the best-characterized members of this subfamily of virulence regulators (19, 27, 55). Rns has been shown to recognize two DNA binding sites upstream of the transcription start site of the coo operon and three sequence motifs—one upstream and two downstream of the rns transcriptional start site (27). It has also been shown that Rns can activate the cognate promoters of other AraC-like proteins, such as VirF, FapR, AggR, and CfaR, which, in turn, can replace Rns for the activation of the coo and rns promoters. However, Rns was not interchangeable with PerA (25).
The DNA sequences bound by AraC-like proteins involved in the regulation of virulence factors have been characterized for only few members of the subfamily, such as Rns (26), VirF of Yersinia spp. (54), UreR (47), ToxT (55), and, more recently, HilC and HilD (31). The two HTH motifs at the C terminus of these proteins seem to make contact with two adjacent major groove regions along one face of the DNA helix (26, 36). However, it is still not clear whether these proteins act as monomers or dimers.
Considering the important role of PerA in EPEC pathogenesis, in this work, we have determined the PerA-binding sequence of bfpA and perA by analyzing protein-DNA interactions and thus have proposed a consensus motif. An additional binding sequence was identified within the bfpA structural gene, which was not required for bfpA activation. In addition, the analysis of protein-protein interactions generated evidence that PerA acts as a monomer to activate the transcription of the bfpA and the perA promoters.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Overnight cultures were grown at 37°C in Luria-Bertani (LB) broth medium (38). Dulbecco's modified Eagle's medium (DMEM) containing 0.45% (wt/vol) glucose and l-glutamine (584 mg/liter), without sodium pyruvate (Gibco Life Technologies), and supplemented with pyridoxal (4 μg/ml), was used for growth at 37°C. When necessary, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 30 μg/ml; gentamicin, 5 μg/ml; tetracycline, 15 μg/ml; chloramphenicol, 30 μg/ml; and streptomycin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains E2348/69 | Wild-type EPEC O127:H26, Nalr | J. B. Kaper |
| E2348/69 perA::Km | E2348/69 with a nonpolar insertional mutation in perA | Unpublished results |
| JPN15 | EAF plasmid-cured derivative of E2348/69, Nalr | J. B. Kaper |
| MC4100 | F−araD139 (argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbcR | Laboratory strain |
| BL21/pLys21 | F−ompT (lon) hsdSB(rB mB) gal dcm (λDE3) | Invitrogen |
| KY2263 | MC4100 clpPX-lon::Cm | 51 |
| KY2266 | MC4100 clpPX-lon::Cm hslVU::Tc | 51 |
| BB2395 | MC4100 lon146::mini-Tn10 Tcr | 51 |
| SU101 | E. coli JL 1434 lexA71::Tn5 (Def) sulA211 Δ(lacIPOZYA)169/F′ lacIqlacZΔM15::Tn9 (op+/op+) | 8 |
| Plasmids | ||
| pCST | pACYC184 derivative carrying perA (bfpT) | 23 |
| pMalC2xa | Vector for constructing MBP fusions | New England Biolabs |
| pMALT2 | pMalC2xa derivative expressing MBP-PerA | This work |
| pKK232-8 | pBR322 derivative containing a promoterless cat gene | Pharmacia Biotech |
| pCAT201 | pKK232-8 derivative containing the bfpA-cat transcriptional fusion from nucleotides −201 to +76 | 5 |
| pCAT201 + 27 | pKK232-8 derivative containing the bfpA-cat transcriptional fusion from nucleotides −201 to +27 | Unpublished results |
| pSR660 | Vector encoding the LexA DBD sequence used for homodimerization studies; TcroriV | 7 |
| pMI660-PerA | pSR660 derivative carrying the lexADBD-perA fusion | This work |
| pRS658-CAT | pSR660 derivative carrying the lexADBD-cat fusion | 7 |
| pRS658-HNS | pSR660 derivative carrying the lexADBD-hns fusion | Unpublished results |
| pMA660-AraC | pSR660 derivative carrying the lexADBD-araC fusion | This work |
Molecular biology techniques.
DNA manipulations were performed according to standard protocols (38). Restriction and DNA-modifying enzymes were obtained from Roche, New England Biolabs, or Gibco BRL and used according to the manufacturer's instructions. Radiolabeled nucleotides ([γ-32P]dATP at 3,000 Ci mmol−1) were purchased from Amersham Corp. Oligonucleotides were purchased from BioSynthesis or provided by the Oligonucleotide Synthesis Facility at Instituto de Biotecnologia, UNAM, Cuernavaca, Mexico. The sequences of all oligonucleotides not reported here are available upon request. PCRs were performed in volumes of 100 or 50 μl with Taq Gold polymerase (Perkin-Elmer) according to the manufacturer's instructions. Double-stranded DNA sequencing of the plasmids generated in this work was carried out by the dideoxy-chain termination procedure with a Thermo Sequenase cycle sequencing kit according to the manufacturer's instructions (Amersham, Inc.).
Expression and purification of MBP-PerA.
Expression and purification of the maltose-binding protein (MBP)-PerA fusion were conducted as described previously (26). Briefly, the PerA expression plasmid pMAL-T2 was constructed by amplifying the perA gene from plasmid pCST with Taq Gold polymerase and cloned in frame at the 3′ end of the malE gene in pMALC2xa (New England Biolabs) by using the BamHI and PstI sites. Strain BL21/pLys7 transformed with pMAL-T2 was grown in LB medium with 0.2% glucose and 100 μg of ampicillin per ml at 30°C in an agitated water bath. The expression of MBP-PerA was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.3 mM when the culture reached an optical density at 600 nm (OD600) of 0.5 to 0.7, and then the culture was incubated for another 3 h. The bacterial cells were collected by centrifugation at 4°C, and the pellet was washed once with ice-cold column buffer (10 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol) and concentrated 100-fold with the same buffer. Cells were sonicated at 4°C by five 30-s pulses with 30-s resting cycles. Crude extract was obtained by centrifugation at 14,000 × g for 30 min at 4°C.
MBP-PerA was bound to an amylose column equilibrated with column buffer at room temperature and eluted with the same buffer supplemented with 10 mM maltose. The concentration of the purified protein was determined by the method of Bradford and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide).
Antibodies.
Rabbit polyclonal antibodies against a synthetic peptide (PerACOOH) with the sequence NH2-GCKKYNGVYSITQGTLP-COOH, corresponding to the last 15 amino acids of the carboxy terminus of PerA (positions 260 to 274) plus a glycine and cysteine that were added at the N-termini to improve the cross-linking reaction, were produced as follows. Peptide PerACOOH was coupled to bovine serum albumin (BSA) by disulfide cross-linking with SPDP (N-succinimidyl 3-[2-pyridyldithio] propionate]) (Pierce), as described by Alagón and King (1). New Zealand rabbits were immunized weekly with 200 μg of the BSA-peptide complex. After 13 weeks, the rabbits were bled, and the titers of antibody specific for the peptide were determined by enzyme-linked immunosorbent assay (ELISA). Western blot analysis demonstrated that the antipeptide antibodies were able to specifically recognize the full PerA protein. Anti-MBP and anti-LexA antibodies were purchased from New England Biolabs and Invitrogen, respectively. Anti-BfpA antibodies were kindly provided by Jorge Girón.
Adherence assay.
Adherence assays were performed as described previously (35). Briefly, bacteria were grown overnight in LB medium at 37°C with agitation. The next day, 5 ml of fresh DMEM supplemented with 1% LB medium was inoculated with a 1/50 dilution of the LB medium overnight and incubated at 37°C for 2 to 3 h (to an OD600 of approximately 0.6 to 0.8). Cell monolayers of HeLa cells, grown in DMEM on glass coverslips in a 24-well microplate (Costar), were infected with 100 μl of the bacterial culture and incubated at 37°C on a 5% CO2 atmosphere for 3 h. Unattached bacteria were removed by being washed three times with phosphate-buffered saline (PBS) solution, fixed with cold methanol, stained with Giemsa, observed under an optical microscope (Nikon), and photographed.
Western blotting.
Overnight bacterial cultures in LB medium were subcultured into DMEM and incubated in an agitated water bath at 37°C to an A600 of 1.0. One milliliter of each bacterial suspension was pelleted and resuspended in Laemmli buffer. Samples were boiled, subjected to SDS-PAGE (10% polyacrylamide), and transferred to 0.22-μm-pore-size nitrocellulose membranes (Amersham, United Kingdom). Membranes were blocked with 5% nonfat milk and incubated with anti-BfpA, anti-PerA, anti-LexA, or anti-MBP antibodies. Membranes were washed with Tris-buffered saline (TBS)-0.05% Tween 20, immunostained with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit antibody (Biomeda), and developed with Chemiglow chemiluminescence reagents according to the manufacturer (Alpha Innotech). Bands were detected with an Alpha-Imager imaging system. Native polyacrylamide gels were transferred in a semidry electrophoresis unit (Bio-Rad) to 0.22-μm-pore-diameter nitrocellulose membranes according to the manufacturer's instructions and processed as described above.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out as follows. PCR-amplified DNA fragments were incubated in binding buffer (25 mM HEPES [pH 7.9], 40 mM KCl, 3 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 5% glycerol) with different concentrations of MBP-PerA in a final volume of 20 to 25 μl at room temperature for 20 min. Samples were loaded onto 0.25× Tris-borate-EDTA (TBE)-2.5% agarose or native 6% acrylamide gels and run at 120 V at room temperature. Gels were stained with ethidium bromide and visualized with an Alpha-Imager UV transilluminator (Alpha Innotech).
Double-stranded oligonucleotide shift assays were performed as previously described (39). Briefly, 100 ng of each primer was 5′ end labeled with [γ-32P]dATP and T4 polynucleotide kinase (Amersham). The same amount of the corresponding complementary oligonucleotide was annealed to its radioactive partner by boiling them together and slowly cooling to room temperature. Double-stranded molecules were purified with Centricon filters (Millipore), and radioactive counts were determined with a Beckman coulter counter. EMSAs were carried out as described above, except that samples were loaded onto a 6% native polyacrylamide gel and run at room temperature with TGE buffer (50 mM Tris-HCl [pH 8.5], 384 nM glycine, 2 mM EDTA) at 300 V (37). Gels were vacuum dried and exposed in a PhosphorImager and/or exposed to X-ray films.
DNase I protection assay.
DNase I protection assays were done as described in references 26 and 32. PCR fragments carrying residues −201 to +26 of bfpA and −155 to +81 of perA were 32P labeled on the 5′ end of the bottom strand. About 100,000 cpm of the probes was preincubated at room temperature with increasing concentrations of MBP-PerA in the same binding buffer used for EMSAs. After 20 min, DNase I (0.003 U) (Roche) in dilution buffer (8 mM Tris-HCl [pH 7.9], 40 mM MgSO4, 4 mM CaCl2, 40 mM KCl, 2 mM EDTA [pH 8.0], 24% glycerol) was added to the mixture, which was incubated at room temperature for 2 min. The reaction was quenched by adding 300 μl of stop solution (570 mM ammonium acetate, 80% ethanol, 50 μg of carrier tRNA per ml). The DNA was precipitated, dried, and suspended in 8 μl of stop/loading buffer (45 mM Tris-borate [pH 8.0], 1 mM EDTA, 80% formamide). Samples were denatured at 85°C for 5 min and resolved by electrophoresis through an 8% polyacrylamide sequencing gel. Gels were vacuum dried and exposed to X-ray films or visualized with a PhosphorImager (Molecular Dynamics). Sequencing reactions performed with the primer corresponding to the 5′ end of the footprinting fragments were included as size markers.
Construction of the LexA-PerA fusion for protein-protein interaction assays.
The LexA-based genetic system used here to monitor protein-protein interactions in an E. coli background was described previously (7, 8). The PerA gene was PCR amplified with primers T-B2-190F and T-KpI-R by using Taq Gold polymerase with plasmid pCST (Table 1) as a template. The resulting fragment was cloned in-frame as a BglII-KpnI fragment into the LexA DNA binding domain (DBD)-coding sequence in pSR660, generating plasmid pMI660-PerA. Plasmids pSR658-HNS (V. H. Bustamante et al., unpublished results), pSR658-CAT (7), and pMA660-AraC (Table 1) were used as positive controls. These plasmids respectively encode LexADBD fusion proteins to H-NS, an oligomeric nucleoid-associated protein involved in both transcription regulation and DNA compaction (52); to chloramphenicol acetyltransferase (CAT), a homotrimeric enzyme (7); and to AraC, a dimeric transcriptional regulator involved in arabinose catabolism that represents the prototype protein of the AraC family (13, 17, 43) These plasmids were introduced into E. coli SU101, which was used as the reporter strain to monitor the ability of the hybrid proteins to form functional LexADBD dimers, with the capacity to repress the expression of a chromosomal sulA::lacZ fusion (8). Strains were grown overnight in LB medium plus 15 μg of tetracycline per ml at 37°C and subcultured the next day in LB medium supplemented with 1 mM IPTG. Samples were collected at OD600s of 0.6, 0.8, 1.0, and 1.2 and used for β-galactosidase assays as described before (32).
CAT assay.
The CAT assay was performed as previously described (6, 35).
RESULTS
Purification of PerA.
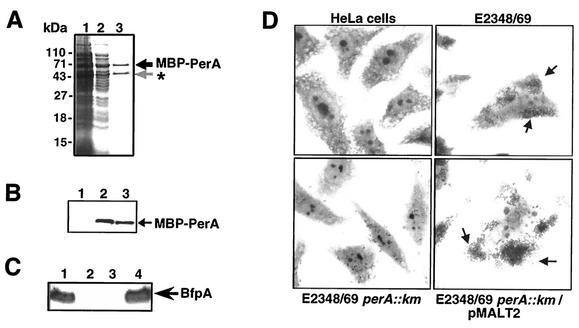
We have previously shown that expression of both bfpA and perA requires the PerA protein and a 40-bp-long AT-rich motif that is located upstream of their respective promoters (5, 23). To precisely define the PerA binding sites in the bfpA and perA promoters, PerA was fused to the MBP to facilitate its purification, by cloning the perA gene into pMALC2xa, generating pMAL-T2 (see Materials and Methods). This strategy has been used successfully for the purification of other AraC-like proteins, such as Rns and ExsA, involved in the regulation of virulence determinants in other bacteria (18, 26). Upon IPTG induction in E. coli BL21/pMAL-T2, the protein was purified throughout an amylose resin column, and two bands were observed in the eluted fractions (Fig. 1A). According to its apparent molecular mass, the first band, of approximately 71 kDa, corresponded to the full fusion protein, while the second, of ca. 51 kDa, corresponded to a truncated product that contains MBP and a portion of the N-terminus domain of PerA. The integrity of the MBP-PerA fusion protein in the first band was corroborated by Western blot analysis with rabbit polyclonal antibodies directed against the last 15 residues of the C-terminal sequence of PerA (Fig. 1B). These antibodies only recognized the upper band, indicating that the entire PerA sequence is expressed at the C terminus of the MBP. In contrast, the short band corresponded to a truncated version of the fusion, which was recognized only by anti-MBP antibodies (data not shown). This product was also generated in other E. coli strains, including EPEC and E. coli strains carrying mutations in major proteases such as Lon and ClpP (51), and in the presence of three protein inhibitors during the purification process (data not shown). Whether this processing is an artifact due to protein overexpression or to posttranscriptional events that control PerA levels in the cell is under investigation.
FIG. 1.
Purification and characterization of the MBP-PerA fusion. (A) PAGE of samples corresponding to different steps of the purification process. Lanes: 1, IPTG-induced cells; 2, crude extract after sonication and centrifugation; and 3, amylose column-purified protein. (B) Detection of MBP-PerA fusion by Western blotting with anti-PerA polyclonal antibodies. Lanes: 1, noninduced cells; 2, crude extract of IPTG-induced cells after sonication and centrifugation; 3, amylose column-purified protein. (C) Complementation of an EPEC E2348/69 perA::Km mutant with plasmid pMALT2 encoding MBP-PerA. Shown are Western blotting results with anti-BfpA antibodies. Lanes: 1, EPEC E2348/69; 2, JPN15; 3, perA::Km; 4, perA::Km/pMALT2. (D) LA assay on HeLa cells.
Cleavage with protease factor Xa generated two bands: one corresponding to MBP and the other corresponding to PerA, as tested by Western blotting with anti-MBP and anti-PerA antibodies (data not shown). However, the band corresponding to PerA was highly insoluble and unstable upon purification. Previous attempts to purify PerA by other strategies were unsuccessful, mainly due to its high insolubility, a characteristic shared by other AraC-like proteins (13). The purified MBP-PerA protein was soluble and stable upon storage at −70°C.
The ability of the recombinant MBP-PerA protein to correctly activate bfpA expression was tested in EPEC strain E2348/69 perA::Km, a strain lacking a functional perA gene (Table 1). First, the expression of BfpA, the structural component of BFP, was tested by Western blotting with extracts of EPEC strains E2348/69 wt, E2348/69 perA::Km, JPN15 (a strain lacking the EAF plasmid), and E2348/69 perA::Km/pMAL-T2. As shown in Fig. 1C, plasmid pMAL-T2 was able to complement the perA mutant for expression of BfpA. Moreover, an adherence assay on Hep-2 cells demonstrated that MBP-PerA was also able to restore the LA phenotype in the perA mutant strain (Fig. 1D).
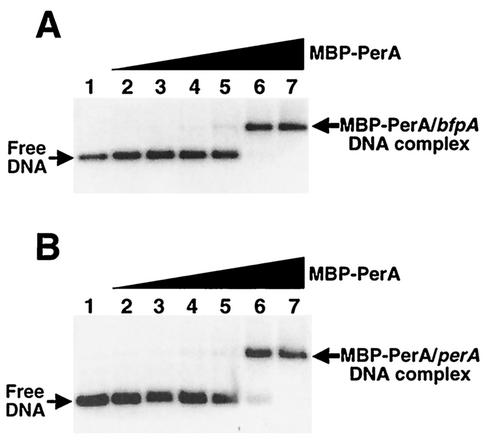
Furthermore, the DNA binding activity of the purified MBP-PerA was evaluated by EMSA (Fig. 2) with PCR products containing the bfpA (positions −214 to −15) and perA (positions −155 to +21) upstream regions that were previously shown to be essential for the PerA-dependent activation of both genes (5, 23). In both cases, DNA-protein complexes were formed at MBP-PerA concentrations of 2.16 μg (1.5 μM), indicating that the purified protein also maintained its DNA binding activity in vitro (Fig. 2). In addition, Western blot analysis of the shifted products with anti-PerA antibodies demonstrated that MBP-PerA was bound to the fragment. These complexes were further shifted when anti-PerA antibodies were included in the binding reaction (data not shown). These results demonstrated that MBP-PerA is functional and is appropriate for further in vitro studies.
FIG. 2.
Binding of MBP-PerA to bfpA and perA. EMSAs with a bfpA fragment from positions −214 to −15 (A) and a perA fragment from positions −155 to +21 (B) were carried out with 0, 0.11, 0.21, 0.43, 0.87, 2.16, and 4.3 μg (0, 74 nM, 148 nM, 298 nM, 596 nM, 1.5 μM, and 3 μM, respectively) of purified MBP-PerA (lanes 1 to 7). Samples were resolved in 2.5% agarose gels, stained with ethidium bromide, and photographed with an Alpha-Imager.
Mapping of PerA binding sites.
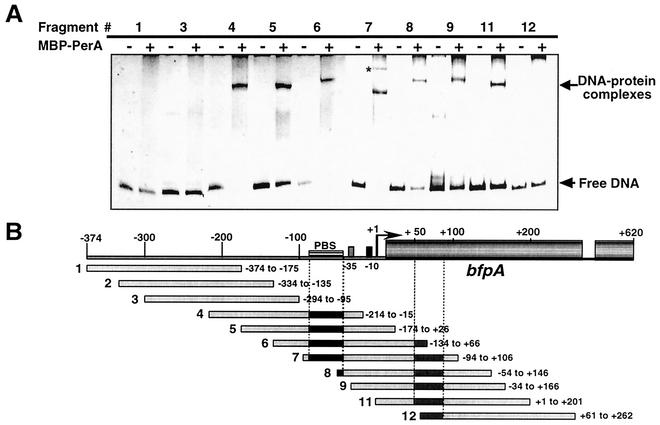
In order to delimit the PerA binding motif on the bfpA regulatory region, EMSAs were performed with overlapping 200-bp-long PCR fragments that span upstream and downstream bfpA sequence from positions −374 to +262 with respect to the transcription start point (Fig. 3). About 2.2 μg of purified MBP-PerA was mixed with approximately 50 ng of each PCR product, and the reaction mixtures were subjected to electrophoresis in 6% native polyacrylamide gels. In these experiments, fragments 4 to 11 formed one major shifted DNA-protein complex, with the interesting exception of fragment 7 (positions −94 to +106), which formed two complexes showing a distinctive mobility pattern (Fig. 3). Fragments 4 to 7 contained the previously proposed PerA binding sequence (5) and thus were expected to be shifted by PerA. However, fragments 8 to 11 were unexpectedly shifted, producing a complex similar to that observed for fragments containing the PerA binding sequence (Fig. 3A) (see below). The pattern observed for fragment 7 and the complex formed by MBP-PerA with fragments 8 to 11, suggested that a second binding site was present between the transcriptional start point and position +106 (Fig. 3B). In contrast, fragments 1 to 3 and 12, which do not contain a putative or full-length PerA-binding sequence, were not bound by MBP-PerA, a result that also supported the specificity of MBP-PerA binding to fragments containing a binding site.
FIG. 3.
Mapping of the PerA binding site on the bfpA regulatory region by EMSA. (A) Overlapping 200-bp-long PCR fragments that span the bfpA upstream and downstream promoter region from positions −374 to +262 were incubated with (+) or without (−) 2.16 μg (1.5 μM) of purified MBP-PerA. Samples were resolved in 6% native polyacrylamide gels and visualized by ethidium bromide staining. The asterisk denotes the second complex formed with fragment 7. (B) Schematic representation of the PCR fragments spanning the bfpA regulatory and structural regions. The fragment number is indicated to the left, and the sequence range covered for each fragment is indicated to the right. The approximate locations of the putative PerA binding sites, upstream and downstream of the bfpA promoter, are indicated by black and gray boxes, respectively. PBS, previously proposed PerA binding sequence
Similar experiments with fragments spanning the perA sequence from positions −161 to +425 allowed us to delimit the putative PerA binding sequence within the region previously proposed to contain this sequence motif (23) and to suggest the presence of a second PerA binding site located in the perA structural sequence downstream of position +180 (data not shown).
Analysis of PerA binding to mutant bfpA regulatory sequences.
We have previously reported that a single nucleotide deletion or insertion at different positions upstream from the promoter, but within the region needed for PerA-dependent activation, decreased bfpA expression to less than 2% (5). In order to define whether these mutations affected PerA affinity for its binding site, EMSAs were performed with 200-bp PCR fragments carrying either a single deletion or insertion at the 10-A tract between positions −65 and −74. Retardation of these fragments was equivalent to that observed for the wild-type sequence (data not shown), supporting the hypothesis that single insertions or deletions do not affect binding, but rather disrupt the correct orientation and alignment between the PerA binding site and the promoter, probably affecting PerA-RNA polymerase interactions (5).
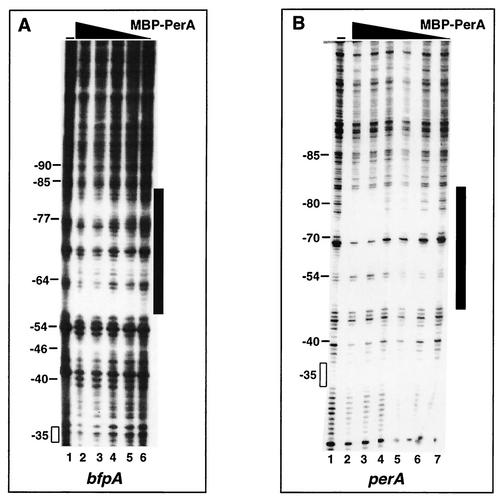
DNase I footprinting analysis.
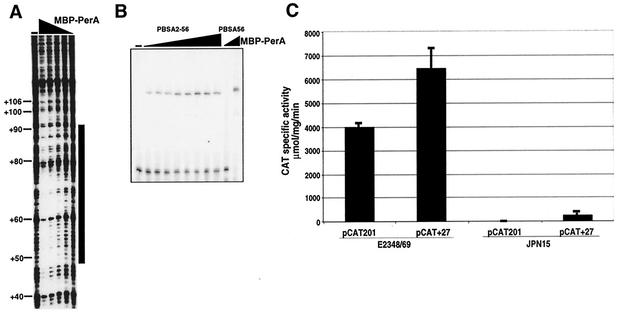
To determine more precisely the sequence recognized by PerA on both promoters, DNase I protection assays (Fig. 4) were performed with radiolabeled fragments that included the putative PerA binding sequences described above and increasing amounts of MBP-PerA, in the range of 37 nM to 1.5 μM for perA and 158 nM to 2.2 μM for bfpA. MBP-PerA protected the sequence located between positions −83 and −56 in bfpA (Fig. 4A), which is consistent with the results described above and those described previously (5). For perA, the protected region, located approximately between positions −81 and −47 (Fig. 4B), included a sequence motif that was previously shown to be important for the PerA-dependent activation of perA (23). Part of this motif is located upstream from the homology region previously suggested to constitute the PerA binding site (5).
FIG. 4.
DNase I protection of the bfpA (A) and perA (B) regulatory regions by MBP-PerA. Increasing amounts of MBP-PerA were mixed with 100,000 cpm of a 32P-end-labeled DNA fragment corresponding to positions −201 to +26 of bfpA and −155 to +81 of perA and treated with 0.003 U of DNase I. Samples were subjected to electrophoresis on an 8% polyacrylamide sequencing gel. The −35 promoter sequence and other upstream positions are indicated on the left; they were determined by running in parallel sequencing reactions with the same fragments (data not shown). The protected regions are indicated by vertical black bars. The black triangles above the gels represent increasing amounts of MBP-PerA. (A) Lanes: 1, DNA alone; 6 to 2, 159 nM, 316 nM, 634 nM, 1.26 μM, and 2.2 μM, respectively. (B) Lanes: 1, DNA alone; 7 to 2, 37 nM, 74 nM, 148 nM, 295 nM, 753 nM, and 1.5 μM, respectively.
PerA binds to double-stranded oligonucleotides containing the putative PerA binding sites.
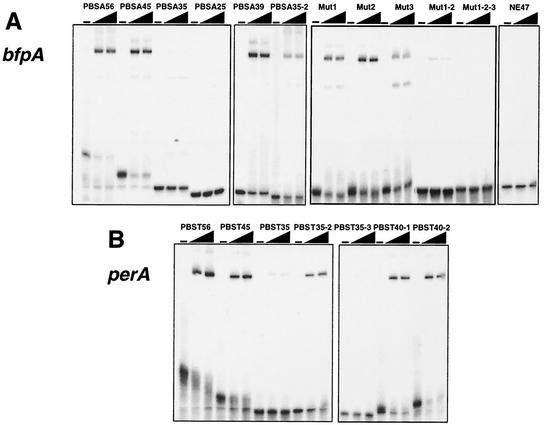
To further characterize the PerA binding motifs, EMSAs were performed with double-stranded oligonucleotides of different lengths, which were designed according to the results described above (Fig. 5 and Tables 2 and 3).
FIG. 5.
PerA binds to double-stranded oligonucleotides containing the putative PerA binding site. EMSAs were performed by incubating 32P-labeled double-stranded oligonucleotides of different lengths containing bfpA (A) or perA (B) sequences (as described in Tables 2 and 3, respectively), with 1.06 and 2.1 μM MBP-PerA. Control reactions with DNA alone were included as controls. A nonspecific 47-mer oligonucleotide containing an unrelated sequence was included as a negative control. In addition, control reactions with 50- and 100-fold molar excesses of the unlabeled 45-mer (PBSA45) were included to demonstrate the specificity of the interaction (data not shown). Samples were subjected to electrophoresis on a 6% native polyacrylamide gel.
TABLE 2.
Binding to double-stranded oligonucleotides corresponding to the bfpA regulatory regiona
| Probe | Sequence relative to bfpA promoter region | Relative shift (%)b |
|---|---|---|
| −98−90−85−77−64−54−46−40−35 |||||||| | ||
| GTACTGGGGGGGACGGAAATATATAAAAAAAAAAGAAAAAAAGATTATTTTTTTTCTTGGTGCTTGCGTGTCTT | ||
| PBSA56 | GGGGACGGAAATATATAAAAAAAAAGAAAAAAAGATTATTTTTTTTCTTGGTGCT | 100 |
| PBSA45 | GGGGACGGAAATATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 100 |
| PBSA35 | GGGGACGGAAATATATAAAAAAAAAAGAAAAAAAG | 19 |
| PBSA25 | GGGGACGGAAATATATAAAAAAAAA | <1 |
| PBSA2/2 | AGAAAAAAAGATTATTTTTTTTCTTGG | 0 |
| PBSA39 | GGAAATATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 100 |
| PBSA35-2 | ATATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 47 |
| Mut1 | GGGGACAACCCGATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 70 |
| Mut2 | GGGGACGGAAATATCGCCCCAAAAAAGAAAAAAAGATTATTTTTT | 82 |
| Mut3 | GGGGACGGAAATATATAAAAAAAAAAGAAAGGCCAATTATTTTTT | 43 |
| Mut1-2 | GGGGACAACCCGATCGCCCCAAAAAAGAAAAAAAGATTATTTTTT | 21 |
| Mut1-2-3 | GGGGACAACCCGATCGCCCCAAAAAAGAAAGGCCAATTATTTTTT | <1 |
| 5′Mut | TCTTCTGGAAATATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 75 |
| PBSAmG64 | GGGGACGGAAATATATAAAAAAAAAAAAAAAAAAGATTATTTTTT | 100 |
| PBSAmG84 | GGGGACAGAAATATATAAAAAAAAAAGAAAAAAAGATTATTTTTT | 100 |
| PBSA2-56c | TTATGGTTTCTAAAATCATGAATAAGAAATACGAAAAAGGTCTGTCTTTGATTGAA | 53d |
| NE34e | TCGAGCTCTCTTCTGTTCTGCTTGACTTCCTAGG | <1 |
| NE47e | GATCAGTCCTTGAATGGTGTGAAGTAAAAGTGCCTTCAAAGAATCCC | 4 |
Modified sequences are underlined. The boldface letters indicate the location of the minimal sequence required for PerA binding.
Estimated by EMSA relative to PBSA45.
Sequence corresponding to positions +38 to +93 of bfpA.
Relative to PBSA56.
Random sequences.
TABLE 3.
Relative binding to double-stranded oligonucleotides corresponding to the perA regulatory regiona
| Probe | Sequence relative to perA promoter region | Relative shift (%)b |
|---|---|---|
| −81−75−66−54−40−35 ||||| | ||
| CAACACCTTGAAAAATATCAGTAAATTTTTAAAAAAAAAGCATAAAAGTATTGATTATTTACAGAG | ||
| PBST56 | CAACACCTTGAAAAATATCAGTAAATTTTTAAAAAAAAAGCATAAAAGTATTGATT | 100 |
| PBST45 | CAACACCTTGAAAAATATCAGTAAATTTTTAAAAAAAAAGCATAA | 100 |
| PBST35 | CAACACCTTGAAAAATATCAGTAAATTTTTAAAAA | 8 |
| PBST40-1 | CCTTGAAAAATATCAGTAAATTTTTAAAAAAAAAGCATAA | 100 |
| PBST35-2 | AAAAATATCAGTAAATTTTTAAAAAAAAAGCATAA | 50 |
| PBST40-2 | GAAAAATATCAGTAAATTTTTAAAAAAAAAGCATAAAAGT | 100 |
| PBST35-3 | GTAAATTTTTAAAAAAAAAGCATAAAAGTATTGAT | 0 |
Modified sequences are underlined. The boldface letters indicate the location of the minimal sequence required for PerA binding.
Estimated by EMSA relative to PBST45.
MBP-PerA binding to a 45-bp DNA probe (PBSA45) (positions −90 to −46) (Fig. 5A) that contained the DNase I-protected region of the bfpA promoter resulted in the formation of a major retarded band. Most of the probe was retarded at an MBP-PerA concentration of 1.06 μM. This binding was specifically competed by an excess of cold probe (not shown). In addition, a nonspecific probe was not shifted by the highest concentration of protein used in these experiments (Fig. 5A). An 11-bp extension, added to the 3′ end to create the oligonucleotide PBSA56 (positions −90 to −35) (Table 2), did not significantly increase PerA binding for this site (Fig. 5A). In contrast, elimination of 10 bp at the 3′ end of PBSA45 to create PBSA35 (positions −90 to −56) reduced PerA binding by about 81%, and a further 10-bp reduction at the 3′ end to create PBSA25 (positions −90 to −66) abolished PerA binding (Table 2 and Fig. 5A). Similarly, a 27-bp probe, PBSA2/2 (positions −65 to −39), which contains 20 bp of the 3′ end of the original PBSA45 plus an extension of 7 bp, did not show any binding (Table 2) (data not shown).
Elimination of 5′ bases with respect to PBSA45 down to position −84 (probe PBSA39) did not affect binding significantly; however, a further 5′ deletion down to position −80 (probe PBSA35-2) reduced binding by about 53% (Table 2 and Fig. 5A). These data suggested that PerA binds efficiently to a sequence located between positions −84 and −45 in the bfpA promoter and that around 39 to 45 bp is required for efficient binding in vitro.
We then tested the role in PerA binding of particular nucleotide positions, by designing probes carrying different modifications within the region that constitutes the binding site. Based on the sequence of PBSA45, positions −90 to −85, −84 to −79, −76 to −71, and −60 to −56 were substituted with random sequences to create probes 5′ Mut, Mut1, Mut2, and Mut3, respectively (Table 2). These changes reduced PerA binding by 25, 30, 18, and 57%, respectively (Table 2 and Fig. 5A). For probes in which changes in Mut1 and -2 (Mut1-2) or Mut1, -2, and -3 (Mut1-2-3) were combined, binding was reduced by about 79% or almost abolished (Table 2 and Fig. 5A), while a single point mutation at position −84 (PBSAmG84) or −64 (PBSAmG64) did not affect PerA binding (Table 2) (data not shown). Two unspecific probes (NE34 and NE47) were used as negative controls for these assays, and for both cases, no shifted band was observed (Table 2 and Fig. 5A) (data not shown).
Changes at the 5′ end of the sequence (5′ Mut, Mut1, and Mut2) only caused a moderate reduction in binding (Fig. 5A and Table 2), suggesting that these motifs are not essential but may contribute to create the appropriate structure for the efficient interaction between PerA and the DNA. However, the combination of these changes had a more dramatic effect on binding, especially when combined with the changes in the sequence motif modified in Mut3, which seems to play a more relevant role in PerA binding (Table 2). In this regard, we have previously shown that the elimination of the sequence motif modified in Mut1, in the context of a bfpA-cat transcriptional fusion (pCAT77), caused an 84% reduction in CAT activity with respect to a bfpA-cat fusion carrying a fragment up to position −85 (5). Likewise, single insertions or deletions that disrupt the appropriate orientation of these motifs between each other or between them and the promoter had dramatic effects by almost abolishing bfpA expression (5). Overall, these results indicate that PerA binds to a sequence located between positions −83 and −49 (Table 2), which shows some flexibility for in vitro binding. However, they also support the notion that the appropriate distribution of particular motifs allows the establishment of functional protein-DNA interactions that permit the transcriptional activation of this promoter.
Similar experiments were performed with double-stranded oligonucleotides containing the putative PerA binding site identified for perA. A 45-bp probe (PBST45) (Table 3) that spans positions −84 to −40 was retarded at an MBP-PerA concentration of 1.06 μM, which was slightly more efficient than the retardation shown at the same concentration for PBSA45 (data not shown). As for the binding site in bfpA, a perA probe with a 3′ extension down to position −30 (PBST56) (Table 3), did not significantly increase PerA binding; in contrast, binding of a 35-bp probe (PBST35), lacking 10 bp at the 3′ end with respect to the sequence of PBST45 (positions −84 to −50), was severely reduced (Table 3 and Fig. 5B). Furthermore, 40-bp-long probes carrying positions −79 to −40 (PBST40-1) and −75 to −36 (PBST40-2), which still included the DNase I-protected region, were efficiently retarded by PerA (Table 3 and Fig. 5B), while binding to a 35-bp probe (PBST35-2) lacking 5′ positions −84 to −75 was reduced by 50% (Table 3 and Fig. 5B). Finally, a 35-bp probe (PBST35-3) containing slightly more than the 3′ half of the site (positions −64 to −30) was not bound by the protein (Table 3 and Fig. 5B). These results demonstrated the relevance of the sequence located between positions −75 and −40 for PerA binding and are also in agreement with a deletion analysis showing that a deletion down to position −66 almost abolished the expression of a perA-cat fusion (23).
As for other AraC-like proteins, here we have shown that, in order to activate transcription, PerA binds to AT-rich regions of about 40 bp located between positions −84 and −46 at the bfpA promoter and between −75 and −40 at the perA promoter. The comparison of these two motifs allows us to propose a consensus PerA DNA binding sequence and a revised sequence alignment between the bfpA and perA regulatory regions (Fig. 6). Although they share an AT-rich composition, the consensus PerA DNA binding sequence does not have homology with the consensus sequences proposed for other AraC-like proteins that are involved in virulence gene regulation, such as Rns and ToxT (19, 26, 55).
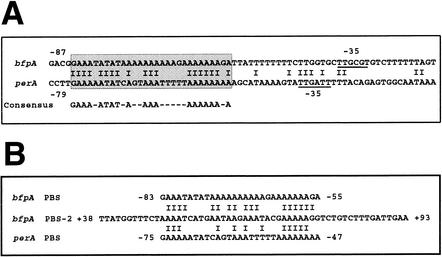
FIG. 6.
PerA binding consensus sequence. (A) Nucleotide sequence alignment of the bfpA (upper line) and perA (lower line) regulatory regions. The boxed sequence denotes the 29-bp-long region that shares 66% identity between bfpA and perA, is required for PerA binding (this study), and has been previously shown to be required for the PerA-dependent activation of both promoters (5, 23). The proposed consensus PerA binding sequence is shown below the alignment. The −35 promoter sequence of both bfpA and perA is underlined. (B). Comparison of the second PerA binding sequence identified at the bfpA structural gene. Positions relative to the transcriptional start site are indicated to the left and right of each sequence.
Identification of the second PerA binding site in the structural sequence of bfpA.
As described above, EMSAs with PCR-overlapping fragments spanning bfpA regulatory and structural sequences allowed the identification of a second PerA binding site at the 5′ end of the bfpA coding sequence, approximately between positions +1 and +106 (Fig. 3). In order to map this site, we performed footprinting analysis of this region with purified MBP-PerA. In agreement with the EMSA shown in Fig. 3, residues between positions +45 and +100 were protected from DNase I digestion (Fig. 7A). To confirm this finding, we performed EMSAs with increasing amounts of MBP-PerA and a 56-bp-long double-stranded oligonucleotide (PBSA2-56) (positions +38 to +93 of bfpA) that encompasses most of the protected sequence (Table 2 and Fig. 7B). This region contains a sequence motif between positions +49 and +77 that shares 59 and 41% identity with the PerA binding sequences found in bfpA and perA, respectively (Fig. 6B). For comparison, PBSA56 carrying the PerA binding site identified upstream from the bfpA promoter (Table 2) was included in the same experiment. PBSA2-56 was also shifted by MBP-PerA, but less efficiently than the retardation observed for PBSA56 (about 50%). In order to determine the role of this second binding site, we analyzed the transcriptional activity of a bfpA-cat fusion containing a bfpA fragment from positions −201 to +27 (pCAT + 27), which lacks this sequence motif, and a fusion containing a fragment from positions −201 to +76 (pCAT201), which partially contains this motif. As shown in Fig. 7C, activation of both fusions was mainly PerA dependent, because they were highly active in the wild-type EPEC strain, but not in an EAF-negative strain, indicating that this second binding-site was not required for activation.
FIG. 7.
Characterization of the second binding site on bfpA. A) The DNase I protection assay of a fragment containing the sequence from positions −54 to +166 was carried out as described in the legend to Fig. 4. The black triangle above the lanes indicates decreasing amounts of MBP-PerA (0, 2.6 μM, 1.7 μM, 880 nM, 528 nM, 176 nM, and 88 nM). (B) EMSA of a 56-mer double-stranded oligonucleotide containing the bfpA sequence between positions +38 and +93 (PBSA2-56) (Table 2). The triangle above the lanes indicates increasing amounts of MBP-PerA (0, 133 nM, 238 nM, 475 nM, 958 nM, 1.85 μM, 5.04 μM, 6.6 μM, and 8.4 μM). Probe PBSA56 was included as a control in the absence or presence of 1.85 μM MBP-PerA. (C) The second PerA binding sequence is not required for bfpA expression. EPEC strains E2348/69 (wild type) and JPN15 (pEAF cured) were transformed with either pCAT201 or pCAT + 27, which carry transcriptional fusions between the reporter cat gene and bfpA regulatory fragments from positions −201 to +76 and −201 to +27, respectively. The CAT specific activity was determined from samples collected from bacterial cultures grown on DMEM at an OD600 of 1.0. The results are the average of three different experiments.
Although neither the role nor the mechanism for PerA binding at a second site was elucidated in this study, we cannot rule out the possibility that such a site has a function under specific conditions. Additional binding sites, downstream of the transcriptional start site, have been described for other AraC-like regulators involved in the regulation of virulence factors (25). For Rns, one of the additional sites located within the rns structural gene was essential for the Rns-dependent activation of its own promoter (27).
PerA does not form dimers as a functional LexA-PerA fusion protein.
It is not known whether the active form of AraC family members that regulate virulence factors is a dimer or a monomer. Dimerization has been shown to be important for the activity of AraC family members that regulate transcription in response to a chemical signal such as AraC, which is active in the presence of arabinose (20, 43), or UreR, which is active in the presence of urea (33). Dimerization has also been suggested based on genetic evidence for Rns-like proteins such as Rns (26), VirF of Shigella flexneri (34), and ToxT of V. cholerae (19). The dimerization capacity of AraC and UreR correlates with the presence of critical leucines that are part of predictable coiled-coil domains at the amino-terminus domain. A coiled-coil domain is a primary structural element in proteins that mediates homo- or hetero-oligomeric interactions (21). Analysis of the PerA sequence with different programs that allow the prediction of potential coiled-coil domains did not reveal the presence of any predictable dimerization domain at its N terminus (data not shown), suggesting that PerA did not have the capacity to form dimers. The apparent lack of symmetry of the sequence motifs required for PerA binding (Fig. 6) further suggested that PerA binds as a monomer instead of as a dimer.
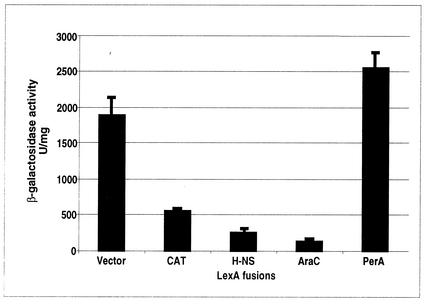
In order to assess PerA dimerization, a LexA-based genetic system was used to evaluate potential protein-protein interactions between monomers of PerA. As positive controls, the N-terminal DBD of LexA was fused to the full-length sequences of various proteins, whose dimerization or oligomerization capacity had been tested before. H-NS and AraC are two transcriptional regulatory proteins in which the active form that binds to DNA and regulates transcription is an oligomer or a dimer (22, 52), and CAT is a homotrimeric enzyme previously used as a positive control in this assay (7). The full-length PerA protein was also fused in frame to the distal part of the LexA DBD, creating plasmid pMI660PerA (Table 1). This plasmid was first transformed into an EPEC E2348/69 perA-negative strain to determine if the LexA-PerA hybrid protein still retained the transcriptional activator properties of wild-type PerA. The hybrid protein restored the expression of BfpA, as observed by Western blotting and the development of the LA phenotype (data not shown), indicating that PerA was still functional when fused to the LexA DBD.
The H-NS, CAT, and AraC control hybrid proteins, which were expected to form functional LexA dimers due to their capacity to oligomerize, repressed the expression of a sulA::lacZ fusion in the E. coli reporter strain SU101 (Fig. 8). The repressor activity of the LexA-AraC fusion protein demonstrated that this assay was useful to assess the dimerization capabilities of proteins belonging to this family of transcriptional activators. In contrast, the LexA-PerA hybrid did not show any repressor activity, strongly suggesting that PerA does not dimerize (Fig. 8).
FIG. 8.
A LexA-PerA fusion does not form dimers. The ability of the full-length PerA protein fused to the LexA DBD to dimerize was evaluated by measuring the activity of a sulA::lacZ fusion in E. coli SU101. The β-galactosidase activity was determined from samples obtained from bacterial cultures grown on LB medium supplemented with 1 mM IPTG at an OD600 of 0.8. The LexA-AraC, LexA-H-NS, and LexA-CAT fusions were used as positive controls. The results are the average of three different experiments.
DISCUSSION
PerA plays a critical role in the regulation of virulence gene expression in EPEC (14, 23, 24, 49). Its essential role has been supported by the observation that a functional perA gene was required for full virulence in human volunteers (3) in addition to its high degree of conservation in different clinical isolates belonging to classical EPEC serotypes associated with human infections (30).
Despite its proposed pleiotropic role in the regulation of virulence gene expression in EPEC, the transcriptional activation of only two promoter regions, bfpA and perA, has been shown to be completely dependent on PerA (5, 23, 35, 49). In contrast, its role in the activation of other virulence genes, such as ler or trcA, seems to be modulatory, because the absence of PerA in an EPEC background has only a mild effect on their expression (6, 24, 50).
As for other AraC-like proteins involved in the regulation of virulence factors, the biochemical characterization of PerA has been hampered by difficulties in purifying a soluble protein (13). In some cases, this obstacle has been overcome by purifying a chimeric protein in which the AraC homologue is fused to a protein or peptide that facilitates its purification and increases its solubility and stability (26, 31, 55). In this study, we have been able to analyze the DNA binding properties of PerA by using a purified MBP-PerA fusion that retains the transcriptional activator features of wild-type PerA in vivo and shows DNA binding activity in vitro (Fig. 1 and 2).
MBP-PerA shifted PCR fragments containing the minimal sequence previously shown to be required for the PerA-dependent activation of bfpA, the gene encoding the BFP structural subunit and the first part of the bfp operon, as well as that of its own gene, previously shown to be autoregulated (5, 23). PerA protected sites between positions −83 and −56 in bfpA and −81 and −47 in perA. Double-stranded oligonucleotides of different lengths, spanning the protected sequences, were used in EMSAs to determine the minimal sequence required by PerA for efficient DNA binding in vitro. For both promoters, around 40 bp was required for efficient shifting. The nucleotide alignment of the defined minimal sequences allowed the proposal of a consensus PerA binding motif, which consisted of an AT-rich 29-bp-long sequence in which 19 positions were identical between the two promoters (Fig. 6). AraC-like proteins recognize sequence motifs that range from 17 to 60 bp. MarA, Rob, and SoxS, three AraC-like proteins involved in stress response, interact as monomers with a 17- to 19-bp-long motif in two adjacent segments of the major groove (16, 36). AraC binds two 17-bp-long inverted repeats that are separated by 6 bp (17). Similarly, XylS, MelR, and RhaS have been shown to bind sequences between 33 to 56 bp that generally include two binding sites separated by a variable number of bases (2, 4, 15, 46). The binding site for Rns and related virulence gene regulators, such as VirFSf, CfaR, and AggR, is contained in an AT-rich 30-bp-long sequence (25, 26). Despite the sequence similarities at the HTH motifs between PerA and other virulence regulators, such as Rns, VirF, or ToxT, no clear sequence similarity was found between the PerA binding site and the binding sites described for these proteins (19, 26, 27, 54, 55). This offers an explanation for why PerA was not able to substitute for Rns to activate the coo and rns promoters and why Rns could not substitute for PerA to activate the perA promoter (25).
In perA, the PerA binding motif is located about one helix turn upstream from the −35 promoter sequence, while in bfpA it is located about two helix turns upstream from the promoter (Fig. 6). The functional significance of this distribution for the activation of both promoters is being further analyzed in the context of potential interactions between PerA and RNA polymerase or additional regulatory proteins. In this regard, we have recently observed that bfpA is not expressed in the absence of integration host factor (IHF) (Y. Martínez-Laguna et al., unpublished observations), a global regulatory protein that has also been involved in the positive regulation of ler expression (12).
The lack of apparent symmetry in the sequence motif bound by PerA and the observation that it forms one major DNA-protein complex by EMSA suggested that it acts as a monomer and not as a dimer, as has been shown for other AraC-like proteins (13). This possibility was tested with a LexA-based hybrid bacterial system that allows the monitoring of protein-protein interactions (7, 8). In order to further corroborate the functionality of the assay, two DNA binding proteins, H-NS and AraC, and an enzyme, CAT, were used as positive controls because their dimerization or multimerization properties have been well documented (7, 17, 52). In all three cases, the hybrid proteins allowed the formation of functional LexA dimers that inhibited the expression of the sulA::lacZ reporter fusion. In particular, the result obtained with the LexA-AraC fusion demonstrated that the LexA-based system is useful to evaluate the dimerization or multimerization properties of other members of the AraC family of transcriptional activators (Fig. 8). In contrast, a LexA-PerA fusion that still conserved the ability to activate bfpA transcription and BFP production in an EPEC perA mutant strain (data not shown) was unable to repress the reporter fusion, strongly suggesting that PerA does not form dimers. Furthermore, expression of the N-terminal domain of PerA from an inducible promoter did not exert a dominant-negative effect on the expression of the bfpA-cat fusion or on the expression of the BfpA subunit in wild-type EPEC, as evaluated by CAT assays and Western blotting (data not shown). This observation supports the notion that PerA does not form dimers to activate transcription.
Based on these results, it is likely that PerA binds to its target sequences upstream of the bfpA and perA promoters as a monomer, probably following the interaction model based on the crystal structure of MarA bound to its target DNA (36), which has been also proposed for Rns (26). In this model, both HTH motifs form specific interactions with two adjacent segments of the major groove along one face of the DNA helix, via nonidentical nucleotide contacts. Dimerization of AraC-like proteins involved in virulence gene control has been proposed: for example, for VirF and ToxT, based on dominant-negative phenotypes observed with mutants that cannot bind DNA (19, 34). At this point, we cannot rule out the possibility that PerA dimerizes upon binding to its target DNA sequence, a mechanism that has been proposed for MelR (4).
The HTH2 motif is the region with the highest similarity between the AraC-like proteins and a consensus sequence derived from the alignment of more than 100 proteins, constituting the AraC family signature (13). The high degree of conservation of this motif has been used as an argument to propose that its function is common for all the AraC-like proteins, which is likely to involve interactions with RNA polymerase. In this regard, we have previously observed that the insertion or deletion of a single nucleotide between the bfpA promoter and the putative PerA binding site abolished bfpA transcription. This suggested that changes in the orientation of the binding site and the promoter affect the proper presentation of a critical protein surface that allows PerA-RNA polymerase interactions that are important to promote transcriptional activation (5). These insertions or deletions did not reduce the affinity of PerA for PCR fragments containing these modifications (data not shown), further supporting the notion that they rather disrupt the appropriate contacts between PerA and RNA polymerase.
In the case of AraC, binding of a sugar effector molecule (arabinose) to the N-terminus domain of the protein induces a structural change that produces the activator form of the AraC dimer (29, 40, 43). For other members of the family, binding of different molecules, such as rhamnose for RhaS, urea for UreR, or alkylbenzoates for XylS, induces similar conformational changes that allow transcriptional activation of their target genes. Thus, the presence of these molecules constitutes the regulatory signals (13). The PerA-mediated expression of bfpA and perA in EPEC is negatively modulated by temperatures above or below 37°C, by the presence of ammonium, and by growth in rich medium (23, 35). Other AraC-like proteins involved in the regulation of virulence factors have also been shown to respond to environmental cues, such as temperature. However, no effector molecules controlling the activity of the members of this subfamily of AraC-like proteins have been identified. It remains to be determined whether proteins such as PerA also undergo conformational changes in response to environmental or physicochemical signals that shift the protein from a constitutively active to inactive form.
Acknowledgments
We particularly thank A. Alagón, V. H. Bustamante, E. Calva, R. A. Edwards, G. Munson, N. Olivares, G. Pedraza, and Y. Rosenstein for advice and helpful discussions. We are also grateful to M. G. Sosa for constructing plasmid pMAL-T2, A. Vázquez for constructing the EPEC perA::Km strain, O. Rodríguez for providing HeLa cells for the adherence assays, J. A. Girón for the BfpA antibodies, R. P. Silver for the LexA-based system, V. H. Bustamante for the plasmid encoding the LexA-HNS fusion and pCAT + 27, E. Mata for animal care, and E. López-Bustos for oligonucleotide synthesis.
This research was supported by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT 33115-N) and the Universidad Nacional Autónoma de México (DGAPA IN-217201) and by a Howard Hughes International Research Scholar Award to J.L.P. J.A.I. was supported by a Ph.D. Fellowship from CONACYT (no. 86036) and by the Dirección General de Estudios de Posgrado (DGEP), UNAM.
REFERENCES
- 1.Alagon, A. C., and T. P. King. 1980. Activation of polysaccharides with 2-iminothiolane and its uses. Biochemistry 19:4341-4345. [DOI] [PubMed] [Google Scholar]
- 2.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 181:5185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 4.Bourgerie, S. J., C. M. Michan, M. S. Thomas, S. J. Busby, and E. I. Hyde. 1997. DNA binding and DNA bending by the MelR transcription activator protein from Escherichia coli. Nucleic Acids Res. 25:1685-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante, V. H., E. Calva, and J. L. Puente. 1998. Analysis of cis-acting elements required for bfpA expression in enteropathogenic Escherichia coli. J. Bacteriol. 180:3013-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 7.Daines, D. A., and R. P. Silver. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182:5267-5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, S. J., V. Sperandio, J. A. Girón, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 11.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 13.Gallegos, M.-T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Perez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marques. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286-2290. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, K. L., and R. E. Wolf, Jr. 2001. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 40:1141-1154. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson, W., and R. Schleif. 1985. A dimer of AraC protein contacts three adjacent major groove regions of the araI DNA site. Proc. Natl. Acad. Sci. USA 82:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hovey, A. K., and D. W. Frank. 1995. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 177:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulbert, R. R., and R. K. Taylor. 2002. Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter. J. Bacteriol. 184:5533-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRonde-LeBlanc, N., and C. Wolberger. 2000. Characterization of the oligomeric states of wild type and mutant AraC. Biochemistry 39:11593-11601. [DOI] [PubMed] [Google Scholar]
- 21.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 22.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 24.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 25.Munson, G. P., L. G. Holcomb, and J. R. Scott. 2001. Novel group of virulence activators within the AraC family that are not restricted to upstream binding sites. Infect. Immun. 69:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 181:2110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 28.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niland, P., R. Huhne, and B. Muller-Hill. 1996. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 264:667-674. [DOI] [PubMed] [Google Scholar]
- 30.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olekhnovich, I. N., and R. J. Kadner. 2002. DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:4148-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oropeza, R., C. L. Sampieri, J. L. Puente, and E. Calva. 1999. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR. Mol. Microbiol. 32:243-252. [DOI] [PubMed] [Google Scholar]
- 33.Poore, C. A., C. Coker, J. D. Dattelbaum, and H. L. T. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 183:4526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porter, M. E., and C. J. Dorman. 2002. In vivo DNA-binding and oligomerization properties of the Shigella flexneri AraC-like transcriptional regulator VirF as identified by random and site-specific mutagenesis. J. Bacteriol. 184:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puente, J. L., D. Bieber, S. W. Ramer, W. Murray, and G. K. Schoolnik. 1996. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol. Microbiol. 20:87-100. [DOI] [PubMed] [Google Scholar]
- 36.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roder, K., and M. Schweizer. 2001. Running-buffer composition influences DNA-protein and protein-protein complexes detected by electrophoretic mobility-shift assay (EMSA). Biotechnol. Appl. Biochem. 33:209-214. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santana, M. A., G. Pedraza-Alva, N. Olivares-Zavaleta, V. Madrid-Marina, V. Horejsi, S. J. Burakoff, and Y. Rosenstein. 2000. CD43-mediated signals induce DNA binding activity of AP-1, NF-AT, and NFkappa B transcription factors in human T lymphocytes. J. Biol. Chem. 275:31460-31468. [DOI] [PubMed] [Google Scholar]
- 40.Saviola, B., R. Seabold, and R. F. Schleif. 1998. Arm-domain interactions in AraC. J. Mol. Biol. 278:539-548. [DOI] [PubMed] [Google Scholar]
- 41.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 42.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C.-Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 44.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 46.Tamai, E., T. A. Belyaeva, S. J. Busby, and T. Tsuchiya. 2000. Mutations that increase the activity of the promoter of the Escherichia coli melibiose operon improve the binding of MelR, a transcription activator triggered by melibiose. J. Biol. Chem. 275:17058-17063. [DOI] [PubMed] [Google Scholar]
- 47.Thomas, V. J., and C. M. Collins. 1999. Identification of UreR binding sites in the Enterobacteriaceae plasmid-encoded and Proteus mirabilis urease gene operons. Mol. Microbiol. 31:1417-1428. [DOI] [PubMed] [Google Scholar]
- 48.Tobe, T., T. Hayashi, C.-G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 50.Tobe, T., I. Tatsuno, E. Katayama, C. Y. Wu, G. K. Schoolnik, and C. Sasakawa. 1999. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a BfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol. Microbiol. 33:741-752. [DOI] [PubMed] [Google Scholar]
- 51.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397-413. [DOI] [PubMed] [Google Scholar]
- 52.Ueguchi, C., C. Seto, T. Suzuki, and T. Mizuno. 1997. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 274:145-151. [DOI] [PubMed] [Google Scholar]
- 53.Vallance, B. A., and B. B. Finlay. 2000. Exploitation of host cells by enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 97:8799-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wattiau, P., and G. R. Cornelis. 1994. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J. Bacteriol. 176:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]