Abstract
Pseudomonas aeruginosa PAO1 secretes a siderophore, pyoverdinePAO, which contains a short peptide attached to a dihydroxyquinoline moiety. Synthesis of this peptide is thought to be catalyzed by nonribosomal peptide synthetases, one of which is encoded by the pvdD gene. The first module of pvdD was overexpressed in Escherichia coli, and the protein product was purified. l-Threonine, one of the amino acid residues in pyoverdinePAO, was an effective substrate for the recombinant protein in ATP-PPi exchange assays, showing that PvdD has peptide synthetase activity. Other amino acids, including d-threonine, l-serine, and l-allo-threonine, were not effective substrates, indicating that PvdD has a high degree of substrate specificity. A three-dimensional modeling approach enabled us to identify amino acids that are likely to be critical in determining the substrate specificity of PvdD and to explore the likely basis of the high substrate selectivity. The approach described here may be useful for analysis of other peptide synthetases.
Secondary metabolites are produced by a very wide range of microorganisms and have a diverse range of functions, including siderophore, antibiotic, immunosuppressant, biosurfactant, herbicidal, and antiviral functions (11, 23, 32). Many secondary metabolites are peptide based, and the structural diversity of these metabolites is due in part to the presence of a large number of unusual and nonproteinogenic amino acid residues (reviewed in references 9, 19, 24, and 45). More than 300 of these residues have been identified to date, and they include pseudo, hydroxy, N-methylated, and d amino acids. The presence of atypical residues in the compounds is usually enabled by a nonribosomal mechanism of peptide synthesis.
Nonribosomal peptide assembly is catalyzed by large multimodular enzymes known as nonribosomal peptide synthetases (NRPSs), and each module governs insertion of a single amino acid in the peptide product (reviewed in references 9 and 20). Each NRPS module is comprised of several semiautonomous domains. Together, these domains provide active sites for recognition and activation of an amino acid substrate (adenylation or A domains) and subsequent incorporation of the adenylated substrate into a peptide product (thiolation or T domains and condensation or C domains). The modules may also contain domains that modify the substrate by N methylation or epimerization prior to incorporation or that contain a thioesterase motif for cleavage of a nascent peptide from the multimodular complex. NRPS modules act in a coordinated fashion to produce a peptide product. This process has been termed the multiple carrier model (39), and it consists of an integrated and stepwise series of amino- to carboxy-terminal transpeptidation reactions.
A domains, which catalyze the adenylation of cognate amino acid substrates, are central to nonribosomal peptide synthesis. Peptide synthetase A domains are typically about 550 amino acid residues long and have 30 to 60% sequence identity (37). An alignment of their sequences shows that there are 10 regions (A domain motif sequences) that are particularly highly conserved and that these regions are interspersed with regions with lower or no sequence similarity (44). Peptide synthetase A domains exhibit approximately 15 to 30% identity with members of the acyl-coenzyme A synthase and luciferase families of enzymes (44). Enzymes in these families also catalyze the adenylation of carboxy substrates and together with the NRPSs form a superfamily of adenylating enzymes. The structures of two enzymes belonging to this superfamily have been determined; these enzymes are firefly luciferase (7) and the A domain of the NRPS GrsA from Bacillus brevis (8). The overall topology of the GrsA A domain is very similar to that of luciferase, despite the fact that the two proteins exhibit only 16% identity at the primary sequence level. NRPS A domains exhibit much greater identity with one another, so it can reasonably be concluded that the GrsA structure represents a prototype for all peptide synthetase A domains (20). This is particularly likely to be true for the active sites, and almost all 10 of the highly conserved A domain motif sequences are clustered around the AMP and phenylalanine substrate-binding sites in the GrsA structure.
Stachelhaus et al. (38) and Challis et al. (5) created data banks of over 150 A domain sequences. Alignments of these sequences showed that other residues around the active site, distinct from the 10 conserved motif sequences, are highly conserved among A domains that recognize the same amino acid substrates. These residues are likely to be responsible for substrate recognition. These alignments were used to generate consensus codes, which allowed the substrate specificity of uncharacterized NRPS modules to be inferred from sequence data. Stachelhaus et al. (38) demonstrated the validity of this approach by targeted alteration of residues predicted to determine the substrate specificity of GrsA, showing that alteration of only two residues in the active site was sufficient to cause adenylation of l-leucine instead of d-phenylalanine. Subsequently, Eppelmann et al. precisely altered the substrate specificity of two surfactin synthetase modules by similar site-directed mutagenesis of substrate-determining residues (14). Challis et al. (5) took the prediction-based approach a step further by using the GrsA structural model to generate a two-dimensional representation of the amino acid-binding pocket. They then superimposed the side chains of the predicted substrate-coding residues over each of the corresponding GrsA side chains in the binding pocket to create a two-dimensional model of the binding pocket for each substrate examined. This allowed a more detailed structural analysis of the residues that are predicted to play key roles in substrate recognition.
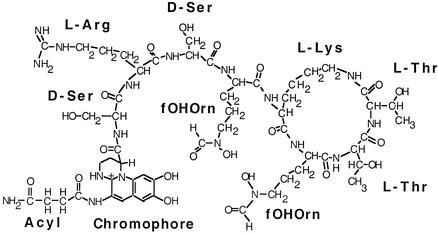
The analysis of amino acid residues in the inferred substrate-binding sites of NRPS A domains has enhanced our understanding of these enzymes. The aim of the research described in this paper was to utilize and extend this approach, in conjunction with biochemical methods, in order to analyze the activity of an NRPS designated PvdD that is required for synthesis of the siderophore pyoverdine by Pseudomonas aeruginosa. P. aeruginosa is a ubiquitous human pathogen and causes a wide range of infections (22). Pyoverdines, which are mixed hydroxamate-catecholate-type siderophores, are required for the virulence of this organism (31, 41). The pyoverdines synthesized by all strains of P. aeruginosa have an identical dihydroxyquinoline group, and attached to this group is a strain-specific peptide that is comprised of seven or eight amino acid residues and includes several atypical amino acids (30). PyoverdinePAO, secreted by P. aeruginosa strain PAO1, contains an eight-member partially cyclic peptide (Fig. 1). The pvdD gene is required for synthesis of pyoverdine (28). Analysis of the pvdD sequence showed that PvdD has two NRPS modules that have 91% sequence identity and 95% sequence similarity in the C-A-T domain repeats. The sequence identity is particularly high for the two substrate-determining A domains, and 521 of 523 amino acid residues are identical when the modules are aligned with each other. Merriman et al. (28) proposed that PvdD governs the incorporation of the two consecutive l-threonine residues at the C terminus of the pyoverdine peptide (d-Ser-l-Arg-d-Ser-fOHOrn-l-Lys-fOHOrn-l-Thr-l-Thr, where fOHOrn is l-N5-formyl-N5-hydroxyornithine).
FIG. 1.
Structure of pyoverdinePAO produced by P. aeruginosa PAO1 (1). The positions of amino acid residues, the dihydroxyquinoline chromophore, and an acyl group are shown. The acyl group can be a succinyl, α-ketoglutaryl, or succinamide residue. fOHOrn, l-N5-formyl-N5-hydroxyornithine.
Here we describe purification of a recombinant PvdD module and enzyme assays performed with the recombinant protein that showed that l-threonine is indeed a substrate for PvdD. The substrate specificity of the enzyme and the basis of this specificity were examined by performing biochemical assays and using a structural modeling approach.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Escherichia coli BL21 [F− ompT hsdSB(rB− mB−) gal dcm; Novagen] was grown in Luria broth (LB) (Gibco BRL). P. aeruginosa PAO1 (18) was cultured in brain heart infusion broth (Gibco BRL). The expression plasmid pPROEX1 was obtained from Gibco BRL as part of the Life Technologies protein expression system. Construction of plasmid pPROEX::mod1 is described below.
DNA methods.
Chromosomal DNA was prepared from P. aeruginosa PAO1 by using the rapid genomic miniprep method (6). The first module of pvdD was amplified from this DNA by using the Expand High Fidelity PCR system (Roche Molecular Biochemicals) and the following primers: 5′-GGGGGAATTCGGTAGGTGCAAGCACTC-3′ and 5′-GGGGGAGCTCAGGTAGCCAGCAACGAATG-3′. The restriction enzymes EcoRI and SstI (introduced sites are indicated by underlining in the primer sequences) were used to directionally clone the amplified module in frame into pPROEX1 by standard methods (33). The resulting plasmid (pPROEX1::mod1) was transformed into E. coli BL21.
Purification of His-PvdD.
A 5-ml culture of BL21(pPROEX1::mod1) was grown overnight at 37°C in LB containing ampicillin (50 μg/ml). The overnight culture was used to inoculate 500 ml of LB containing ampicillin (50 μg/ml), glycine betaine (2.5 mM), and sorbitol (1 M). This culture was grown to an optical density at 600 nm (OD600) of 0.5 at 18°C (about 2 days), and then isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.6 mM and the bacteria were incubated at 18°C until the OD600 was 1.5 (one additional day). Cells were harvested by centrifugation (10,000 × g, 10 min), and the pellet was frozen at −70°C until it was required. For protein purification, the cells were resuspended in 10 ml of lysis buffer (50 mM Tris-HCl [pH 8.5], 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [added fresh]) and sonicated with an MSE sonicator (15 10-s bursts at 10-s intervals, until 80% cell lysis was obtained, as estimated by the change in OD600). Lysates were centrifuged at 10,000 × g for 30 min to remove cellular debris. To analyze the insoluble fractions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the pellets were resuspended to obtain approximately 10% (wt/vol) solutions in 20 mM Tris-HCl (pH 8.0). Soluble fractions were analyzed directly. The recombinant His-PvdD, which comprises the first module of PvdD fused to a hexahistidine affinity tag by a peptide linker, was purified from the soluble fraction by using Ni-nitrilotriacetic acid (NTA) resin in conjunction with the protocol described by Life Technologies. All column purification steps were performed at 4°C in a temperature-controlled environment. Contaminating proteins were removed as described below.
ATP-PPi exchange assays.
ATP-PPi exchange assays were performed by using a modification of the protocol described by Lee and Lipmann (21). The reaction mixture was prepared by combining one part each of a 20 mM amino acid substrate solution, an ATP buffer stock solution (10 mM ATP, 50 mM MgCl2, 2.5 mM EDTA, 5 mM dithiothreitol, 100 mM triethanolamine buffer [pH 7.7], 25% [wt/vol] sucrose), a 0.4% (wt/vol) bovine serum albumin solution in 10 mM NaOH, and a 10 mM PPi solution (pH 7.7) containing 20 μCi of 32PPi (0.15%). The 32PPi was added immediately prior to use to avoid precipitation as Mg2P2O7. The exchange reaction was initiated by incubating 50 μl of a 50-μg/ml His-PvdD solution with 200 μl of reaction medium in a 1.5-ml microcentrifuge tube at 37°C and was allowed to proceed for 15 min. For the negative control 50 μl of a 50 mM Tris-HCl (pH 8.5) solution was added in place of His-PvdD. The reaction was stopped by adding 500 μl of an activated charcoal suspension (50 ml [wet volume] of acid-washed activated charcoal, 250 ml of 14% perchloric acid containing 400 mM PPi, and enough distilled H2O to bring the volume to 1 liter) to the microcentrifuge tube. Following mixing, the charcoal was collected by passing the mixture through a 3-ml syringe attached to a Millipore filter containing 0.45-μm-pore-size filter paper. While contained within the filter, the charcoal was washed three times with 3 ml of PPi (100 mM, adjusted to pH 8.0 with HCl) and two times with 3 ml of distilled water. The ATP was then eluted from the charcoal with two 3-ml washes with an elution solution containing methanol and NH3 (1:1, vol/vol) (43) into a scintillation vial containing 9 ml of water. Cerenkov counts were obtained directly with this vial by using an LKB Rackbeta liquid scintillation counter. The background count was considered to be the lowest data point recorded for the no-enzyme-no-substrate control; this base value was subtracted from all other values.
Protein modeling.
The sequence of the PvdD A domain was aligned with that of GrsA by using a gapped ClustalW alignment, and putative substrate pocket-lining residues of PvdD were identified by using the strategies of Stachelhaus et al. (38) and Challis et al. (5). Next, the PvdD primary sequence was threaded through the GrsA structural model (PDB ID Code 1AMU) by using the program Swiss PDB viewer Deep View such that the PvdD residues suggested by the alignment to correspond to equivalent residues in the GrsA primary sequence were overlaid onto the α carbon positions of the corresponding GrsA residues. The model resulting from the gapped alignment of GrsA with PvdD contained regions in which residues were not matched with partners from the opposite sequence. These gaps and insertions mostly occurred in loop regions; none occurred within 18 Å of the active site, and they were removed from the PvdD model. This structural model was then loaded into XtalView (27), and the active site residues of the two structures were compared. The l-threonine substrate of PvdD was inserted into the new model in an orientation identical to that of the corresponding l-phenylalanine substrate of GrsA (8). Diagrams of these models were made by using the Raster3D interface of XtalView (29). Additional diagrams were produced by using PyMOL.
RESULTS
Expression and purification of a soluble PvdD module.
The first module of PvdD was purified so that we could study its substrate specificity. Partial NRPS modules comprising the A and T domains or the A domain alone exhibit tendencies toward inclusion body formation (10, 12, 16). Therefore, the entire peptide synthetase module was purified, because we thought that interactions between the A and T domains and the C domain might play a part in influencing the solubility of the module.
The first module of pvdD was amplified by PCR, and the product was cloned into pPROEX1. The resulting plasmid (pPROEX::mod1) was transformed into E. coli BL21. Addition of IPTG to a culture of these bacteria resulted in production of a protein estimated to have a molecular mass of ∼114 kDa (data not shown); this compared well with the predicted mass of the recombinant His-tagged PvdD module (121 kDa). N-terminal protein sequencing of the first six residues of this protein gave the expected MGHHHH signal, confirming that the correct protein (His-PvdD) had been produced.
BL21(pPROEX::mod1) cells were induced to make His-PvdD, grown in LB for 3 h at 37°C, sonicated, and centrifuged. Virtually all of the His-PvdD protein was in the insoluble fraction (data not shown). Previous researchers had great difficulty obtaining biologically active peptide synthetase domains or modules from inclusion body aggregates, and only one group reported successful renaturation of a functional synthetase (16), so it was preferable to develop a procedure for expressing His-PvdD in a soluble form. Several methods have been described that increase the likelihood that recombinant proteins will be expressed in E. coli in a soluble and active form. Two of these methods are induction of expression at a low temperature (35) and growth in the presence of the osmoregulator glycine betaine with a high concentration of sorbitol in the growth medium (3). Cells were grown in LB with or without betaine and sorbitol, and His-PvdD production was induced over a range of temperatures. Growth of cells at 18°C in the presence of betaine and sorbitol were the only conditions that gave significant amounts of soluble His-PvdD (data not shown).
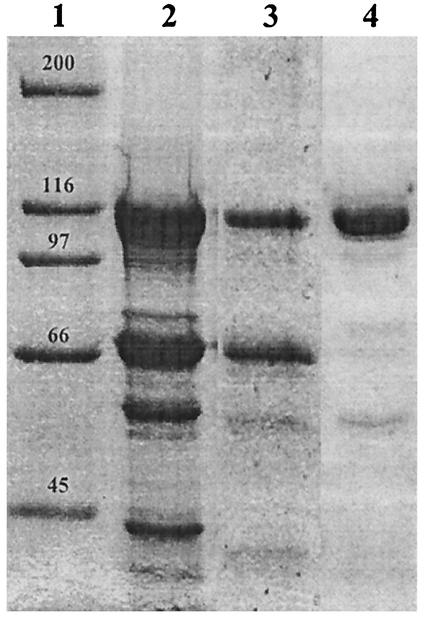
Soluble His-PvdD was purified by using a nickel-containing resin (Ni-NTA). Fractions containing His-PvdD were pooled and concentrated. There were three major contaminating species, which had molecular masses of approximately 69, 57, and 42 kDa (Fig. 2, lane 2). The pooled material was reapplied to a fresh Ni-NTA column, and this step significantly reduced the amounts of the two smaller contaminants but not the amount of the ∼69-kDa protein (Fig. 2, lane 3). N-terminal sequencing of this protein showed that the eight N-terminal residues were MGKIIGID, the same residues as those in the 70-kDa DnaK protein of E. coli. DnaK is a member of the Hsp70 family of chaperone proteins (reviewed in reference 26), is often copurified with recombinant proteins that are overexpressed in E. coli, and on occasion may interfere with the optimal folding of these proteins (36).
FIG. 2.
Affinity purification of His-PvdD. E. coli BL21(pPROEX::mod1) was grown at 18°C in LB containing betaine and sorbitol and was induced to express His-PvdD as described in the text. The cells were sonicated, and His-PvdD was purified by using a nickel affinity column. Protein preparations obtained following a single passage (lane 2), following two passages (lane 3), and following an ATP incubation step (lane 4) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane 1 contained Bio-Rad broad-range standard markers.
The concentrated pooled fraction was incubated for 10 min at 37°C with ATP (2 mM), a treatment that causes DnaK to dissociate from other proteins (36). Following a second passage through an Ni-NTA column, the DnaK band was absent from the repurified fractions (Fig. 2, lane 4). The fractions containing the largest amounts of His-PvdD were pooled, concentrated, and analyzed by ATP-PPi exchange assays.
Determination of the substrate specificity of PvdD by ATP-PPi exchange.
ATP-PPi exchange assays were performed by using the protocol of Lee and Lipmann (21). These assays exploit the equilibrium nature of the aminoacylation reaction, which can be summarized as follows: E + S + ATP ⇆ [E-S-AMP] + PPi, where E is the A domain of an NRPS enzyme, S is a substrate activated by the A domain of the NRPS enzyme, and [E-S-AMP] is the enzyme-aminoacyl adenylate complex. This is an equilibrium reaction, so if radiolabeled pyrophosphate is present, then labeled phosphate groups are introduced into ATP, but only if a suitable amino acid substrate is present.
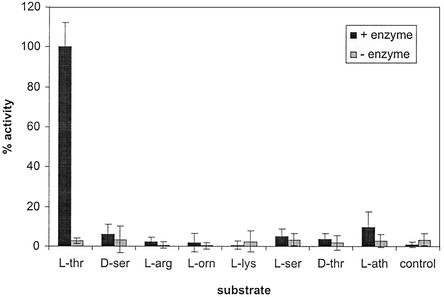
Exchange assays were performed with each of the amino acids present in pyoverdine (l-ornithine was used in place of l-N5-formyl-N5-hydroxyornithine, because the latter compound could not be obtained commercially). His-PvdD activated only l-threonine (Fig. 3), which is consistent with the proposal that PvdD incorporates l-threonine residues into pyoverdine. The specificity of this enzyme was examined by carrying out ATP-PPi exchange assays in the presence of amino acids that have structural similarities to l-threonine (l-serine, d-threonine, and l-allo-threonine). The results (Fig. 3) indicated that His-PvdD has a high level of specificity for l-threonine.
FIG. 3.
ATP-PPi exchange activity of His-PvdD. ATP-PPi exchange activity was measured with different amino acids. The highest exchange activity observed was defined as 100%. The error bars indicate one standard deviation. L-thr, l-threonine; D-ser, d-serine; L-arg, l-arginine; L-orn, l-ornithine; L-lys, l-lysine; L-ser, l-serine; D-thr, d-threonine; L-ath, l-allo-threonine.
Structural modeling of the PvdD substrate-binding pocket.
A three-dimensional structural modeling approach was developed to investigate the basis of the substrate specificity of PvdD. In this modeling approach we used the crystal structure of the GrsA A domain (8) and took advantage of the likely overall structural similarities of GrsA and PvdD. It was assumed that the structures of the two enzymes are very similar around the active sites. The sequences of GrsA and PvdD, along with those of other threonine-activating enzymes, were aligned (Fig. 4). Residues in the active site of GrsA were then replaced with the corresponding residues from the PvdD sequence, as described in Materials and Methods. The basis of this approach is that the overall structures of NRPSs are likely to be highly conserved around the active sites of the enzymes, with residues lining the active site pocket providing substrate specificity in the context of very similar structures; any major structural differences would be likely to be in parts of the proteins that are located away from the active sites. The following observations support this approach: (i) the GrsA structure is extremely similar at the tertiary level to the structure of firefly luciferase (8), despite the fact that these proteins exhibit only 16% identity at the amino acid level, and GrsA and PvdD are 38% identical and hence are likely to be at least as similar at the structural level; (ii) the regularly spaced A1 to A10 motif sequences that are highly conserved in all peptide synthetases (44) are primarily clustered around the active site in the GrsA structure; (iii) the K517 and D235 residues of GrsA, which are involved in α-carboxyl and α-amino substrate group binding, are essentially invariant in all peptide synthetases, suggesting that all A domains present their cognate substrate residues to a discriminating pocket region in a conserved fashion; and (iv) as determined by other researchers, substrate specificity is correlated with residues predicted to line the active sites (5, 14, 38), implying that the substrate-specifying pocket regions are structurally similar.
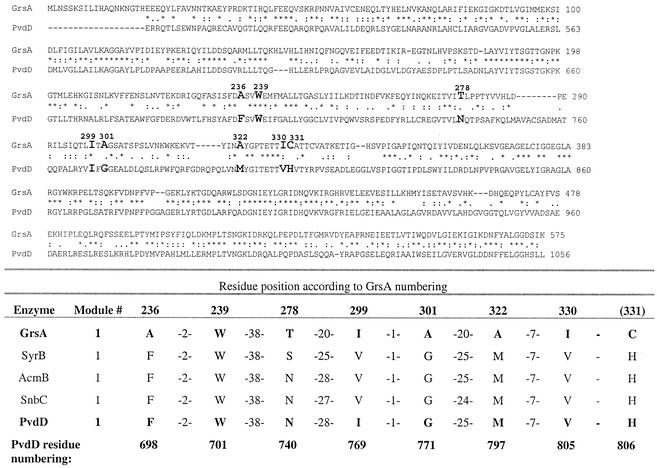
FIG. 4.
Identification of substrate-determining residues of PvdD. The sequence of the A domain of the first module of PvdD was aligned with that of GrsA. An asterisk indicates identical residues, a colon indicates residues with a high level of chemical similarity, and a period indicates residues with a lower level of chemical similarity. The eight residues of PvdD that are predicted to determine the amino acid substrate specificity (the coding residues [5, 38]) are indicated by boldface type in the alignment. Residue 331 (GrsA numbering) was considered to be a coding residue by Stachelhaus et al. (38) but not by Challis et al. (5). The predicted coding residues of three other l-threonine-activating A domains are from SyrB, a syringomycin synthetase from Pseudomonas syringae (15); AcmB, an actinomycin synthetase from Streptomyces chrysomallus (34); and SnbC, a pristinamycin synthetase from Streptomyces pristinaespiralis (42). Threonine activation was demonstrated by ATP-PPi exchange for SnbC and SyrB. For AcmB, assignment of threonine as the substrate was inferred from the linear order of the modules in the chromosome according to the nearly invariant colinearity rule (reviewed in reference 24).
In the resulting model, all of the gaps and insertions in PvdD compared to the GrsA sequence (Fig. 4) are on the surface of the protein. No gaps or insertions were found in the immediate proximity of the active site, and the nearest insertion in the PvdD model was more than 18 Å from the substrates. Twenty-eight amino acid residues surround the active site of PvdD in the model. Eight of these residues are the predicted substrate-determining residues shown in Fig. 4. The remaining 20 active site residues are predicted to interact with AMP, Mg2+, and the substrate α-amino and α-carboxyl groups. Seventeen of these residues are identical to the corresponding residues in GrsA, and one is similar (Ile to Val). Another two residues are predicted to interact with AMP through the main chain carbonyl groups, and the side chains do not contribute to substrate binding. The extremely high level of sequence identity between the active site of GrsA and the predicted active site of PvdD further supports the validity of the modeling approach used.
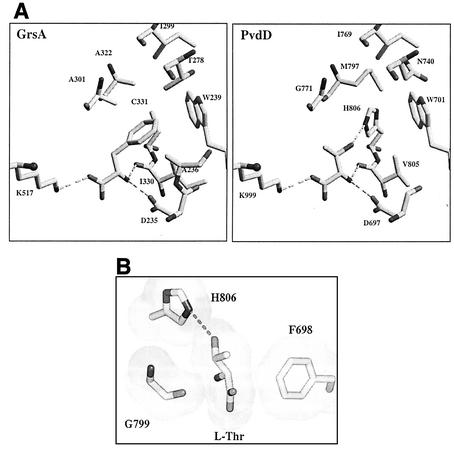
The PvdD model was then transferred into XtalView, and the l-phenylalanine substrate from the GrsA model was altered to l-threonine. The α-amino and α-carboxyl groups and the α and β carbons of the l-threonine substrate were maintained in locations identical to the locations of the equivalent groups and atoms of the l-phenylalanine substrate in the GrsA model (Fig. 5A). The l-threonine side chain was then rotated around the β carbon to investigate possible interactions with the pocket-lining residues. Deviation from the GrsA configuration of pocket-lining side chains was minimized in order to maintain the integrity of the modeling approach. In the resulting model of PvdD, the locations of the side chain α and β carbons of each of the pocket-lining residues were identical to those of the equivalent residues in GrsA. The general orientation of the side chains was also maintained, with one significant exception. In the GrsA structure, C331 is directed out from the pocket, away from the phenylalanine substrate, but the corresponding residue in PvdD, H806, is likely to play an extremely important substrate-selecting role, forming a discriminating hydrogen bond with the threonine side chain hydroxyl (Fig. 5). Bringing the imidazole ring of this histidine residue into the active site by rotation around the α-β carbon bond (relative to GrsA) allows a nitrogen atom of the side chain ring to hydrogen bond with the side chain oxygen atom of the threonine, with an estimated bond distance of 2.7 Å. The resulting rotamer is well within the normal range for histidine residues (13), and adoption of this conformation is probably promoted and stabilized by the side chains of M797, W701, and V805, which are predicted to take up the space that all other common histidine rotamers could occupy.
FIG. 5.
Model explaining the substrate specificity of PvdD. (A) Phenylalanine-binding site of GrsA (8) and predicted threonine-binding site of PvdD. Carbon and sulfur atoms are represented by light grey bars, and oxygen and nitrogen are represented by dark grey bars. The view is from the same angle in both cases, and the substrate amino acids are in the middle of each frame. All 10 residues that line the amino acid substrate-binding pocket are shown in the GrsA structure, but residue F698 has been removed from the foreground of the PvdD structure for clarity. The cosubstrate AMP molecule is predicted to occupy much of the left side of each frame. Three hydrogen bonds that are predicted to stabilize the substrate amino and carboxyl groups in the active sites are indicated by dashed lines. A substrate-determining hydrogen bond is also predicted to be formed between the threonine side chain hydroxyl and a nitrogen atom in the side chain of H806 of PvdD. (B) View rotated 90o with respect to the view shown in panel A. The van der Waals surfaces of l-threonine, G799, and the side chains of F698 and H806 of PvdD are shown. The substrate l-threonine is predicted to be sandwiched between F698 and the main-chain carbonyl of G799. Modeling of insertion of other isomers of threonine (d-threonine and l-allo-threonine) into the predicted PvdD substrate-binding pocket showed that while these isomers could form hydrogen bonds with D697, V805, and K999 (as shown for l-threonine in panel A), they would be unable to form a hydrogen bond with H806 without being unacceptably close to F698 or G799 (data not shown).
GrsA activates both the l and d stereoisomers of phenylalanine (37), and this lack of stereospecificity can be attributed to the symmetrical nature of the phenylalanine side chain (8). By contrast, PvdD exhibits strong stereoselectivity for l-threonine over d-threonine or l-allo-threonine (Fig. 3). Different isomers of threonine were modeled into the proposed structure of the active site of PvdD, and in our model the H806 residue along with F698 and G799 determines the stereospecificity of this enzyme (Fig. 5B).
DISCUSSION
A number of researchers have encountered difficulties in purifying peptide synthetase domains following expression in E. coli. In this study, soluble recombinant PvdD was purified following overexpression in E. coli grown at a suboptimal temperature in the presence of glycine betaine and a high concentration of sorbitol. The mechanisms by which these conditions promote the solubility of overexpressed proteins in E. coli are poorly understood, but the suggested explanations include (i) a reduction in the rate at which newly synthesized protein is released from the ribosome into the cytoplasm, (ii) alteration of the conditions in which the protein must fold, and (iii) environmental stress which induces the production of Hsp chaperones that assist in protein folding (2, 17). Our approach may have general application for the purification of peptide synthetase enzymes for functional characterization studies. Recently, it has been demonstrated that coexpression of the E. coli chaperones GroES/EL can sometimes assist in the purification of overexpressed peptide synthetase proteins from E. coli (40), and it may be that this approach could be combined with the protocol described here to enable purification of particularly recalcitrant enzymes.
The copurification of DnaK may indicate that the His-PvdD protein was initially improperly folded, as one intracellular function of DnaK is the targeting of misfolded proteins for degradation (4). Consistent with this idea, ATP-PPi exchange activity was not detected with l-threonine as the substrate when an enzyme preparation that included DnaK was employed in assays (data not shown). Moreover, it may be that association with DnaK inhibited complete folding of His-PvdD. Incubation of the preparation with ATP removed the contaminating DnaK species and perhaps promoted correct folding of the His-PvdD protein in the process.
Many peptide synthetases have a high degree of substrate specificity with only a single known amino acid substrate, whereas others have a lower degree of substrate specificity and can adenylate (activate) more than one amino acid substrate at discernible levels (reviewed in references 24 and 45). His-PvdD (and, by extension, PvdD itself) has high substrate specificity for l-threonine compared to the other amino acids that were tested. It is possible that PvdD activates the other substrates that were investigated but at a level that was too low to detect in our assay system. For example, there may have been slight activity with l-allo-threonine (Fig. 3), but the value was within the margin of error for the ATP-PPi exchange assays. The l-threonine-activating NRPSs SyrB and SnbC also have very high specificities for l-threonine (15, 42). VibF from Vibrio cholerae is an NRPS that catalyzes adenylation of l-threonine and also adenylates l-serine and l-cysteine, but only at about 1/100 of the rate for l-threonine (25).
A three-dimensional structural modeling approach was developed to generate a model of the PvdD active site region and to investigate the basis of the high substrate specificity of PvdD. In the absence of a crystal structure for PvdD, this model allows some understanding of the structural basis of the action of this enzyme. The two-dimensional model of Challis et al. (5) proposes that N740 of PvdD is involved in a key interaction with the l-threonine side chain hydroxyl. The three-dimensional model (Fig. 5) argues against this, since N740 is almost certainly too distant to interact directly with the threonine substrate. Furthermore, a serine residue is present at the corresponding position in SyrB (Fig. 4). Instead, it appears that the H806 residue is the key stabilizing and selective residue for substrate binding in PvdD. The cysteine in the equivalent position in GrsA does not appear to play an important role in substrate selection by this enzyme, but the three-dimensional modeling approach indicates that residues in this position play a role in substrate recognition for some peptide synthetases. Given the highly conserved nature of the predicted substrate-determining residues of other l-threonine-activating peptide synthetases (Fig. 4) (5, 38), the PvdD model could provide an accurate representation of the active site region for most l-threonine-activating A domains. The only exception is VibF (25), which has a completely different configuration of predicted substrate-determining residues (MFVAGLIW, compared with FWNIGMVH for PvdD [Fig. 4]) and presumably employs an unrelated mechanism of substrate recognition.
The PvdD model was also used to investigate the high substrate specificity of PvdD. As threonine residues are asymmetrical, PvdD should bind different stereoisomers in quite different orientations, but discriminating features of the substrate-binding pocket should restrict the access of all isomers except l-threonine to the active site. Identification of potential substrate-determining residues is an important outcome of the modeling approach and provides a basis for site-directed mutagenesis studies. The modeling approach could also be applied to other peptide synthetase A domains and could, for example, provide supporting evidence for assignments of substrate specificity or assist in the development of inhibitor molecules targeted against peptide synthetases. For example, an inability to produce pyoverdine has been correlated with substantially diminished virulence in P. aeruginosa strains (31, 41), so the development of inhibitors of PvdD could have important medical implications.
In conclusion, the experimental data and modeling approaches described here provide complementary insights into the nature of substrate selection by PvdD. The possibility of carrying out rational modification of active sites of peptide synthetases to enable incorporation of different amino acids into peptides has been demonstrated for GrsA (38) and two surfactin synthetases (14). The approaches that we describe here may allow this approach to be extended to a wide range of peptide synthetases, thereby enabling generation of a wide range of modified peptide products.
Acknowledgments
D.A. was supported by a postgraduate scholarship awarded by the Health Research Council of New Zealand. This research was supported in part by a grant from the University of Otago.
REFERENCES
- 1.Abdallah, M. A. 1991. Pyoverdines and pseudobactins, p. 139-153. In G. Winkelmann (ed.), CRC handbook of microbial iron chelates. CRC Press, Boca Raton, Fla.
- 2.Barth, S., M. Huhn, B. Matthey, A. Klimka, E. A. Galinski, and A. Engert. 2000. Compatible-solute-supported periplasmic expression of functional recombinant proteins under stress conditions. Appl. Environ. Microbiol. 66:1572-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell, J. R., and R. Horgan. 1991. A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett. 295:10-12. [DOI] [PubMed] [Google Scholar]
- 4.Buchberger, A., and B. Bukau. 1997. Escherichia coli DnaK, p. 22-24. In M. J. Gething (ed.), Guidebook to molecular chaperones and protein-folding catalysts. Sambrook and Tooze, Oxford University Press, Oxford, United Kingdom.
- 5.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., and T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti, E., N. P. Franks, and P. Brick. 1996. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure 4:287-298. [DOI] [PubMed] [Google Scholar]
- 8.Conti, E., T. Stachelhaus, M. A. Marahiel, and P. Brick. 1997. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 16:4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Crecy-Lagard, V., V. Blanc, P. Gil, L. Naudin, S. Lorenzon, A. Famechon, N. Bamas-Jacques, J. Crouzet, and D. Thibaut. 1997. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. J. Bacteriol. 179:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demain, A. L. 1992. Microbial secondary metabolism: a new theoretical frontier for academia, a new opportunity for industry, p. 3-23. In D. J. Chadwick and J. Whelan (ed.), Secondary metabolites: their function and evolution, vol. 171. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 12.Dieckmann, R., Y. O. Lee, H. van Liempt, H. von Dohren, and H. Kleinkauf. 1995. Expression of an active adenylate-forming domain of peptide synthetases corresponding to acyl-CoA-synthetases. FEBS Lett. 357:212-216. [DOI] [PubMed] [Google Scholar]
- 13.Engh, R. A., and R. Huber. 1991. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. Sect. A 47:392-400. [Google Scholar]
- 14.Eppelmann, K., T. Stachelhaus, and M. A. Marahiel. 2002. Exploitation of the selectivity-conferring code of nonribosomal peptide synthetases for the rational design of novel peptide antibiotics. Biochemistry 41:9718-9726. [DOI] [PubMed] [Google Scholar]
- 15.Guenzi, E., G. Galli, I. Grgurina, D. C. Gross, and G. Grandi. 1998. Characterization of the syringomycin synthetase gene cluster. A link between prokaryotic and eukaryotic peptide synthetases. J. Biol. Chem. 273:32857-32863. [DOI] [PubMed] [Google Scholar]
- 16.Haese, A., R. Pieper, T. von Ostrowski, and R. Zocher. 1994. Bacterial expression of catalytically active fragments of the multifunctional enzyme enniatin synthetase. J. Mol. Biol. 243:116-122. [DOI] [PubMed] [Google Scholar]
- 17.Hockney, R. C. 1994. Recent developments in heterologous protein production in Escherichia coli. Trends Biotechnol. 12:456-463. [DOI] [PubMed] [Google Scholar]
- 18.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 19.Kleinkauf, H., and H. von Dohren. 1990. Nonribosomal biosynthesis of peptide antibiotics. Eur. J. Biochem. 192:1-15. [DOI] [PubMed] [Google Scholar]
- 20.Konz, D., and M. A. Marahiel. 1999. How do peptide synthetases generate structural diversity? Chem. Biol. 6:R39-R48. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S. G., and F. Lipmann. 1975. Tyrocidine synthetase system. Methods Enzymol. 43:585-602. [DOI] [PubMed] [Google Scholar]
- 22.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 23.Maplestone, R. A., M. J. Stone, and D. H. Williams. 1992. The evolutionary role of secondary metabolites—a review. Gene 115:151-157. [DOI] [PubMed] [Google Scholar]
- 24.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, C. G., M. D. Burkart, T. A. Keating, and C. T. Walsh. 2001. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry 40:10655-10663. [DOI] [PubMed] [Google Scholar]
- 26.Mayer, M. P., and B. Bukau. 1998. Hsp70 chaperone systems: diversity of cellular functions and mechanism of action. Biol. Chem. 379:261-268. [PubMed] [Google Scholar]
- 27.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 28.Merriman, T. R., M. E. Merriman, and I. L. Lamont. 1995. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J. Bacteriol. 177:252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrit, E. A., and M. Murphy. 1994. Raster3D version 2.0: a program for photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, J., A. Stintzi, D. D. Vos, P. Cornelis, R. Tappe, K. Taraz, and H. Budzikiewicz. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143:35-43. [DOI] [PubMed] [Google Scholar]
- 31.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffitt, M. C., and B. A. Neilan. 2000. The expansion of mechanistic and organismic diversity associated with non-ribosomal peptides. FEMS Microbiol. Lett. 191:159-167. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., D. W. Russell, and N. Irwin. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Schauwecker, F., F. Pfennig, W. Schroder, and U. Keller. 1998. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. J. Bacteriol. 180:2468-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schein, C. H. 1989. Production of soluble recombinant proteins in bacteria. Bio/Technology 7:1141-1147. [Google Scholar]
- 36.Sherman, M., and A. L. Goldberg. 1992. Involvement of the chaperonin DnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 11:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J. Biol. Chem. 270:6163-6169. [DOI] [PubMed] [Google Scholar]
- 38.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 39.Stein, T., J. Vater, V. Kruft, A. Otto, B. Wittmann-Liebold, P. Franke, M. Panico, R. McDowell, and H. R. Morris. 1996. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 271:15428-15435. [DOI] [PubMed] [Google Scholar]
- 40.Symmank, H., W. Saenger, and F. Bernhard. 1999. Analysis of engineered multifunctional peptide synthetases. Enzymatic characterization of surfactin synthetase domains in hybrid bimodular systems. J. Biol. Chem. 274:21581-21588. [DOI] [PubMed] [Google Scholar]
- 41.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thibaut, D., D. Bisch, N. Ratet, L. Maton, M. Couder, L. Debussche, and F. Blanche. 1997. Purification of peptide synthetases involved in pristinamycin I biosynthesis. J. Bacteriol. 179:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuboi, K. K., and T. D. Price. 1959. Isolation, detection and measure of microgram quantities of labeled tissue nucleotides. Arch. Biochem. Biophys. 81:223-237. [DOI] [PubMed] [Google Scholar]
- 44.Turgay, K., M. Krause, and M. A. Marahiel. 1992. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol. Microbiol. 6:529-546. [DOI] [PubMed] [Google Scholar]
- 45.von Dohren, H., U. Keller, J. Vater, and R. Zocher. 1997. Multifunctional peptide synthetases. Chem. Rev. 97:2675-2705. [DOI] [PubMed] [Google Scholar]