Abstract
Mature Pseudomonas aeruginosa biofilms form complex three-dimensional architecture and are tolerant of antibiotics and other antimicrobial compounds. In this work, an in vivo expression technology system, originally designed to study virulence-associated genes in complex mammalian environments, was used to identify genes up-regulated in P. aeruginosa grown to a mature (5-day) biofilm. Five unique cloned promoters unable to promote in vitro growth in the absence of purines after recovery from the biofilm environment were identified. The open reading frames downstream of the cloned promoter regions were identified, and knockout mutants were generated. Insertional mutation of PA5065, a homologue of Escherichia coli ubiB, was lethal, while inactivation of PA0240 (a porin homologue), PA3710 (a putative alcohol dehydrogenase), and PA3782 (a homologue of the Streptomyces griseus developmental regulator adpA) had no effect on planktonic growth but caused defects in biofilm formation in static and flowing systems. In competition experiments, mutants demonstrated reduced fitness compared with the parent strain, comprising less than 0.0001% of total biofilm cells after 5 days. Therefore, using in-biofilm expression technology, we have identified novel genes that do not affect planktonic growth but are important for biofilm formation, development, and fitness.
Formation and maturation of a bacterial biofilm, a surface-attached, highly structured, single- or multispecies population enmeshed in self-produced exopolysaccharide, are complex processes resembling the development of a multicellular organism (37). Biofilms are prevalent in disease caused by Pseudomonas aeruginosa, a common opportunistic and nosocomial pathogen and the leading cause of morbidity and mortality in cystic fibrosis (CF) patients (59). P. aeruginosa biofilms play a role in the pathogenesis of ventilator-associated pneumonia, urinary and peritoneal dialysis catheter infections, bacterial keratitis, otitis externa, and burn wound infections (12). For this reason, P. aeruginosa is one of the most widely studied model bacteria in biofilm research.
P. aeruginosa biofilm formation occurs when planktonic bacteria encounter a surface and attach to it using various adhesins, including flagella, type IV pili, and fimbriae (39, 60). The bacteria colonize the surface, forming microcolonies that mature into distinctive three-dimensional mushroom-like structures. The genetic basis for this elaborate morphogenesis is currently unknown. One of the most distinctive and challenging features of mature biofilms is their remarkable tolerance of antibiotics (6, 12). Although many theories have been proposed to explain biofilm-mediated antibiotic resistance (including diffusion limitation, slow growth rates, and expression of drug-metabolizing enzymes), none are able to account for all of the resistance seen in a typical biofilm (5-7, 10, 20, 31, 34, 69). This residual resistance has led to the concept of a biofilm phenotype (23), a physiological state in which altered gene expression leads to cooperative development of biofilm architecture and expression of increased tolerance. One component of tolerance was recently postulated to be loss of the capacity to undergo programmed cell death upon exposure to concentrations of antibiotics well above the conventional MIC (6, 31, 57).
Although the switch between the planktonic and the biofilm phenotypes may be regulated in a global fashion, little is known about how bacteria sense surface contact or attachment and respond at the genetic level. Quorum sensing has been shown to modulate the transformation of cells to the biofilm phenotype in P. aeruginosa (21). Deletion of lasI, the structural gene for N-(3-oxydodecanoyl)-l-homoserine lactone synthetase, causes loss of exopolysaccharide production and defects in biofilm formation (14). P. aeruginosa has been shown to produce both N-butyryl-l-acyl HSL and N-(3-oxydodecanoyl)-l-homoserine lactone in the CF lung (19, 55). The range of genes influenced by quorum-sensing regulation and their relation to biofilm formation are under intensive investigation in a number of laboratories (12).
A small number of regulatory factors involved in Pseudomonas biofilm development have been identified. Parkins and colleagues (42) showed that the P. aeruginosa global virulence regulator GacA is involved in biofilm development, in that cells lacking GacA adhered to surfaces but failed to develop into mature biofilm structures. Another regulatory factor, Crc (catabolite repression control protein), was shown to influence biofilm formation through its effects on type IV pili expression (38). Studies in both Escherichia coli and P. aeruginosa demonstrated that the stationary-phase sigma factor RpoS may play a role in biofilm physiology, although opposite effects were reported (1, 68). More recently, a two-component-type regulatory gene, pvrR, controlling a phenotypic switch between wild-type morphology and a rough small colony variant with increased biofilm-forming capacity and antibiotic tolerance, has also been identified (16).
On a broader scale, Kolter and colleagues (39, 40, 45) used a simple microtiter plate model to identify transposon mutants unable to initiate biofilm formation in two species of Pseudomonas and E. coli. They identified both pili and flagella as being important for the early stage of biofilm formation. In E. coli, a study of biofilm gene expression showed that of 446 transposon-mediated lacZ fusions examined, 38% were differentially expressed by at least twofold between biofilm and planktonic cells grown in microtiter plates (47). These fusions represented classes of genes responsive to osmolality, oxygen limitation, and increased cell densities, suggesting that environmental features play a significant role in regulation of the phenotype in biofilm growth. The above studies were limited by use of transposons that can introduce polar mutations and the use of a static model of biofilm growth, in which gradients of oxygen and other factors may form across the biofilm, influencing the range of genes identified.
Subsequent studies of biofilm gene expression at the proteome and RNA levels have used biofilms grown in flowthrough systems (41). Whiteley and colleagues (65) used DNA microarrays to compare gene expression of biofilm and planktonic P. aeruginosa PAO1 grown either in chemostats or in once-flowthrough tubing and identified only 73 genes (representing approximately 1% of the total genome) whose expression varied twofold or greater. The genes identified were divided evenly between those that were up-regulated and down-regulated under conditions of biofilm growth, and approximately one-third were hypothetical open reading frames (ORFs) (65). Sauer and colleagues (50) used a combination of reporter gene, microscopy, and two-dimensional gel electrophoresis analyses of P. aeruginosa grown in a once-flowthrough tubing system to follow changes in gene and protein expression throughout various stages of biofilm development. They reported that, depending on the stage of biofilm maturation, up to 56% of resolvable proteins demonstrated altered levels of expression compared with those in chemostat-grown planktonic cells. Therefore, the type, number, and magnitude of expression changes detected vary with the experimental system employed, and multiple approaches are necessary to fully appreciate the changes in bacterial physiology induced by growth as a biofilm.
We chose to use in vivo expression technology (IVET) to identify P. aeruginosa genes induced during biofilm formation and development. IVET was originally developed to study putative virulence genes induced in pathogenic bacteria during growth in the complex environment of the host (56). In IVET, the complex environment of interest is used to screen a reporter gene library and to select for those clones in which gene expression is on. The P. aeruginosa IVET system used in this study is based on complementation of de novo adenine biosynthesis by randomly cloned promoters able to drive expression of a promoterless purEK operon (63). This P. aeruginosa IVET system was used previously by Jin and colleagues to identify genes specifically induced in mouse models of acute peritonitis and burn wound infection (24, 63) and by respiratory mucus from CF patients (62). Since biofilms, like mammals, are complex and heterogeneous growth environments (49), we reasoned that IVET could be useful for the identification of genes important for P. aeruginosa biofilm development.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. The P. aeruginosa purEK deletion strain PAK-AR2 and the PAK IVET library were described previously (63) and were the generous gift of Shouguang Jin (University of Florida). PAK-AR2 is resistant to streptomycin and spectinomycin, and the integrated IVET plasmid confers tetracycline and piperacillin resistance upon clones of the library. Growth media included Davis minimal media with 0.1% glucose (DMM; Difco), supplemented where necessary with 50 μg of adenine/ml (DMM+A); Luria-Bertani (LB) broth and agar (Difco); and Pseudomonas isolation agar (PIA; Difco). Antibiotics were used where indicated at the following concentrations: ampicillin, 50 μg/ml for E. coli; carbenicillin, 150 μg/ml for P. aeruginosa; gentamicin, 15 μg/ml for E. coli and 200 μg/ml for P. aeruginosa; piperacillin, 50 μg/ml for P. aeruginosa; spectinomycin, 200 μg/ml for P. aeruginosa; streptomycin, 200 μg/ml for P. aeruginosa; and tetracycline, 15 μg/ml for E. coli and 50 μg/ml for P. aeruginosa.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristics | Reference or Source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type, serotype O5 | 25 |
| PAK | Wild type, serotype O6; background for IVET | 62 |
| PAK-AR2 | PAK lacking the purEK gene; Spr Smr | 62 |
| 9.27A | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 9.27B | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 9.27C | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 9.27D | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 9.27E | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 9.29 | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| 3.15 | Biofilm-induced IVET clone; Tcr Pipr Spr Smr | This study |
| PA0240::Gmr | PAO1 with insertional mutation in PA0240 | This study |
| PA3710::Gmr | PAO1 with insertional mutation in PA3710 | This study |
| PA3782::Gmr | PAO1 with insertional mutation in PA3782 | This study |
| PA5065::Gmr/pUCP26-PA5065 | PAO1 with insertional mutation in PA5065, expressing PA5065 in trans from pUCP26 lac promoter | This study |
| PA5065::Gmr/pUCP20-ubiBEc | PAO1 with insertional mutation in PA5065, expressing E. coli ubiB in trans from pUCP20 lac promoter | This study |
| E. coli | ||
| JM109 | endA1 gyrA96 hsdR17(rK− mK+), mcrB+recA1 relA1 supE44 thi-1 Δ(lac-proAB) F′(traD36 proAB lacIZΔM15) | 71 |
| SM10 | thi-1 thr leu tonA lacY supE recA RP4-2-Tc::Mu Kmr | 54 |
| TOP10 | mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU | Invitrogen |
| Plasmids | ||
| pCR2.1-TOPO | Topoisomerase-linked T/A cloning vector; Apr Kmr | Invitrogen |
| pUCP20 | pUC18-derived broad-host-range vector; Apr Pipr | H. Schweizer |
| pUCP26 | pUC18-derived broad-host-range vector; Tcr | 64 |
| pUCGM | Source of Gmr cassette; Apr Gmr | 51 |
| pEX18Ap | Gene replacement vector; oriT+sacB+ Apr | 27 |
| pUCP26-PA5065 | Construct used for in-trans expression of PA5065 in PA5065::Gmr insertional mutant | This study |
| pUCP20-ubiBEc | Construct used for in-trans expression of E. coli ubiB in PA5065::Gmr insertional mutant | This study |
| pEX18Ap-PA5065::Gmr | Construct used for generation of PA5065::Gmr insertional mutation | This study |
| pEX18Ap-0240::Gmr | Construct used for generation of PA0240::Gmr insertional mutation | This study |
| pEX18Ap-3710::Gmr | Construct used for generation of PA3710::Gmr insertional mutation | This study |
| pEX18Ap-3782::Gmr | Construct used for generation of PA3782::Gmr insertional mutation | This study |
Growth of biofilms individually and in competition.
For single-strain biofilms, bacteria were grown overnight in 5 ml of DMM+A at 37°C with agitation (150 rpm). The cultures were diluted to a 0.5 McFarland standard (∼107cells/ml), and then 2.5 ml was subcultured into 250 ml of DMM+A. The culture was passed through sterile silicone 5-mm-diameter tubing at 50 ml/h for 4 h, allowing bacterial attachment. The flow from the culture was shut off, and DMM alone was flowed through the system at 50 ml/h for 5 days (120 h). In the literature, flow rates for perfused biofilm systems range from 10 to 100 ml/h (3). In our experiments, a flow rate of 50 ml/h was used to ensure that planktonic bacteria were removed from the system and that adenine remaining from the inoculating culture was removed.
On the fifth day, 5 cm of silicone tube was retrieved and rinsed with phosphate-buffered saline (PBS), pH 7.1, to remove planktonic cells. The biofilm was scraped with a sterile spatula into 5 ml of PBS. The suspension was vortexed for 30 s to break up cellular clumps, and serial tenfold dilutions of the suspension were plated onto DMM+A agar to isolate single colonies.
For competition assays, two strains were grown individually overnight in 5 ml of DMM+A. The cultures were diluted to a 0.5 McFarland standard, and 2.5 ml of each was subcultured into 250 ml of DMM+A. The mixed culture was grown as a biofilm for 5 days as above. Sections of tubing, 5 cm in length, were sampled at 24, 48, and 96 h. The tubing was rinsed and the biofilm was harvested as described above. Serial tenfold dilutions were plated in parallel on LB agar and LB agar with either streptomycin (200 μg/ml) and spectinomycin (200 μg/ml) (for PAK-AR2) or gentamicin (200 μg/ml) (for knockout mutants), as described below. The relative composition of the biofilms was determined by subtraction of colony counts obtained on selective medium (mutants) from total counts obtained on nonselective LB plates.
Growth of the IVET library as a biofilm.
The original IVET library consisted of a minimum of 2 × 104 pooled colonies (63). A subculture of the library sent to us was passaged once and frozen at −80°C. An aliquot of the IVET library freezer stock was used to inoculate 5 ml of DMM+A and incubated overnight at 37°C with agitation (150 rpm). The media was supplemented with adenine to alleviate the purine auxotrophy and ensure survival of all cointegrates. The 5-ml culture was then subcultured into 250 ml of DMM+A containing spectinomycin (200 μg/ml), streptomycin (200 μg/ml), carbenicillin (150 μg/ml), and tetracycline (100 μg/ml) and grown for 4 h at 37°C. Carbenicillin was used in initial experiments; we later switched to piperacillin, a more effective and less expensive antipseudomonal beta-lactam antibiotic. After growth as a biofilm as described above, harvested cells were diluted and plated on DMM+A agar to obtain single colonies.
Southern blot of the IVET isolates and PAK.
To generate a probe for Southern blot analysis, a 968-bp internal fragment of purK was amplified with primers purKup (5′-AAAATCGGTGTCATCGGTGG-3′) and purKdown (5′-GTAGTGATGCAGGTGGCAGT-3′), gel purified (QIAEXII; Qiagen), and labeled by random priming (DIG DNA labeling kit; Roche). Chromosomal DNA was prepared from each of the seven IVET isolates and PAK by using DNAZOL (Invitrogen). Prehybridization, hybridization, and detection were carried out in accordance with the manufacturer's directions. The hybridized DNA probe was detected by using an anti-DIG polyclonal antibody conjugated to alkaline phosphatase and its chemiluminescent substrate AMPPD [0.235 mM 3-(2′-spiroadamantane)-4-methoxy-4(3′-phosphoryloxy)-phenyl-1,2-dioxetane] (Roche) followed by exposure to X-ray film (Kodak). X-ray films were developed with an automated developer (X-OMAT 480 RA Processor; Kodak).
Identification of the cloned promoters upstream of purEK.
To identify the cloned promoter regions driving purEK expression, a single primer (purEKreverse; 5′-TGCGCTCATCGTCGTGCCTC-3′) based on the DNA sequence between the BglII cloning site for the IVET library and the 5′ end of purEK was used for touch-down PCR (15), using annealing temperatures from 60°C to 40°C, decreasing 1°C per cycle. An additional 20 cycles at an annealing temperature of 40°C were carried out. Extension time for all cycles was 72°C for 2 min. The PCR was separated on a 0.7% agarose gel, the largest and/or most prominent band was excised, and the DNA was extracted with a gel purification kit (QIAEXII).
The purified DNA was cloned into TOPO-pCR2.1 (Invitrogen) as per the manufacturer's instructions and transformed into E. coli TOP10. A rapid screening protocol was used to identify plasmids containing inserts (18). Purified plasmid DNA (5 μg; QIAprep spin miniprep kit; Qiagen) was sequenced at the molecular core facility of York University (North York, Ontario, Canada) by using a nested primer (IVETsequence; 5′-GTGCCTCTCTCAAATGCG-3′) to obtain the sequence of the DNA immediately upstream of purEK. Sequences were compared to the P. aeruginosa PAO1 genome (http://www.pseudomonas.com) (58). The putative promoter-containing sequence was located, and ORFs in the same orientation as purEK were identified. BlastP (4) analysis using conceptual products from translation of these ORFs was then used to search all protein sequences in GenBank. To examine the distribution of specific ORFs among various strains of P. aeruginosa, PCR amplification of two of the identified sequences, PA3710 and PA3782, was carried out with template DNA from 15 clinical strains of P. aeruginosa. Using primers 3782 up (5′-ACCGCATCAGTCCGTTCCAC-3′) and down (5′-AAATCCTCC GCCCACATCC-3′) and 3710 up (5′-GGCGAATGGCGGGTAGAA-3′) and down (5′-GGCAGGGTGGAAGATGGTGG-3′), PA3782 and PA3710 were PCR amplified in a multiplex reaction.
Generation of knockout mutants.
Chromosomal knockout mutants were created as described previously (8). Briefly, the gene of interest was amplified by PCR and cloned into the nonreplicative vector pEX18Ap containing the sacB gene for sucrose counterselection. A gentamicin resistance cassette was inserted into a unique site within the gene of interest, and the construct was transformed into the donor strain E. coli SM10 for conjugal transfer into PAO1. After overnight incubation on LB plates, mating mixtures were plated on PIA containing 200 μg of gentamicin/ml. Gmr colonies were then plated on LB agar containing 200 μg of gentamicin/ml and 5% sucrose to identify isolates that had lost the vector-associated sacB gene (resistant to sucrose). To confirm loss of the vector-associated DNA, Gmr sucrose-resistant isolates were plated in parallel on LB agar containing 200 μg of gentamicin/ml and LB agar containing piperacillin (15 μg/ml) to identify clones lacking the Pipr marker associated with pEX18Ap.
Confirming lethality of the PA5065 mutation.
Electrocompetent PAO1 was prepared as described previously (8) and transformed with pUCP26-PA5065 (Tetr). Next, E. coli SM10-pEX18Ap-PA5065::Gmr was mated with PAO1-pUCP26-PA5065 as described above. The mating mixture was plated on PIA containing 50 μg of tetracycline/ml and 200 μg of gentamicin/ml to counterselect the E. coli donor strain. Resulting colonies were plated in parallel on LB agar containing 200 μg of gentamicin/ml and 50 μg of tetracycline/ml and on LB agar containing 15 μg of piperacillin/mg. All colonies grew on LB agar containing 200 μg of gentamicin/ml, and all were Pips. Plasmid purification and restriction digests were used to confirm that an intact copy of PA5065 was present on pUCP26. To show that the chromosome contained the disrupted copy of PA5065, primers were designed to amplify the Gmr cassette, namely, Gm up (5′-AAGCCTGTTCGGTTCGTAA-3′) and down (5′-TTCTTCCCGTATGCCCAA-3′). The expected amplicon of 700 bp was produced, confirming that the cassette was present in the chromosome.
The E. coli ubiBEc gene was PCR amplified by using primers ubiEEc up (5′-AGAATTCCG TGCTGTTGATGCCCTGA-3′) and ubiEEc down (5′-TTAAGCTTGCCGAGCTTTTTGGTGCC-3′). The product was cloned into the β-lactamase-encoding cloning vector pUCP20, which has the same origin of replication as pUCP26, making them incompatible. Purified pUCP20 and pUCP20-ubiBEc were transformed in parallel into PAO1 PA5065::Gmr/pUCP26-PA5065, and the transformants were plated on LB agar containing 15 μg of piperacillin/ml. Plasmids were purified from P. aeruginosa transformants and used to transform E. coli JM109, which was plated separately onto LB agar containing 15 μg of tetracycline/ml and LB agar containing 50 μg of ampicillin/ml. Plasmid DNA was purified from the E. coli transformants and digested and analyzed by electrophoresis, as described above.
Growth curves.
Growth curves of PAO1 and mutant strains PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr were generated by standardizing overnight cultures grown in DMM to 0.5 McFarland (∼107cells/ml). Each strain was inoculated in triplicate as a 1/10 dilution (40 μl into 400 μl of DMM) and grown in an automated shaking incubator-spectrophotometer (Bioscreen-C; Lab System). Each strain was grown at 37°C with agitation, with turbidity readings at 600 nm procured every 20 min over a 24-h period.
Static biofilm formation assay.
The assay of O'Toole and Kolter (39) was used to measure biofilm formation under static conditions. Overnight cultures of PAO1, PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr strains were standardized to 107 CFU/ml and inoculated 1/20 in 200 μl of DMM in a 96-well microtiter plate that was incubated overnight at room temperature. Eight wells per experiment were inoculated for each strain. After removal of planktonic cells, the wells were rinsed with tap water, and biofilms were stained with 200 μl of 1% crystal violet (CV). After 15 min, the plate was washed with tap water, and retained CV was solubilized with 200 μl of 95% ethyl alcohol. The turbidity at 600 nm of planktonic cells (growth control) and CV were measured by using an automated 96-well plate reader (VersaMAX microplate reader), and the data were compiled with Softmax PRO software (Molecular Dynamics).
SEM of biofilms.
PAO1, PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr were grown as individual biofilms in DMM at room temperature. Two-centimeter sections of tubing were rinsed with PBS and fixed in 5 ml of 2% glutaraldehyde. Samples were processed in the scanning electron microscopy (SEM) department at the Hospital for Sick Children, Toronto, Ontario, Canada. After fixation in OsO4 for 1 h at room temperature, specimens were washed three times in PBS followed by a rinse in double-distilled H2O. The samples were dehydrated in a graded ethanol series, critical-point dried, and mounted on SEM stubs for viewing with a JEOL 820 SEM and Iridium image capturing software.
CSLM of biofilms.
PAO1, PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr were grown as individual biofilms in a commercially available flow cell (Stovall) in DMM at room temperature. After 48 h, biofilms were stained with BacLight live/dead stain (Molecular Probes), and images were acquired with a Zeiss confocal scanning laser microscope (CSLM) and processed with Zeiss LSM510 software.
Motility assays.
Bacterial strains were tested for swimming motility by inoculation on 0.3% agar plates (48) and for twitching motility by stab assay using 1% agar plates (52). Assays were performed in triplicate, and the resulting zones of motility were measured.
Antibiotic susceptibility testing.
The planktonic MIC of each antibiotic for each strain was obtained (35). Biofilms of each strain were then formed on a polystyrene peg lid (Nunc) in a 96-well microtiter plate filled with Mueller Hinton broth (MHB) and cells (107 cells/ml) overnight at 37°C with shaking (9, 57). Subsequently, the pegs were rinsed twice with sterile PBS and placed in a fresh microtiter plate containing 10-fold serial dilutions in MHB of each antimicrobial agent and then incubated overnight at 37°C. After the incubation period, the pegs were rinsed twice in PBS and placed in another microtiter plate filled with MHB. Biofilms were removed by sonication for 10 min in a water bath sonicator (Branson Ultrasonics Corporation). Dislodged cells were enumerated by serial dilution and plate counts.
RESULTS
Use of IVET to identify genes induced by biofilm growth.
To determine whether a purEK-based IVET selection system (63) was applicable to the biofilm environment, the ability of the purEK deletion strain, PAK-AR2, to form a biofilm on a silicone substratum in the absence of purines was compared to that of the wild-type PAK. As reported previously (62), PAK-AR2 was able to survive in the absence of adenine. AR2 formed a biofilm on silicone tubing, but cell counts (CFU/cm2) were 4 log units lower than those obtained for PAK.
To determine if biofilm growth would select for purEK-expressing strains and to determine the length of selection time required, a competition assay of PAK versus PAK-AR2 was performed. By 96 h, a biofilm that was inoculated with equal amounts of PAK and PAK-AR2 consisted solely of PAK (not shown). This experiment showed that in a competitive situation, a non-purine-producing strain does not survive the selection phase, even in coculture with an adenine-producing strain. Therefore, growth of the IVET library for 5 days in the absence of purines should enrich for clones that contain active promoters upstream of the integrated purEK operon while depleting those unable to produce adenine.
The IVET library was grown as a biofilm on a silicone substratum for 5 days without adenine, harvested, and diluted to obtain single colonies on DMM+A plates. Seven isolates, numbered 9.27A to E, 9.29, and 3.15, that grew poorly on DMM compared to DMM+A were identified. Due to the use of agar plates for screening, promoters induced in response to surface contact or attachment may not be distinguished by this screen. Each of the isolates alone formed a biofilm, with cell counts approximately 2 log units lower than that of the wild type but 2 log units higher than that of PAK-AR2 alone.
Molecular characterization of cloned promoters and downstream genes.
Southern hybridization with a labeled purEK probe was used to screen for unique clones. Clones 9.27B and C were identical, whereas the remaining clones had unique restriction patterns (not shown), showing that six different integrants were present among the seven isolates.
DNA containing the putative promoter of interest upstream of purEK was amplified by touch-down PCR, cloned, and sequenced. The resulting sequences were used to locate the promoter regions on the PAO1 genome (58) and to identify downstream ORFs. BlastP (4) analysis with the conceptual products from translation of ORFs downstream of the cloned promoter and in the same orientation as purEK was performed (Table 2). PA designations from the P. aeruginosa genome project (http://www.pseudomonas.com) were used to identify the ORFs of interest. Although the P. aeruginosa PAK genome has not been sequenced, the cloned DNA shared ≥95% nucleotide identity with the PAO1 genome sequence, and no sequences were identified which were not present in PAO1. Also, PCR amplification of identified ORFs using chromosomal DNA from 15 randomly chosen P. aeruginosa clinical isolates as templates yielded amplicons identical in size to those from PAK in each case (not shown). Therefore, the genes of interest appear to be conserved in P. aeruginosa.
TABLE 2.
Putative biofilm-induced proteins and their homologues
| IVET clone ORF (no. of nucleotides) | Size (aa) | Selected homologuesa | Function | % Iden- tityc | % Simi- larityd | GenBank accession no. |
|---|---|---|---|---|---|---|
| 9.27A PA0240 (1266) | 421 | PhaK-like protein, P. putida | Putative porin | 71 | 82 | AF031417 |
| Pflu_p_1331, Pseudomonas fluorescens | Hypothetical protein | 69 | 81 | AABA01000097 | ||
| Psyr0433, Pseudomonas syringae pv. syringae B728a | Hypothetical protein | AABH01000001 | ||||
| BenF, P. putida | Benzoate-specific porin-like protein | 42 | 61 | AF218267 | ||
| 9.29 PA2247 (1233) | 410 | BkdA1, P. putida | 2-Oxoisovalerate dehydrogenase (alpha subunit) | 79 | 88 | AAA65614 |
| BkdA1, Mesorhizobium loti | 2-Oxoisovalerate dehydrogenase (alpha subunit) | 64 | 76 | AP003004 | ||
| 9.27B and C PA3710 (1674) | 557 | Pflu_p_4963, P. fluorescens | Hypothetical protein | 76 | 85 | AABA01000160 |
| SMc03116, Sinorhizobium meliloti | Putative alcohol dehydrogenase protein | 63 | 76 | AL591792 | ||
| CC1278, C. crescentus | Oxidoreductase, GMC family | 55 | 68 | AE005802 | ||
| AlkJ, P. putida | Aliphatic alcohol dehydrogenase | 40 | 57 | AJ233397 | ||
| 3.15 PA3782 (954) | 317 | Z0442, E. coli O157:H7 | Transcriptional regulator | 48 | 67 | CAB87229 |
| mlr2766, M. loti | Transcriptional regulator | 49 | 63 | BAB49814 | ||
| AdpA, S. griseus | AraC-like transcriptional regulator; streptomycin and aerial mycelium production | 48 | 62 | BAA86265 | ||
| AdpA, S. coelicolorb | AraC-like transcriptional regulator | 47 | 62 | CAB87229 | ||
| 9.27D and E PA5065 (1602) | 542 | UbiB, E. coli | Ubiquinone biosynthesis | 56 | 70 | D65188 |
| UbiB, Vibrio choleraeb | Ubiquinone biosynthesis | 55 | 69 | AAF93263 | ||
| UbiB (AarF), Providencia stuartii | Ubiquinone biosynthesis | 59 | 74 | AAB96577 | ||
| UbiB, Neisseria meningitidis | Ubiquinone biosynthesis | 57 | 72 | AAF40987 |
Homologous proteins, identity, and similarity scores were determined by the BLASTp program at NCBI.
Hypothetical protein.
Identical amino acids.
Identical plus conserved amino acids.
Strain 9.27A contained the promoter upstream of PA0240. As predicted from Southern blot analysis, strains 9.27B and 9.27C contained the same cloned fragment, upstream of PA3710. Strains 9.27D and 9.27E each contained the same promoter region cloned on two different-sized fragments, yielding unique patterns for each on Southern blot analysis. This promoter region lay upstream of an apparent operon containing ubiE-PA5064-PA5065 (ubiB). Strain 9.29 contained the promoter region upstream of PA2247, the first gene in a putative four-gene operon (bkdA1, bkdA2, bkdB, and lpdV) that is identical in organization to the bkd operon of Pseudomonas putida (32, 33). The last strain, 3.15, contained the promoter region upstream of PA3782, encoding a putative AraC-type transcriptional regulator homologous to AdpA of Streptomyces griseus and Streptomyces coelicolor. AdpA is a key developmental regulator required for the formation of aerial mycelium and secondary metabolites, including streptomycin (36, 70).
Inactivation of PA5065 is lethal.
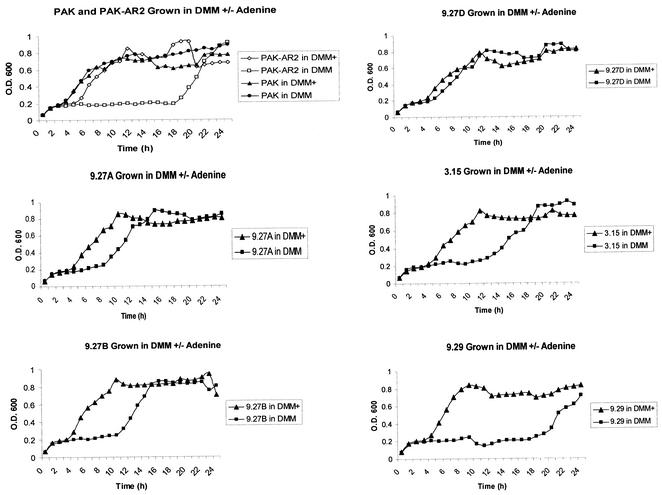
Growth curve analysis, with and without adenine, of the purEK deletion strain AR2 and the IVET clones recovered from the biofilm was performed. PAK-AR2, as noted above, could grow in the absence of adenine but had a lag phase of approximately 19 h (Fig. 1). One of the five IVET clones, 9.29, had a growth curve identical to that of AR2 (Fig. 1), suggesting that it lacked a functional promoter upstream of the integrated purEK gene. Therefore, this strain was not further analyzed. The remaining IVET clones had lag phases intermediate between those of PAK and PAK-AR2 (Fig. 1).
FIG. 1.
Growth curves of recovered IVET clones. Planktonic growth of PAK, PAK-AR2, and IVET clones (PAK-AR2 with integrated IVET plasmids) recovered after 5 days in a biofilm was compared in DMM and DMM+A. Since strains 9.27B and C were identical, only 9.27B is shown. Note that strain 9.29 has a growth curve similar to that of AR2.
To study the role of these genes in biofilm development, insertional mutants were generated. The mutants were generated in P. aeruginosa strain PAO1 instead of PAK because of the availability of flanking sequence information from the PAO1 genome (58) and the conservation of the genes of interest between the two strains. Despite repeated attempts, a PA5065::Gmr mutant could not be generated, suggesting that the gene was essential for survival of PAO1. To confirm the lethality of such a mutation, supporting experiments were conducted. Similar experiments were used previously in PAO1 to demonstrate that inactivation of the lipopolysaccharide core kinase WaaP led to death of the cells (61).
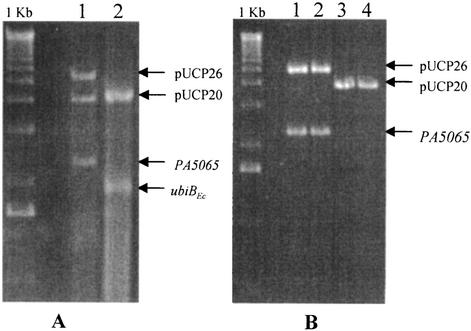
As outlined in Materials and Methods, PAO1 was first transformed with a plasmid encoding PA5065, and the chromosomal copy of the gene was inactivated by insertion of a gentamicin cassette. We then attempted to cure the plasmid-encoded copy of PA5065 by using either an incompatible plasmid vector with no insert or a plasmid with a cloned copy of ubiBEc (formerly yigR) of E. coli, a homologue of PA5065. Although pUCP26-PA5065 was readily displaced by pUCP20-ubiBEc, the vector pUCP20 alone was unable to displace pUCP26-PA5065. The PA5065::Gmr strain retained both plasmids despite piperacillin selection for pUCP20 and the exclusionary pressure of incompatible plasmids (Fig. 2). Plasmid DNA isolated from this strain was used to transform E. coli, yielding both Tetr and Ampr transformants, confirming that both pUCP20 and pUCP26-PA5065 were present (Fig. 2). Therefore, ubiBEc is a functional homologue of PA5065, and expression of PA5065 or a functional homologue in trans was essential for survival if the chromosomal copy of PA5065 had been inactivated.
FIG. 2.
Analysis of plasmids isolated from recombinant PA5065::Gmr insertional mutants. (A) Plasmid DNA was isolated from the recombinant PA5065::Gmr/pUCP26-PA5065 strain after transformation with pUCP20 (lane 1) or pUCP20-ubiBEc (lane 2) and digested with EcoRI and HindIII to release the inserts. pUCP26-PA5065 was isolated in tandem with pUCP20 (lane 1) despite counterselection, but it was lost if pUCP20-ubiBEc was present in the cell (lane 2). (B) The plasmid DNA isolated from PA5065::Gmr/pUCP26-PA5065 was used to transform E. coli JM109, which was grown on plates containing either 15 μg of tetracycline/ml or 50 μg of ampicillin/ml. Analysis of plasmid DNA isolated from the resulting Tetr (lanes 1 and 2) and Ampr (lanes 3 and 4) recombinant E. coli strains by digestion with EcoRI and HindIII confirmed that both plasmids were originally present in the recombinant P. aeruginosa mutant despite counterselection against pUCP26-PA5065.
Characterization of PA0240, PA3710, and PA3782 knockout mutants.
Knockout mutants in the remaining three genes, PA0240, PA3710, and PA3782, were generated successfully. Planktonic growth curves of the parental PAO1 strain and the PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr mutants were compared to test whether these putative biofilm genes had an effect on nonbiofilm growth. The growth curves for all four strains were identical (not shown), suggesting that the inactivated genes were not important for planktonic growth.
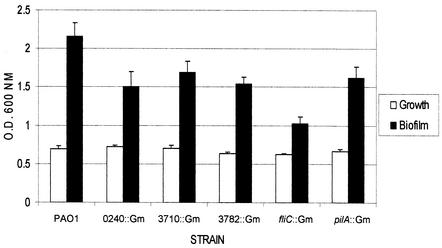
To assess the effect of the mutations on biofilm formation under static conditions, a microtiter plate assay (40) was performed. Pilin and flagellin mutants, which were previously demonstrated to be defective in biofilm formation (40), were used as negative controls. Relative to the wild type, each of the biofilm mutants showed reduced biofilm formation (Fig. 3) of a magnitude similar to that of the pilA mutant. Loss of pilA has previously been shown to reduce biofilm formation to 80% or less of that of the wild type in static assays (40, 60). The swimming (flagella) and twitching (pili) motilities of the mutants were similar to that of the parent strain (not shown), showing that loss of motility was not responsible for decreased biofilm formation.
FIG. 3.
Static biofilm formation assay. The abilities of PAO1 and its isogenic mutants to form biofilms in a static assay were compared. The amount of planktonic growth in each well as measured by the turbidity (reported here as optical density [O.D.]) at 600 nm is shown in white bars, while the amount of biofilm is shown as the absorbance of bound CV at 600 nm in black bars. Pilin (pilA) and flagellin (fliC) mutants were used as negative controls.
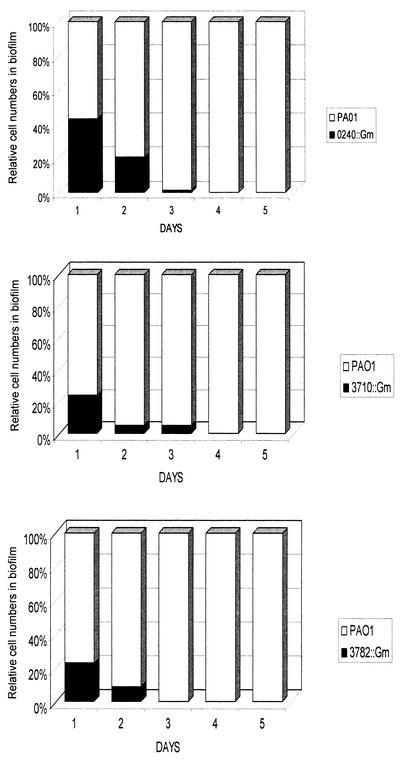
Biofilm formation was then investigated in a flowing system. The wild type and mutants were grown as individual biofilms for 5 days in silicone tubing. The mutants were able to form biofilms but gave cell counts approximately 1 to 2 log units less than PAO1. We then tested the fitness of the mutants compared with the parent strain during biofilm growth by using competition experiments. Equal quantities of PAO1 and each mutant were cocultured as a biofilm for 5 days. The biofilms were sampled at 24-h intervals, and the relative quantities of each strain were determined by comparing total plate counts (representing both strains) to counts obtained from selective (200 μg of gentamicin/ml) plates (representing the Gmr insertional mutants). In each case, the mutants were less fit and by the fifth day comprised less than 0.0001% of the total number of bacteria in the mixed biofilms (Fig. 4).
FIG. 4.
Biofilm competition assay. Equal quantities of PAO1 and its isogenic mutants were grown together as a biofilm for 5 days. A section of tubing was removed daily, and the total number of cells, as well as the number of Gmr cells, was enumerated by serial dilution and plate count. All three mutants were less fit than the wild type and were reduced to 0.0001% or less of total cells after 5 days.
Effect of mutations on biofilm morphology.
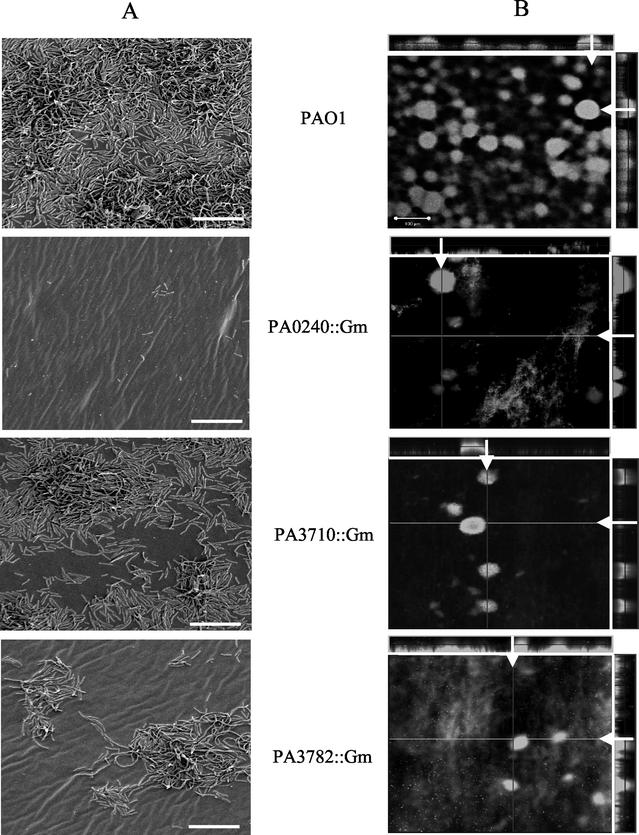
CSLM and SEM were used to visualize morphology of single-strain biofilms. Both techniques generated similar results despite the differences in substrata (glass versus silicone) and flow characteristics. The PAO1 wild-type strain formed characteristic microcolonies that later develop into mushroom-like structures present in a mature biofilm (Fig. 5). In contrast, SEM analysis showed that PA0240::Gmr colonized the substratum poorly, and this strain demonstrated poor microcolony formation in CSLM analysis. Similarly, SEM analysis showed that PA3782::Gmr colonized the surface poorly, forming atypical microcolonies with elongated cells (Fig. 5), possibly reflecting other defects that may have resulted from inactivation of this putative transcriptional regulator. In CSLM, only small microcolonies were present, as well as a patchy colonization of the substratum (Fig. 5). The PA3710::Gmr mutant was intermediate in its ability to form a biofilm by both SEM and CSLM analysis. While it formed microcolonies, it did not cover the substratum as completely as the wild type (Fig. 5). These experiments showed that all three mutants formed abnormal biofilms compared with the parent strain.
FIG. 5.
Biofilm morphology of mutants. (A) SEM of P. aeruginosa grown in silicone tubing. Each individual strain was grown under flow in DMM+ media for 24 h (longer growth times resulted in thicker biofilms that collapsed during dehydration, making structure difficult to see). Magnification, 1,000×; bars, 10 μm. (B) CSLM of BacLight live/dead stained 48-h biofilms. Sagittal (x-z) sections through the biofilm are indicated by white arrows. Note that defects in biofilm formation are consistent despite differences in imaging techniques (SEM versus CSLM), substrata (silicone versus glass), and length of incubation (24 versus 48 h), showing that these defects are not specific to cultivation conditions. Magnification, 100×; bar, 100 μm.
Antibiotic resistance of biofilm mutants.
One of the hallmarks of biofilm-grown cells is an increased tolerance of antimicrobial agents compared with planktonic cells of the same strain. Therefore, the antibiotic resistance of biofilm mutants PA0240::Gmr, PA3710::Gmr, and PA3782::Gmr was compared with that of the PAO1 wild type by using a peg lid system (9, 57). While the three mutants made biofilms containing approximately 0.5 log units less cells than the wild type on this substratum (polystyrene), there were no significant differences in the resistance of the biofilms to the antibiotics piperacillin, chloramphenicol, tetracycline, or ciprofloxacin (not shown).
DISCUSSION
Traditionally, bacteria have been regarded as individual organisms and have been grown and studied as homogeneous planktonic populations. However, bacteria in natural environments are usually found as a community of sessile organisms organized in a biofilm (13). In this study, we utilized an IVET system, previously used in vitro to identify genes induced by CF respiratory mucus (62), to study gene expression in 5-day-old P. aeruginosa biofilms grown on a silicone substratum.
Using IVET to study biofilm gene expression was feasible, since a competition study of PAK versus PAK-AR2 showed that after 96 h, PAK-AR2 could not be detected in the biofilm. Therefore, prolonged growth (5 days) in the absence of adenine should select for strains that contain promoters driving purEK expression while selecting against strains containing promoters not active under biofilm conditions. Interestingly, although PAK-AR2 should be unable to grow in the absence of adenine due to the deletion of purEK, biofilm growth experiments showed that it could survive and grow in an adenine-free environment, but to lower densities than the wild type. Similar observations were made by Wang et al. (62) when using this strain for their in vitro studies identifying genes up-regulated by exposure of P. aeruginosa to CF mucus. Growth curve analyses in our laboratory showed that after a reproducible lag phase of approximately 19 h, PAK-AR2 could overcome its auxotrophy and enter log phase growth (Fig. 1). The mechanism by which growth occurs is unknown at this time. The phenomenon is not due to recovery of purEK function, since PAK-AR2 is a deletion mutant. Neither is growth of PAK-AR2 due to acquisition of second-site mutations, since onset of growth occurs at a reproducible time point and cells subcultured into fresh medium show the same lag phase as the original culture. Regardless, biofilm competition assays showed that strains not expressing purEK, such as PAK-AR2, were reduced to undetectable levels in mixed biofilms over the course of the selection period.
Five separate promoter regions in seven isolates exhibiting poor ex biofilm growth without adenine were identified in this study. Although the numbers of independent clones recovered were low, they are consistent with previous work using this library (24, 62, 63), suggesting that the diversity of the library is lower than predicted. Analysis of these putative promoter regions showed no obvious consensus sequences for any of the common bacterial sigma factors, such as RpoD (σ70), RpoS (σ38), or RpoN (σ54), no quorum sensing response elements (lux boxes), nor any shared features that could represent binding sites for biofilm-responsive elements. Disruption of the genes downstream of these promoters resulted in strains that showed defects in biofilm formation on a variety of substrata (polystyrene, silicone, glass) and growth conditions (one static and two flowing systems).
PA5065 is an essential gene that could be inactivated only in the presence of an intact copy of PA5065 or its functional homologue ubiBEc. UbiB is necessary for aerobic synthesis of coenzyme Q (CoQ) and thus is essential to aerobiosis (44). The heterogeneity of O2 tension levels in biofilms has been demonstrated with the aid of microelectrodes (11), and reporter gene fusion studies in E. coli biofilms showed induction of genes that respond to reduced O2 levels (47). Since PA5065 mutations were lethal, we were unable to assess any phenotypes that may be associated with loss of PA5065 function. While ubiB mutations are not lethal for E. coli, ubiBEc mutants have alternate, as-yet unidentified hydroxylation mechanisms used in the anaerobic synthesis of ubiquinone (2). P. aeruginosa, a primarily aerobic bacterium, may lack the alternate ubiquinone biosynthetic pathways found in facultative anaerobes such as E. coli. P. aeruginosa is capable of anaerobic growth using nitrate or nitrite as an electron acceptor, but CoQ is also required in that respiratory pathway (66, 73).
PA3710 is predicted to encode an alcohol dehydrogenase or oxidoreductase with homology to choline dehydrogenases, such as BetA (29, 30), that are involved in osmoregulation. E. coli biofilms have been shown to up-regulate genes involved in the osmotic stress response (47), and the mutations in the osmotic stress-sensing, two-component regulatory system OmpR-EnvZ can stimulate biofilm formation (46). Choline dehydrogenases play a role in the accumulation of compatible solutes such as glycine betaine in the bacterial cytoplasm to reduce the effects of high external osmotic pressures (29, 30). P. aeruginosa has a separate betA homologue (PA5372), and growth of the PA3710 mutant in increasing salt or sucrose concentrations was similar to that of the wild type (not shown). Therefore, the function of this protein remains obscure.
P. aeruginosa encodes a large number of putative porin proteins (58), but the functions of only a few are known. Loss of the putative porin encoded by PA0240 had detrimental effects on the ability of the knockout strain to form normal biofilms and to compete with the wild type in the biofilm environment. The function and potential substrates of this porin are currently unknown. Annotation of the putative outer membrane proteins of P. aeruginosa by F. Brinkman and R. E. Hancock (www.cmdr.ubc.ca/bobh/) designates the product of PA0240 as OpdF, a member of the OprD family of substrate-specific porins. OpdF has homology to OprE, whose synthesis is induced by anaerobic growth conditions. Recent work by Yoon and colleagues (72) showed that the major porin OprF is up-regulated in anaerobic biofilms and that its loss results in poor biofilm formation under anaerobic conditions. Together with identification of ubiB, above, these results suggest that an important environmental signal in biofilms is the presence of anaerobic domains that induce expression of genes responsive to reduced oxygen concentrations. Identification of genes expressed under anaerobic conditions in P. aeruginosa biofilms may be particularly relevant to CF lung biofilms. CF mucus is a hypoxic environment (67), and anaerobic conditions were recently shown to promote biofilm formation by P. aeruginosa (72).
One of the most intriguing genes identified in this study was PA3782, encoding a putative AraC-like transcriptional regulator. The AraC family of transcriptional regulators is involved in regulation of carbon metabolism, production of virulence factors, including fimbriae and adhesins, and response to environmental stressors (22). Streptomyces spp. undergo complex morphological differentiation to form vertical multicellular hyphae terminating in desiccation-resistant spores. S. griseus mutants lacking the PA3782 homologue AdpA are unable to produce streptomycin or form aerial mycelium or spores (36). AdpA has been shown to be a key positive regulator that enhances the expression of several genes, including adsA (encoding an extracytoplasmic function sigma factor) and sgmA (encoding a zinc metalloendopeptidase) (28, 70). In recent years, biofilm maturation has come to be considered a developmental process that shares features with other bacterial morphogenic pathways (37), such as sporulation in gram-positive bacteria (17), fruiting body formation in Myxococcus xanthus (43, 53), and stalked-cell formation in Caulobacter crescentus (26). The PA3782::Gmr mutant was significantly impaired in its ability to form normal biofilms, and in competition with the wild type, PA3782::Gmr comprised less than 0.0001% of the mixed biofilm by the fifth day. Future research efforts will be directed towards identification of genes whose expression is under PA3782 control in order to understand its role in biofilm development.
One of the limitations of applying IVET to study bacterial gene expression is the inability to identify genes that are repressed in the environment of interest. Alternate strategies that would identify genes down-regulated in a biofilm environment include use of DNA microarrays (65), subtractive hybridization (41), or differential-display PCR. All of these techniques rely on comparison of mRNA populations expressed under different growth conditions. Of note, regulatory genes such as PA3782 are typically transcribed at very low levels, and the transcripts may be difficult to detect using these technologies. Therefore, although IVET has its limitations, it may be superior for detection of genes expressed at low levels.
It is interesting that none of the genes identified in our study were identified as upregulated in P. aeruginosa PAO1 biofilms using DNA microarrays (65) or proteome analyses (50), although only a small fraction of numerous proteins with altered expression patterns in the latter study were identified. A reporter-based study of changes in gene expression following attachment of P. putida for 6 h to silicone tubing showed its PA5065 (ubiB) homologue to be down-regulated compared with that of planktonic cells, but the time frame studied was much shorter than our 5-day growth period. The lack of overlap between the results of PAO1 biofilm expression studies to date emphasizes that multiple approaches, including reporter techniques such as IBET, over the course of biofilm development are required to gain a broad understanding of the genetic and phenotypic changes that are occurring.
Acknowledgments
We thank Selva Sinnadurai for excellent technical assistance and Aina Tilups and Mike Woodside for help with SEM and CSLM, respectively.
This work was supported by a Canadian Institutes of Health Research/Canadian Infectious Diseases Society/Rx&D Fellowship to A.F. and operating grants from the University of Toronto Connaught Committee and the Natural Sciences and Engineering Council of Canada (RGPIN 227817-01) to L.L.B.
Footnotes
For a commentary on this article, see page 2687 in this issue.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, K., and I. G. Young. 1978. Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry 17:4750-4755. [DOI] [PubMed] [Google Scholar]
- 3.Allison, D., T. Maira-Litran, and P. Gilbert. 1999. Perfused biofilm fermenters. Methods Enzymol. 310:232-248. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, M. R., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-780. [DOI] [PubMed] [Google Scholar]
- 8.Burrows, L. L., D. F. Charter, and J. S. Lam. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481-495. [DOI] [PubMed] [Google Scholar]
- 9.Ceri, H., M. E. Olson, C. Stremick, R. R. Read, D. Morck, and A. Buret. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cochran, W. L., G. A. McFeters, and P. S. Stewart. 2000. Reduced susceptibility of thin Pseudomonas aeruginosa biofilms to hydrogen peroxide and monochloramine. J. Appl. Microbiol. 88:22-30. [DOI] [PubMed] [Google Scholar]
- 11.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 15.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 17.Dunny, G. M., and B. A. Leonard. 1997. Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51:527-564. [DOI] [PubMed] [Google Scholar]
- 18.Eckhardt, T. 1978. A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584-588. [DOI] [PubMed] [Google Scholar]
- 19.Erickson, D. L., R. Endersby, A. Kirkham, K. Stuber, D. D. Vollman, H. R. Rabin, I. Mitchell, and D. G. Storey. 2002. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect. Immun. 70:1783-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, D. J., D. G. Allison, M. R. Brown, and P. Gilbert. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27:177-184. [DOI] [PubMed] [Google Scholar]
- 21.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, P., J. Das, and I. Foley. 1997. Biofilm susceptibility to antimicrobials. Adv. Dent. Res. 11:160-167. [DOI] [PubMed] [Google Scholar]
- 24.Ha, U., and S. Jin. 1999. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect. Immun. 67:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hancock, R. E., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 28.Kato, J. Y., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strom. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 30.Lamark, T., T. P. Rokenes, J. McDougall, and A. R. Strom. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madhusudhan, K. T., K. L. Hester, V. Friend, and J. R. Sokatch. 1997. Transcriptional activation of the bkd operon of Pseudomonas putida by BkdR. J. Bacteriol. 179:1992-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madhusudhan, K. T., D. Lorenz, and J. R. Sokatch. 1993. The bkdR gene of Pseudomonas putida is required for expression of the bkd operon and encodes a protein related to Lrp of Escherichia coli. J. Bacteriol. 175:3934-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 35.National Committee for Clinical Laboratory Standards. 2000. Performance standard for antimicrobial susceptibility testing. Approved standard M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 36.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 41.Parkins, M. D., M. Altebaeumer, H. Ceri, and D. G. Storey. 2001. Subtractive hybridization-based identification of genes uniquely expressed or hyperexpressed during biofilm growth. Methods Enzymol. 336:76-84. [DOI] [PubMed] [Google Scholar]
- 42.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215-1226. [DOI] [PubMed] [Google Scholar]
- 43.Plamann, L., Y. Li, B. Cantwell, and J. Mayor. 1995. The Myxococcus xanthus asgA gene encodes a novel signal transduction protein required for multicellular development. J. Bacteriol. 177:2014-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon, W. W., D. E. Davis, H. T. Ha, T. Jonassen, P. N. Rather, and C. F. Clarke. 2000. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 46.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasmussen, K., and Z. Lewandowski. 1998. Microelectrode measurements of local mass transport rates in heterogeneous biofilms. Biotechnol. Bioeng. 59:302-309. [DOI] [PubMed] [Google Scholar]
- 50.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 52.Semmler, A. B., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 53.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 54.Simon, R., U. Priefer, and A. Puhler. 1983. A broad-host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1 :784-791.
- 55.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 56.Slauch, J. M., M. J. Mahan, and J. J. Mekalanos. 1994. In vivo expression technology for selection of bacterial genes specifically induced in host tissues. Methods Enzymol. 235:481-492. [DOI] [PubMed] [Google Scholar]
- 57.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 59.Tummler, B., and C. Kiewitz. 1999. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol. Med. Today 5:351-358. [DOI] [PubMed] [Google Scholar]
- 60.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh, A. G., M. J. Matewish, L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 35:718-727. [DOI] [PubMed] [Google Scholar]
- 62.Wang, J., S. Lory, R. Ramphal, and S. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 63.Wang, J., A. Mushegian, S. Lory, and S. Jin. 1996. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434-10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 65.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 66.Wissenbach, U., A. Kroger, and G. Unden. 1990. The specific functions of menaquinone and demethylmenaquinone in anaerobic respiration with fumarate, dimethylsulfoxide, trimethylamine N-oxide and nitrate by Escherichia coli. Arch. Microbiol. 154:60-66. [DOI] [PubMed] [Google Scholar]
- 67.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, K. D., M. J. Franklin, C. H. Park, G. A. McFeters, and P. S. Stewart. 2001. Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously-fed Pseudomonas aeruginosa biofilms. FEMS Microbiol. Lett. 199:67-71. [DOI] [PubMed] [Google Scholar]
- 69.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 70.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 72.Yoon, S. S., R. F. Hennigan, G. M. Hilliard, U. A. Ochsner, K. Parvatiyar, M. C. Kamani, H. L. Allen, T. R. DeKievit, P. R. Gardner, U. Schwab, J. J. Rowe, B. H. Iglewski, T. R. McDermott, R. P. Mason, D. J. Wozniak, R. E. Hancock, M. R. Parsek, T. L. Noah, R. C. Boucher, and D. J. Hassett. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593-603. [DOI] [PubMed] [Google Scholar]
- 73.Zimmermann, A., C. Reimmann, M. Galimand, and D. Haas. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483-1490. [DOI] [PubMed] [Google Scholar]