The plasma membrane of virus-infected cells constitutes a barrier to the egress of newly assembled virus particles but also can serve as a platform that viruses use to facilitate their assembly. In the case of HIV-1, virus assembly is driven by the Gag precursor protein, Pr55Gag (1–3), and it has been established for many years that the association of Pr55Gag with the plasma membrane is an essential step in the virus assembly pathway. Less clear are the precise mechanism by which Pr55Gag binds membrane and the identity of host cell components that dictate the specificity of the membrane-binding process. In most cell types, HIV-1 assembly appears to take place primarily at the plasma membrane, although in some cells, including the physiologically relevant macrophage, assembly may occur in intracellular compartments known as multivesicular bodies (MVBs) (4–7). Recent findings indicate that a particular member of the phosphoinositide family of lipids, phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2], serves an important function in directing Pr55Gag to the plasma membrane (8). In this issue of PNAS, Saad et al. (9) present evidence for a direct interaction between Gag and PI(4,5)P2 and provide a compelling structural model to help explain the role that PI(4,5)P2 plays in promoting the association of Pr55Gag with the plasma membrane. In a broader context, the proposed model suggests a novel mechanism by which phosphoinositides can help stabilize the association of proteins with cellular membranes.
Pr55Gag is composed of several major domains that are cleaved from the Gag precursor by the viral protease during particle release from the cell. These domains are as follows (from N to C terminus): matrix (MA), capsid (CA), nucleocapsid (NC), and p6 (Fig. 1A). Each of these domains serves critical functions in virus assembly and release. For example, MA directs Pr55Gag to membrane via a bipartite motif consisting of an N-terminal, covalently attached myristic acid and a highly basic cluster of amino acid residues (10). The membrane association of a number of myristylated proteins is regulated by a so-called “myristyl switch” mechanism whereby the myristate can adopt either an exposed or a sequestered conformation (11). When the myristate is in the exposed conformation, it promotes membrane association; conversely, when the myristate is sequestered, membrane binding is diminished. A decade ago, it was proposed that the interaction of the MA domain of Pr55Gag with membrane is regulated by a myristyl switch (12), and a substantial amount of evidence supports this model (13–15). Most recently, structural studies from Summers and colleagues (16) demonstrated that the myristate is sequestered when MA is in a monomeric form, which binds membrane poorly. In contrast, when downstream sequences in CA that promote oligomerization are added to MA, the myristate becomes exposed, and membrane binding is substantially increased.
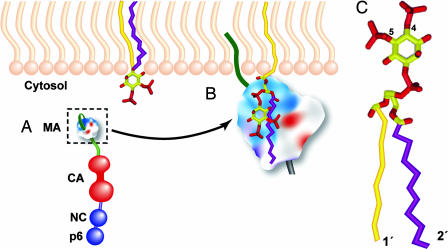
Fig. 1.
Binding of the MA domain of Pr55Gag to PI(4,5)P2. (A) Organization of the Gag precursor protein, Pr55Gag. Matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains are shown. The N-terminal myristic acid moiety (dark green) covalently attached to MA is shown tucked back in its sequestered conformation. The highly basic patch of MA is depicted in blue. (B) PI(4,5)P2 association with the MA domain. PI(4,5)P2 is embedded in the inner (cytosolic) leaflet of the lipid bilayer. The 1′ and 2′ acyl chains of PI(4,5)P2 are shown in yellow and purple, respectively. According to the model proposed by Saad et al. (9), after binding between MA and PI(4,5)P2, the myristic acid (dark green) flips out into the exposed conformation and inserts into the lipid bilayer, and the 2′ acyl chain (purple) is extruded from the membrane and sequestered by MA. (C) Structure of PI(4,5)P2, with phosphates on the 4 and 5 position of the inositol ring (yellow) shown in red and the 1′- and 2′-acyl chains indicated. Note that the 2′ acyl chain is more “kinked” than the 1′ chain, indicating a lower degree of saturation of the 2′ chain.
Intriguingly, the highly basic domain of MA not only promotes the association of Gag with membrane but also confers specificity to this binding. Mutations within these basic residues can reduce the efficiency of virus production by redirecting virus assembly from the plasma membrane to intracellular membrane (17–19). These findings raised the possibility that a component of the plasma membrane interacts with basic residues in MA and, thereby, promotes the targeting of Gag to the cell surface. A candidate for this targeting factor emerged with the demonstration that depletion of plasma membrane PI(4,5)P2 induced the relocalization of HIV-1 assembly to MVBs (8).
The phosphoinositides all are closely related to the parent molecule, phosphatidylinositol (PI), and differ from one another in the number and position of phosphate groups on the six-carbon inositol ring that forms the bulky head group of these lipids (Fig. 1 B and C). The phosphoinositides also differ in their subcellular localization: PI(4,5)P2 and PI(3,4,5)P3 are concentrated predominantly at the plasma membrane, PI(3)P in early endosomes, PI(3,5)P2 in late endosomes, and PI(4)P in the Golgi apparatus (20). Because, in part, of their distinct subcellular localization, the phosphoinositides play a central role in the trafficking of a number of cellular proteins (21, 22), with PI(4,5)P2 clearly implicated in directing proteins to the plasma membrane (22). The role of PI(4,5)P2 in Gag targeting suggests that HIV-1 Gag has evolved to subvert this cellular trafficking pathway to regulate the localization of virus assembly. Interestingly, a previous study reported that molecules structurally related to the phosphoinositides are able to promote efficient assembly of virus-like particles from HIV-1 Gag in vitro (23).
The work of Saad et al. (9) provides a structural framework with which to understand the ability of PI(4,5)P2 to promote membrane binding and regulate the site of HIV-1 assembly. The authors report that myristylated MA (myrMA) binds directly to PI(4,5)P2 and that this binding is highly specific; MA does not interact with several related phosphoinositides, including PI(3,5)P2, which is identical to PI(4,5)P2 except for the location of one of the phosphates on the inositol ring. Apart from PI(4,5)P2, only PI(3,4,5)P3 shows binding to MA, suggesting an important role for both position 4 and 5 phosphates in MA binding. The NMR structure of myrMA bound to PI(4,5)P2 reveals that the inositol head group and the 2′ fatty acid chain of the lipid molecule fit into a hydrophobic cleft in MA (Fig. 1B). The negatively charged phosphates form salt bridges with basic residues in MA. For technical reasons, Saad et al. used forms of PI(4,5)P2 with truncated fatty acyl chains; however, modeling suggests that PI(4,5)P2 with the more physiological 18- to 20-carbon acyl chains would exhibit similar binding conformations (ref. 9; Fig. 1B).
A particularly noteworthy aspect of the Saad et al. (9) structure is that binding of PI(4,5)P2 to myrMA triggers the myristic acid moiety to “flip out” from the sequestered to the exposed conformation. As discussed above, myristate exposure is key to promoting the stable association of MA with membrane. According to these recent findings, PI(4,5)P2 promotes membrane association via two mechanisms: by serving as an anchor for Gag in the lipid bilayer and triggering the myristyl switch that further retains Gag in the membrane (Fig. 1B). Because PI(4,5)P2 is concentrated at the plasma membrane, this model helps explain why depletion of PI(4,5)P2 results in the loss of virus assembly at the cell surface and a redirection of assembly to intracellular membranes. The observed lack of binding between MA and phosphoinositides that are concentrated in intracellular membranes is consistent with the localization of HIV-1 assembly predominantly at the plasma membrane in most cell types.
Another interesting feature of the Saad et al. (9) model involves the association of Gag with lipid raft microdomains. Lipid rafts are cholesterol- and saturated lipid-enriched patches in the plasma membrane that serve as concentration platforms for proteins involved in a number of biological processes, including virus assembly (24, 25). Because the predominant form of PI(4,5)P2 in cells contains a saturated 1′ fatty acid side chain but an unsaturated 2′ side chain (Fig. 1C), PI(4,5)P2 may equilibrate between raft and nonraft domains in the membrane. According to the model proposed by Saad et al. (9), upon binding between myrMA and PI(4,5)P2, the 2′ side chain is extruded from the lipid bilayer and packs against MA, leaving only the highly saturated 1′ fatty acid side chain in the plasma membrane. The removal of the unsaturated 2′ side chain from the lipid bilayer, in theory, could increase the affinity of PI(4,5)P2 and its associated Gag molecule for lipid raft microdomains.
The role of PI(4,5)P2 in regulating the binding of HIV-1 Gag to membrane and the localization of virus assembly suggests several exciting new directions for research. Analysis of phosphoinositide-binding preferences may reveal whether the effect of HIV-1 MA mutations on Gag localization can be explained by a switch in phosphoinisitide binding from PI(4,5)P2 to other members of this family of lipid molecules. Given that residues in the highly basic domain of MA appear to be crucial for PI(4,5)P2 binding (9, 26) and that analogous basic patches are evident in the MA domains of a number of other retroviruses (27), it will be of great interest to determine the importance of PI(4,5)P2 in promoting the assembly of retroviruses other than HIV-1. From a more practical perspective, the structural information provided by Saad et al. (9) may help in the design of inhibitors that disrupt HIV-1 replication by interfering with the interaction between Gag and PI(4,5)P2.
Footnotes
Conflict of interest statement: No conflicts declared.
See companion article on page 11364.
References
- 1.Swanstrom R., Wills J. W. Synthesis, Assembly, and Processing of Viral Proteins. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [PubMed] [Google Scholar]
- 2.Adamson C. S., Jones I. M. Rev. Med. Virol. 2004;14:107–121. doi: 10.1002/rmv.418. [DOI] [PubMed] [Google Scholar]
- 3.Freed E. O. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 4.Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen D. G., Booth A., Gould S. J., Hildreth J. E. J. Biol. Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 6.Pelchen-Matthews A., Kramer B., Marsh M. J. Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ono A., Freed E. O. J. Virol. 2004;78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ono A., Ablan S. D., Lockett S. J., Nagashima K., Freed E. O. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saad J. S., Miller J., Tai J., Kim A., Ghanam R. H., Summers M. F. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou W., Parent L. J., Wills J. W., Resh M. D. J. Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ames J. B., Ishima R., Tanaka T., Gordon J. I., Stryer L., Ikura M. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 12.Zhou W., Resh M. D. J. Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spearman P., Horton R., Ratner L., Kuli-Zade I. J. Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ono A., Freed E. O. J. Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paillart J. C., Gottlinger H. G. J. Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang C., Loeliger E., Luncsford P., Kinde I., Beckett D., Summers M. F. Proc. Natl. Acad. Sci. USA. 2004;101:517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X., Yu X., Lee T. H., Essex M. J. Virol. 1993;67:6387–6394. doi: 10.1128/jvi.67.11.6387-6394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed E. O., Englund G., Martin M. A. J. Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermida-Matsumoto L., Resh M. D. J. Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Matteis M. A., Godi A. Biochim. Biophys. Acta. 2004;1666:264–274. doi: 10.1016/j.bbamem.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Simonsen A., Wurmser A. E., Emr S. D., Stenmark H. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 22.McLaughlin S., Murray D. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 23.Campbell S., Fisher R. J., Towler E. M., Fox S., Issaq H. J., Wolfe T., Phillips L. R., Rein A. Proc. Natl. Acad. Sci. USA. 2001;98:10875–10879. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons K., Toomre D. Nat. Rev. Mol. Cell. Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 25.Ono A., Freed E. O. Adv. Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 26.Shkriabai N., Datta S. A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 27.Murray P. S., Li Z., Wang J., Tang C. L., Honig B., Murray D. Structure (London) 2005;13:1521–1531. doi: 10.1016/j.str.2005.07.010. [DOI] [PubMed] [Google Scholar]